Abstract
Modulation of the activity of the upstream binding factor (UBF) plays a key role in cell cycle-dependent regulation of rRNA synthesis. Activation of rDNA transcription on serum stimulation requires phosphorylation of UBF at serine 484 by G1-specific cyclin-dependent kinase (cdk)/cyclin complexes. After G1 progression UBF is phosphorylated at serine 388 by cdk2/cyclin E and cdk2/cyclin A. Conversion of serine 388 to glycine abolishes UBF activity, whereas substitution by aspartate enhances the transactivating function of UBF. Protein–protein interaction studies reveal that phosphorylation at serine 388 is required for the interaction between RNA polymerase I and UBF. The results suggest that phosphorylation of UBF represents a powerful means of modulating the assembly of the transcription initiation complex in a proliferation- and cell cycle-dependent fashion.
Differential phosphorylation of the upstream finding factor (UBF) is emerging as a focal point for cell cycle-dependent oscillations of rRNA synthetic activity. It was recognized early that the transactivating potential of UBF is modulated by posttranslational modifications. Transcriptional silencing in quiescent or serum-deprived cells correlates with hypophosphorylation of UBF (1–3). Moreover, the activity of UBF may be altered by interaction with cellular proteins. For example, the product of the retinoblastoma susceptibility gene (pRb) has been demonstrated to interact with UBF and inhibit rDNA transcription in vitro (4, 5). These results suggest that UBF is targeted by different signaling pathways during differentiation, proliferation, and cell growth. UBF contains multiple phosphorylation sites that are dispersed throughout the protein with several clustered sites in the C-terminal acidic tail. The phosphorylation state of UBF appears to determine its ability to activate transcription, but not its ability to bind to DNA (1, 2, 6). Thus, phosphorylation-mediated transcriptional activation results from an event following DNA binding, presumably the recruitment of components of the Pol I transcription machinery to the rDNA promoter. Consistent with this, the C-terminal domain of UBF, which is necessary for transcriptional activation, interacts with two subunits of TIF-IB/SL1, namely TBP and TAFI48, and this interaction is regulated by phosphorylation (7). Dephosphorylation abolishes the binding of UBF to TIF-IB/SL1 and prevents transcriptional activation, a finding that underscores the key role for UBF phosphorylation in the control of rRNA synthesis.
Besides the acidic tail, internal regions of UBF are phosphorylated and the pattern of UBF phosphorylation is altered in response to extracellular signals (2). In a recent report we have shown that activation of rDNA transcription on serum stimulation is mediated by sequential phosphorylation of UBF. The initiating event is phosphorylation of UBF at Ser-484, which is mediated by G1-specific cyclin-dependent kinase (cdk)/cyclin complexes (8). We now demonstrate that after progression through G1, UBF is phosphorylated at Ser-388 by cdk2/cyclin E and A. Both in transient transfection assays and reconstituted in vitro transcription systems, Ser-388 is indispensable for the transactivating function of UBF. Substitution of Ser-388 with glycine renders UBF transcriptionally inactive, whereas substitution with aspartate enhances UBF activity. These data reveal a correlation between cell cycle-specific phosphorylation of UBF and cellular rRNA synthetic activity and suggest that both processes are intimately linked.
Materials and Methods
Plasmids.
pMr1930–BamHI/HindIII (BH) represents a fusion between a murine rDNA promoter fragment (from −1930 to +292) and a 3′-terminal BH rDNA fragment separated by pUC sequences (9). pRc/CMV-FLAG-UBF1 has been described (8). Site-directed mutagenesis was performed by overlap extension PCR with oligonucleotides that replace Ser-388 by glycine (forward primer: 5′-GCAAACCACCGGTCCGGCCTCCAAGAAGCC-3′; backward primer: 5′-GGAGGCCGGACCGGTGGTTTGCTTCTTATTGATGTTC-3′) or by aspartate (forward primer: 5′-GCAAACCACCGATCCGGCCTCCAAGAAGCC-3′; backward primer: 5′-GGAGGCCGGATCGGTGGTTTCG-3′) and primers corresponding to the N and C terminus of UBF, respectively. UBF1/WT was cut with MscI and BglII and the fragment encoding aa 316–491 was replaced by the corresponding PCR product to yield mutants UBF1/S388G and UBF1/S388D. The generation of the recombinant baculoviruses carrying the respective mutations was done as described (8).
Cell Culture, Transfections, and RNA Analysis.
NIH 3T3 fibroblasts were synchronized by culturing in medium containing 0.1% FCS for 48 h and then stimulated by addition of fresh medium containing 10% FCS. Sf9 insect cells were cultured at 27°C in TC-100 medium supplemented with 10% FCS. For transient expression, 5 × 105 NIH 3T3 cells were transfected with the rDNA reporter plasmid pMr1930-BH and cellular RNA was analyzed on Northern blots as described (8). After hybridization at 65°C in 50% formamide/5× SSC/50 mM sodium phosphate, pH 6.5/8× Denhardt's/0.5 mg/ml yeast RNA/0.1% SDS, the filters were washed in 0.2× SSC/0.1% SDS at 65°C. The blots were subsequently hybridized with cytochrome c oxidase-specific riboprobes.
Purification of cdk Complexes and in Vitro Kinase Assay.
Sf9 cells were coinfected with baculoviruses encoding the respective cdks and cyclins. After 44 h the cells were lysed in buffer AM-300 [300 mM KCl/20 mM Tris⋅HCl, pH 7.9/5 mM MgCl2/0.1 mM EDTA/10% glycerol/0.5 mM dithioerythritol (DTE)] supplemented with protease inhibitors (0.5 mM PMSF and 2 μg/ml each of pepstatin, leupeptin, and aprotinin) and phosphatase inhibitors (80 mM β-glycerophosphate, 20 mM potassium fluoride, and 1 mM Na-orthovanadate). Cdk/cyclin complexes were immunopurified with α-cdk2 and α-cdc2 antibodies (Santa Cruz Biotechnology). Ten-microliter kinase assays contained 10 μg of the respective substrate peptide in 20 mM Tris⋅HCl (pH 7.9), 60 mM KCl, 8 mM MgCl2, 1 mM DTT, 0.04 mM ATP, and 1 μCi γ-[32P]ATP (1 μCi = 37 GBq). For in vitro phosphorylation, 200 ng of purified FLAG-UBF1 were incubated for 30 min at 30°C with bead-bound cdk2/cyclin E, cdk2/cyclin A, or cdc2/cyclin A in 20 μl of 50 mM Hepes (pH 7.6), 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 1 mM DTT, 0.1% Triton-X100, 10% glycerol, 0.1 mM PMSF, 2 μg/ml of each leupeptin, aprotinin, and pepstatin, 10 mM β-glycerophosphate, 1 mM KF, 0.1 mM sodium orthovanadate, 25 μM ATP, and 20 μCi γ-[32P]ATP.
Tryptic Phosphopeptide Mapping.
NIH 3T3 cells were transfected with expression vectors encoding wild-type or mutant UBF1 and labeled for 10 h in phosphate-free DMEM containing 10% dialyzed FCS and 1 mCi/ml 32P-orthophosphate. Alternatively, in vivo phosphorylation was performed in Sf9 cells infected with baculoviruses expressing FLAG-UBF1, cdk2, and cyclin A (8). Cells were lysed in RIPA buffer (20 mM Tris⋅HCl, pH 8.0/100 mM NaCl/0.5% sodium deoxycholate/0.5% Nonidet P-40/0.5% SDS/10 mM EGTA/20 mM KF/1 mM sodium orthovanadate/10 mM K2HPO4/2 μg/ml of each leupeptin, aprotinin, and pepstatin) and incubated overnight with α-UBF antibodies coupled to protein G-agarose or with α-FLAG M2-agarose (Sigma). Precipitated proteins were separated by 6% SDS/PAGE and processed for tryptic phosphopeptide mapping as described (2).
In Vitro Transcription Assays.
The fractionation scheme for purification of murine Pol I and Pol I-specific transcription factors has been described (10). Standard transcription reactions (25 μl) contained 8 ng pMrWT/NdeI, 4 μl of partially purified Pol I (H-400 fraction), 1 μl of TIF-IA/TIF-IC (poly-L-lysin-agarose fraction), 3 μl of TIF-IB (CM-400 fraction), and 0.2–10 ng of UBF (5). After incubation for 1 h at 30°C, run-off transcripts were analyzed by gel electrophoresis and autoradiography.
Protein–Protein Interaction Assays.
FLAG-tagged wild-type and mutant UBF1 were immobilized on M2-agarose beads (500 ng/μl). As a control, M2-agarose beads saturated with FLAG peptide were used. Eight microliters of packed beads containing equal amounts of immobilized proteins were incubated for 1 h at 4°C with 300 μg of nuclear extract proteins in buffer AM-100 (100 mM KCl/20 mM Tris⋅HCl, pH 7.9/5 mM MgCl2/0.1 mM EDTA/20% glycerol/0.5 mM DTE) supplemented with protease inhibitors, 20 mM β-glycerophosphate, and 0.2% Nonidet P-40. After washing four times in buffer AM-100/0.5% Nonidet P-40, bead-bound Pol I and TIF-IB/SL1 were detected on immunoblots by using purified α–RPA116 or α–TAFI95 antibodies. For dephosphorylation, bead-bound UBF1 (4 μg) was incubated for 30 min at 30°C with 4 units of shrimp alkaline phosphatase (Amersham Pharmacia) in 30 μl of AM-100 and then washed three times in 50 μl of buffer AM-100.
To assay coimmunoprecipitation of UBF1 and Pol I, [35S]methionine-labeled FLAG-tagged UBF1 was synthesized in a coupled in vitro transcription–translation system (Promega). Fifteen microliters of the lysate were incubated with 50 μl of Pol I (H-400 fraction) or 50 μl of TIF-IB (CM-400 fraction), which were essentially free of UBF. After incubation for 2.5 h at 4°C in buffer AM-100/0.2% Nonidet P-40, M2-beads were added and the incubation continued for 2 h at 4°C. The immunoprecipitates were washed three times with IP-buffer and analyzed on Western blots.
For pull-down assays, GST-RPA53 and GST were expressed in E. coli BL21(codon plus) and purified on glutathione-Sepharose. Equal amounts of bead-bound GST and GST-RPA53 were incubated for 4 h at 4°C with 35S-labeled UBF in AM-100/0.2% Nonidet P-40 plus protease inhibitors. After washing four times with binding buffer, bound proteins were analyzed by 8% SDS/PAGE and autoradiography.
DNA Binding Assay.
Binding of UBF to four-way junction DNA was monitored by electrophoretic mobility shift assays as described (5). Briefly, increasing amounts of recombinant UBF were incubated for 15 min at room temperature with 10 fmol of labeled four-way junction DNA in 10 μl of binding buffer containing 8% Ficoll, 100 mM KCl, 5 mM MgCl2, 10 mM Hepes (pH 7.9), 1 mM EDTA, 0.5 mM DTT, and DNA–protein complexes were analyzed by electrophoresis on 6.5% PAA gels in 0.5× TBE at 11 V/cm and 4°C.
Results and Discussion
Cell Cycle-Dependent Alterations of UBF Phosphorylation.
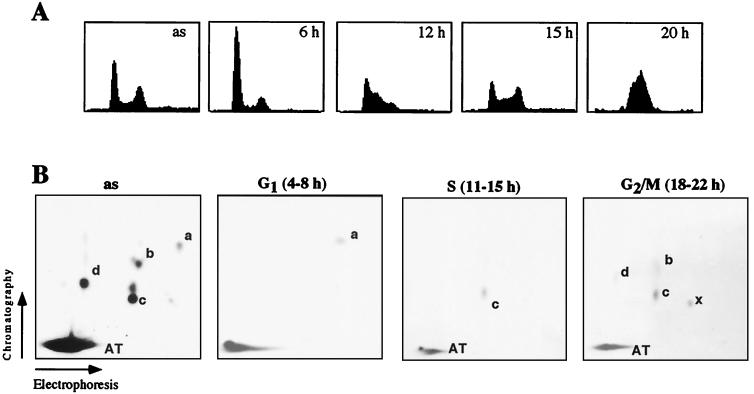
To monitor changes in the pattern of UBF phosphorylation during cell cycle progression, NIH 3T3 fibroblasts were arrested in G0 by serum starvation and released into the cell cycle by serum addition (Fig. 1A). At different times after mitogenic stimulation, the cells were metabolically labeled for 4 h with [32P]orthophosphate, and UBF was immunoprecipitated and subjected to tryptic phosphopeptide mapping (Fig. 1B). Consistent with previous data, most of the label in UBF from asynchronous cells was found in peptide AT, a large tryptic fragment encompassing the acidic tail (aa 675–765) that is targeted by CKII (1, 3, 6). Moreover, four additional peptides (marked a–d) were phosphorylated. As demonstrated in a recent study, phosphorylation of peptide a (aa 481–486) at Ser-484 by G1-specific cdk/cyclin complexes correlates with activation of rDNA transcription during G1 progression (8). Consistent with this, phosphopeptide a, which is underrepresented in UBF from asynchronous cells (Fig. 1B, panel as), is the only tryptic peptide that is labeled in G1-phase cells (panel G1). During S-phase—e.g., 11–15 h after mitogenic stimulation—peptide a is not labeled anymore, but peptide c becomes preferentially phosphorylated (panel S). Phosphorylation of peptide c is not restricted to S-phase but is also observed in G2- or M-phase cells. At these late times peptides b and d, as well as an additional peptide (x) that is hardly detectable in UBF from asynchronous cells, are labeled.
Figure 1.
Phosphorylation pattern of UBF during G1-, S-, and G2/M-phase. (A) Fluorescence-activated cell sorter (FACS) analysis. NIH 3T3 cells were synchronized by serum starvation followed by serum stimulation for 6, 12, 15, and 20 h. Asynchronous cells (as) are shown for comparison. (B) Tryptic phosphopeptide maps. 4 × 105 asynchronous (as) or synchronized NIH 3T3 cells were metabolically labeled for 4 h in the presence of [32P]orthophosphate (2.5 mCi/ml). UBF was precipitated with α-UBF antibodies and analyzed by tryptic phosphopeptide mapping. The autoradiographs were exposed for 5 (as) and 9 days (G1, S, G2/M).
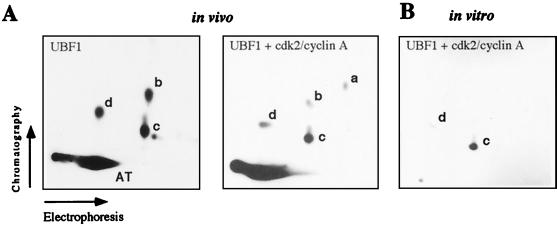
The experiment above suggests that peptide c is targeted by cdk2/cyclin E and/or cdk2/cyclin A. To test this, FLAG-tagged UBF1 was coexpressed in Sf9 cells together with cdk2 and cyclin A, labeled with [32P]orthophosphate, and subjected to tryptic phosphopeptide mapping. The peptide map of UBF coexpressed with cdk2/cyclin A is shown in Fig. 2A. The overall pattern of phosphopeptides resembles that of cellular UBF; however, peptide c was preferentially labeled in cells overexpressing cdk2/cyclin A (Right). A similar pattern was observed when cdk2/cyclin E or cdc2/cyclin A were coexpressed with UBF (data not shown). Moreover, in vitro phosphorylation of recombinant UBF using purified cdk2/cyclin A (Fig. 2B) or cdk2/cyclin E (data not shown) labeled peptide c, indicating that this site of UBF is targeted during late G1- and S-phase.
Figure 2.
Tryptic peptide maps of UBF phosphorylated in vitro and in vivo. (A) Tryptic peptide maps of recombinant UBF1 phosphorylated in vivo. Sf9 cells were infected with baculovirus encoding FLAG-UBF1 (Left) or triple-infected with baculoviruses encoding FLAG-UBF1, cdk2, and cyclin A (Right). After labeling with [32P]orthophosphate, UBF1 was immunoprecipitated and processed for tryptic phosphopeptide mapping. (B) In vitro phosphorylation. Recombinant UBF1 was phosphorylated in vitro with purified cdk2/cyclin A and analyzed by two-dimensional tryptic phosphopeptide mapping.
A Negative Charge at Position 388 Is Required for UBF Activity.
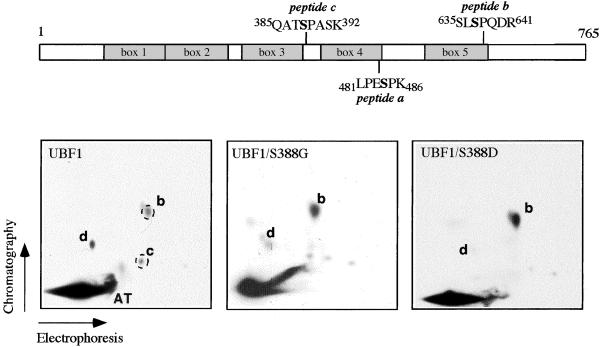
Cyclin-dependent kinases target S/T-P sites, and previous studies have established that UBF is exclusively phosphorylated on serine residues (2). UBF contains three Ser/Pro motifs (Ser-484, Ser-388, and Ser-637) that may be phosphorylated by cdks. To identify which of the serine residues is contained in peptides b and c, synthetic phosphopeptides were tested for comigration with 32P-labeled phosphopeptides from UBF labeled in vivo. This analysis revealed that the phosphopeptide containing Ser-637 comigrated with peptide b, whereas the putative tryptic peptide containing phospho-Ser-388 (QAT-PS-PASK) precisely colocalized with spot c (data not shown). The identity of peptide d is still unknown.
In an attempt to assess the functional importance of peptide c phosphorylation, Ser-388 was replaced by glycine (UBF1/S388G) or aspartic acid (UBF1/S388D). Wild-type and mutant UBF were labeled in vivo and tryptic fingerprint analysis was performed (Fig. 3). Consistent with peptide c containing phospho-Ser-388, this peptide was missing in both mutant proteins, UBF1/S388G (Center) and UBF1/S388D (Right). Interestingly, both mutants showed increased levels of phosphorylation in peptide b and a decrease of peptide d, suggesting an interdependence of phosphorylation at Ser-388 with those at Ser-367 and an unknown residue in peptide d.
Figure 3.
Identification of phospho-Ser-388. NIH 3T3 cells were transfected with 2 μg of pRc/CMV-FLAG-UBF1, pRc/CMV-FLAG-UBF1/S388G, or pRc/CMV-FLAG-UBF1/S388D, and labeled for 8 h with 0.5 mCi/ml [32P]orthophosphate; UBF was immunoprecipitated and analyzed by tryptic phosphopeptide mapping. Two synthetic peptides (Q-A-T-PS-P-A-S-K and S-L-PS-P-Q-D-R) were added before electrophoresis. The position of the synthetic peptides was visualized by fluorescamine staining (dashed circles). A schematic illustration of UBF indicating the positions of phosphopeptides a, b, and c is shown above.
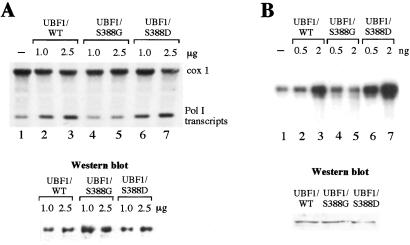
Having established that peptide c contains phospho-Ser-388, the transactivating properties of wild-type UBF and the two mutants were compared. In the experiment shown in Fig. 4A, NIH 3T3 cells were transfected with expression vectors encoding UBF1, UBF1/S388G, and UBF1/S388D, respectively, together with pMr1930-BH, a reporter plasmid harboring an artificial murine rDNA minigene (9). Western blots confirmed that wild-type and mutant UBF were expressed at similar levels (Fig. 4A Lower). Overexpression of wild-type UBF augmented transcription of the reporter plasmid up to 3-fold (Fig. 4A, lanes 2 and 3). In contrast, UBF1/S388G failed to stimulate transcription (lanes 4 and 5), demonstrating the essential role of Ser-388 phosphorylation in UBF function. Significantly, replacement of Ser-388 by aspartic acid did not impair UBF activity but rather enhanced UBF-mediated transcriptional activation (lanes 6 and 7). This suggests that substitution of phosphoserine by a negative charge at position 388 retains UBF activity.
Figure 4.
Ser-388 is required for UBF activity in vivo and in vitro. (A) UBF1/S388G does not activate rDNA transcription in vivo. NIH 3T3 cells were transfected with 10 μg of pMr1930-BH together with 1 and 2.5 μg of pRc/CMV-FLAG-UBF1 (lanes 2 and 3), pRc/CMV-FLAG-UBF1/S388G (lanes 4 and 5), and pRc/CMV-FLAG-UBF1/S388D (lanes 6 and 7). Transcripts from the reporter plasmid were analyzed on Northern blots by using a plasmid-specific riboprobe. The blot was rehybridized with a riboprobe against cytochrome c oxidase (cox 1). To monitor FLAG-UBF1 expression, 10 μg of protein from the transfected cells were subjected to Western blotting by using antibodies against the FLAG epitope (M2). (B) Replacement of Ser-388 by aspartate does not impair UBF1 function. FLAG-tagged UBF1, UBF1/S388G, and UBF1/S388D were immunopurified from NIH 3T3 cells and assayed in a reconstituted transcription system.
Given that phosphorylation on Ser-388 is required for UBF activity, we compared transcriptional activity of wild-type and mutant UBF in a reconstituted transcription system (Fig. 4B). Increasing amounts of wild-type UBF1 strongly augmented transcription (lanes 1–3), whereas UBF1/S388G had no effect (lanes 4 and 5). Remarkably, UBF1/S388D exhibited an even higher transcriptional competence than wild-type UBF1 (lanes 6 and 7). This result suggests a key role for Ser-388 in transcriptional activation and demonstrates that a negative charge at aa 388 is required for UBF1 activity.
Ser-388 Phosphorylation Facilitates the Interaction Between UBF1 and Pol I.
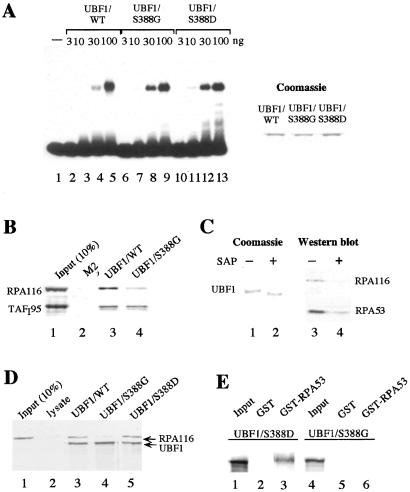
Little is known of how phosphorylation affects UBF activity and why UBF is phosphorylated by different cdks during cell cycle progression. In previous studies, we and others have shown that dephosphorylation does not affect the ability of UBF to bind to the rDNA promoter, the enhancer repeats, or to cruciform DNA (1, 6). These studies, however, did not exclude the possibility that phospho-Ser-388 may have escaped dephosphorylation. To examine whether mutation of Ser-388 may alter the interaction of UBF with DNA, we compared binding of wild-type and mutant UBF to synthetic four-way junction DNA in electrophoretic mobility shift assays (Fig. 5A). Clearly, all three UBF polypeptides bound with similar affinity to the labeled cruciform probe, indicating that phosphorylation does not affect the DNA-binding properties of UBF.
Figure 5.
Phosphorylation at Ser-388 is necessary for association of UBF1 with Pol I. (A) Mutation of Ser-388 does not interfere with DNA binding of UBF. A 32P-labeled cruciform DNA probe was incubated with 3, 10, 30, and 100 ng of FLAG-tagged UBF1/WT (lanes 2–5), UBF1/S388G (lanes 6–9), and UBF1/S388D (lanes 10–13) immunopurified from baculovirus-infected Sf9 cells. UBF-DNA complexes were analyzed on a 6.5% PAA-gel. A Coomassie blue stain of 500 ng of UBF1/WT, UBF1/S388G, and UBF1/S388D is shown at Right. (B) Pull-down experiment. Extracts from FM3A cells were incubated for 1 h at 4°C with either bead-bound FLAG-tagged UBF1/WT, UBF1/S388G, or control beads saturated with the FLAG-epitope peptide as indicated. Bead-bound proteins were analyzed on Western blots by using antibodies against TAFI95 and RPA116. Lane 1 shows 10% of input TAFI95 and RPA116. (C) Phosphatase treatment impairs the interaction between Pol I and UBF1. Bead-bound FLAG-UBF1/WT was preincubated in the absence or presence of shrimp alkaline phosphatase (SAP) for 30 min at 30°C. Bead-bound UBF1 was analyzed by Coomassie staining (lanes 1 and 2) and assayed for interaction with Pol I by using antibodies against RPA116 and RPA53 (lanes 3 and 4). (D) Coimmunoprecipitation experiment. Fifty microliters of Pol I (H-400 fraction) were incubated with 35S-labeled FLAG-tagged UBF1/WT (lane 3), UBF1/S388G (lane 4), or UBF1/S388D (Lane 5), followed by immunoprecipitation with M2− antibodies. An unprogrammed reticulocyte lysate was used as a control (lane 2). UBF was visualized by autoradiography, Pol I was monitored on Western blots by using α–RPA116 antibodies. Lane 1 shows 10% of RPA116 present in the fraction. (E) Interaction between UBF and RPA53. 35S-labeled UBF1/S388D (lanes 1–3) or UBF1/S388G (lanes 4–6) were incubated with immobilized GST (lanes 2 and 5) or GST-RPA53 (lanes 3 and 6). Bead-bound proteins were separated on 8% SDS/PAA gels and visualized by autoradiography. Lanes 1 and 4 show 10% of the UBF used.
Because phosphorylation of Ser-388 does not impair binding of UBF to DNA, we reasoned that a negative charge at aa 388 may facilitate protein–protein interactions required for initiation complex assembly. Previous studies have established that UBF1 interacts with both Pol I and TIF-IB/SL1, the TBP-containing promoter selectivity factor (7, 11–14). To unravel the mechanism of how cdk-dependent phosphorylation at Ser-388 may regulate UBF activity, we studied the interaction of wild-type and mutant UBF with TIF-IB/SL1 and Pol I, respectively. For this, wild-type and mutant UBF1 were immobilized on agarose beads and incubated with nuclear extract from FM3A cells. Association of Pol I and TIF-IB with bead-bound UBF1 was examined on immunoblots by using antibodies against specific subunits of TIF-IB (anti-TAFI95) and Pol I (anti-RPA116). As shown in Fig. 5B, neither Pol I nor TIF-IB was retained on the control matrix containing anti-FLAG antibodies saturated with the FLAG peptide (lane 2). On the other hand, significant levels of TIF-IB were retained at the UBF1-affinity matrix, regardless of whether wild-type or mutant UBF1 were used (lanes 3 and 4). This result demonstrates that Ser-388 is not required for the interaction between UBF1 and TIF-IB. A different result was obtained when the association of UBF1 with Pol I was measured. Whereas Pol I efficiently interacted with wild-type UBF1 (lane 3), binding to UBF1/S388G was significantly reduced (lane 4). This result suggests that UBF1/S388G fails to activate transcription because it is not capable of interacting with Pol I.
To demonstrate the functional relevance of UBF phosphorylation for association with Pol I, bead-bound UBF1 was incubated with shrimp alkaline phosphatase (SAP) and the association of untreated and phosphatase-treated UBF1 with Pol I was compared (Fig. 5C). Phosphatase treatment increased the electrophoretic mobility of UBF1, demonstrating that UBF was efficiently dephosphorylated (Fig. 5C, lanes 1 and 2). Consistent with phosphorylation being required for the interaction between UBF and Pol I, phosphatase treatment markedly reduced the association of Pol I with UBF1 (lanes 3 and 4), underscoring the involvement of UBF phosphorylation in the interaction with Pol I.
The failure of the S388G mutant to associate with Pol I was also demonstrated by coimmunoprecipitation experiments. In the experiment in Fig. 5D, in vitro translated wild-type and mutant FLAG-tagged UBF1 were incubated with a partially purified Pol I fraction that was virtually free of UBF. After immunoprecipitation with anti-FLAG antibodies, associated Pol I was monitored on immunoblots by using antibodies against RPA116, the second largest subunit of Pol I. In support of the pull-down experiment and the in vitro transcription data, Pol I was coimmunoprecipitated with wild-type UBF but not mutant UBF1/S388G (Fig. 5D, lanes 3 and 4). The association of Pol I with UBF1/S388D, on the other hand, was not impaired. This indicates that a negative charge at aa 388 is necessary for the interaction between UBF and Pol I.
UBF is known to interact with RPA53, the 53-kDa subunit of Pol I (12). In a previous paper, we found that UBF contacts the third largest subunit of Pol I (11), which has been estimated to be 62 kDa (15). However, this size estimation turned out to be incorrect. The third largest subunit of mammalian Pol I is RPA53, and a 62-kDa subunit does not exist. To demonstrate the involvement of Ser-388 phosphorylation in the interaction of UBF with this subunit, binding of 35S-labeled UBF1/S388G and UBF1/S388D to immobilized GST-RPA53 and GST, respectively, was monitored. Again, UBF1/S388D was retained on the GST-RPA53 affinity matrix (Fig. 5D, lanes 1–3), whereas no binding of UBF1/S388G to GST-RPA53 was observed (lanes 4–6). Though previously we have demonstrated that a polypeptide harboring aa 92 and 373 of UBF is sufficient for interaction with Pol I (11), we believe that the present experiments, which use full-length UBF, are much more physiological and more conclusive.
Together, these and previous data underscore the requirement of site-specific phosphorylations for different functions of UBF. All phosphorylations identified so far—e.g., at the acidic tail, Ser-484 and Ser-388—appear to be involved in protein–protein interactions that are required for assembly of the initiation complex. This finding implies that changes in the pattern of UBF phosphorylation may have pronounced effects on the cell's biosynthetic activities. Consistent with this, cell cycle-dependent fluctuations of pre-rRNA synthesis have been shown to be mediated by site- and phase-specific phosphorylation of UBF. Pol I transcription is maximal in S and G2, shuts down in mitosis, and recovers in G1. Transcriptional silencing in mitosis is mediated by inactivation of TIF-IB/SL1 by cdc2/cyclin B-dependent phosphorylation (16). Besides TIF-IB/SL1, UBF is mitotically inactivated by an as yet unidentified mechanism (17). We hypothesize that peptide x, a phosphopeptide that is preferentially labeled in mitotic cells, is causally involved in the inactivation of UBF during mitosis. In early G1-phase, the overall rDNA transcriptional activity remains low despite the fact that TIF-IB/SL1 activity has resumed (17). Recovery of cellular rRNA synthesis during G1 progression requires both the removal of inhibitory phosphate group(s) by okadaic-sensitive phosphatase(s) and de novo phosphorylation of UBF at Ser-484 (8, 17). Further transcriptional activation during S-phase correlates with phosphorylation of UBF at Ser-388 by S/G2-phase specific cyclin-dependent kinases. Moreover, during S-phase the total amount of UBF associated with nucleoli increases, indicating that newly synthesized UBF immediately associates with replicating rDNA (18). Our results suggest that phosphorylation at Ser-388 enables UBF to interact with RNA polymerase I, which in turn is a prerequisite for the assembly of transcription initiation complexes at newly replicated rDNA. Although some aspects of this model need further investigation, the elucidation of the temporal and functional nature of these modifications helps in understanding basic principles of rDNA transcriptional regulation during cell cycle progression.
Acknowledgments
We thank M. Hoffmann and N. Wagner for expert technical assistance, P. Jansen-Dürr for providing various expression plasmids encoding human cdks and cyclins, and R. Laskey and J. Pines for baculoviruses encoding cdks and cyclins. This work was supported by the Deutsche Forschungsgemeinschaft (Vo728/1–1 and Priority Program “Cell Cycle”) and the Fond der Chemischen Industrie.
Abbreviations
- UBF
upstream binding factor
- cdk
cyclin-dependent kinase
- BH
BamHI/HindIII
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg H G, Grummt I. EMBO J. 1992;11:2211–2218. doi: 10.1002/j.1460-2075.1992.tb05280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voit R, Kuhn A, Sander E E, Grummt I. Nucleic Acids Res. 1995;23:2593–2599. doi: 10.1093/nar/23.14.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Mahony D J, Xie W Q, Smith S D, Singer H A, Rothblum L I. J Biol Chem. 1992;267:35–38. [PubMed] [Google Scholar]
- 4.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Nature (London) 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 5.Voit R, Schäfer K, Grummt I. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Mahony D J, Smith S D, Xie W Q, Rothblum L I. Nucleic Acids Res. 1992;20:1301–1308. doi: 10.1093/nar/20.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuan J C, Zhai W, Comai L. Mol Cell Biol. 1999;19:2872–2879. doi: 10.1128/mcb.19.4.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voit R, M, Hoffmann M, Grummt I. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grummt I, Maier U, Öhrlein A, Hassouna N, Bachellerie J-P. Cell. 1985;43:801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- 10.Schnapp A, Grummt I. Methods Enzymol. 1996;273:346–359. doi: 10.1016/s0076-6879(96)73023-9. [DOI] [PubMed] [Google Scholar]
- 11.Schnapp G, Santori F, Carles C, Riva M, Grummt I. EMBO J. 1994;13:190–199. doi: 10.1002/j.1460-2075.1994.tb06248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanada K, Song C-Z, Yamamoto K, Yano K, Maeda Y, Yamaguchi K, Muramatsu M. EMBO J. 1996;15:2217–2226. [PMC free article] [PubMed] [Google Scholar]
- 13.Beckmann H, Chen J L, O'Brien T, Tjian R. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 14.Hempel W M, Cavanaugh A H, Hannan R D, Taylor L, Rothblum L I. Mol Cell Biol. 1996;16:557–563. doi: 10.1128/mcb.16.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz L B, Roeder R G. J Biol Chem. 1974;249:5898–5906. [PubMed] [Google Scholar]
- 16.Heix J, Vente A, Voit R, Budde A, Michaelidis T M, Grummt I. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein J, Grummt I. Proc Natl Acad Sci USA. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junéra H R, Mason C, Géraud G, Suja J, Hernandez-Verdun D. Mol Biol Cell. 1997;8:145–156. doi: 10.1091/mbc.8.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]