Abstract
African Americans carrying two apolipoprotein L1 gene (APOL1) renal risk variants have a high risk for nephropathy. However, only a minority develops end-stage renal disease (ESRD). Hence, modifying factors likely contribute to initiation of kidney disease such as endogenous (HIV infection) or exogenous (interferon treatment) environmental modifiers. In this report, genome-wide association studies and a meta-analysis were performed to identify novel loci for non-diabetic ESRD in African Americans and to detect genetic modifiers in APOL1-associated nephropathy. Two African American cohorts were analyzed, 1749 non-diabetic ESRD cases and 1136 controls from Wake Forest and 901 lupus nephritis (LN)ESRD cases and 520 controls with systemic lupus erythematosus but lacking nephropathy from the LNESRD Consortium. Association analyses adjusting for APOL1 G1/G2 renal-risk variants were completed and stratified by APOL1 risk genotype status. Individual genome-wide association studies and meta-analysis results of all 2650 ESRD cases and 1656 controls did not detect significant genome-wide associations with ESRD beyond APOL1. Similarly, no single nucleotide polymorphism showed significant genome-wide evidence of an interaction with APOL1 risk variants. Thus, although variants with small individual effects cannot be ruled out and are likely to exist, our results suggest that APOL1-environment interactions may be of greater clinical importance in triggering nephropathy in African Americans than APOL1 interactions with other single nucleotide polymorphisms.
Keywords: African Americans, APOL1, chronic kidney disease, FSGS, gene-gene interaction, GWAS
Introduction
Apolipoprotein L1 gene (APOL1)-associated nephropathy is the result of an inherited predisposition coupled with exposure to modifying factors that induce a spectrum of progressive chronic kidney diseases (CKD).1;2;3; Infection with HIV and exogenous administration of interferon are environmental factors or “second hits” that trigger nephropathy in individuals possessing two APOL1 renal-risk variants.4;5 These gene-environment interactions cause HIV-associated nephropathy (HIVAN) and focal segmental glomerulosclerosis (FSGS), collapsing variant, respectively. Lupus nephritis (LN) is also characterized by high interferon levels and severe nephropathy or end-stage renal disease (ESRD) and is strongly associated with APOL1.6;7 Other environmental factors likely modify nephropathy risk; inflammatory factors are logical triggers because they stimulate increased expression of APOL1 messenger RNA.8 Odds ratios (ORs) for APOL1 risk genotype association in LN-ESRD and glomerulosclerosis range from ~3 to 7.3.1;7 Although these are large effects for a complex disease, they are at the lower end of the APOL1 disease spectrum (relative to ORs of 17 in FSGS and 29–89 in HIVAN). Based on the relatively lower ORs, we hypothesized that LN-ESRD and hypertension-attributed ESRD would more likely reveal second hits, because effects of the APOL1 genotype were less strong than in FSGS and HIVAN.
The dominating effect of the APOL1 G1 and G2 renal-risk alleles may mask or modify the effects of other CKD risk loci.9 For example, a null variant in the nearby apolipoprotein L3 gene (APOL3) on chromosome 22q reproducibly interacts with APOL1-mediated risk for CKD in targeted analyses.10 Roles for non-APOL genes in ESRD have been sought in a genome-wide association study (GWAS) using pooled DNA, a combined Wake Forest School of Medicine (WFSM)/National Institute of Diabetes and Digestive and Kidney Disease “Family Investigation of Nephropathy and Diabetes” (FIND) report and in the African American Study of Kidney Disease and Hypertension (AASK).11;12 Initial results suggested that variation in or near the podocin (NPHS2), serologically defined colon cancer antigen 8 (SDCCAG8), bone morphogenetic protein 4 (BMP4), and glutathione s-transferase mu 1 genes (GSTM1) might interact with APOL1 to alter risk of CKD.12;13 These potential interactions require additional assessment and replication.
The present analyses sought to identify additional ESRD risk loci or loci whose effect is modified by the APOL1 renal-risk genotype in an expanded set of samples from African Americans including cases with non-diabetic ESRD and controls without nephropathy. This study formally adjusted and stratified by APOL1 G1/G2 risk status to increase power to detect novel ESRD risk-loci and tested for an interaction between APOL1 G1/G2 risk status and single nucleotide polymorphisms (SNPs) across the genome. Explicitly incorporating APOL1 risk genotype status into these analyses might help identify novel pathways. If strong associations do not emerge, results may suggest that the roles of secondary loci and SNP-by-APOL1 interactions are individually less clinically relevant in initiation of renal injury and that environmental triggers interacting with APOL1 are of greater clinical importance.
Results
Sample characteristics
The WFSM non-diabetic ESRD cohort and LN-ESRD cohort had comparable African ancestral proportions and APOL1 genotype frequencies; however, several clinical differences were present (Table 1). Cases in the LN-ESRD cohort exhibited an earlier age at onset of dialysis than did cases in the non-diabetic ESRD cohort (34 years vs. 49 years) and a stronger female bias (88% vs. 41%), both expected clinical findings. Within the WFSM non-diabetic ESRD cohort, cases and controls differed in African ancestral proportions, percent female, and age at ESRD onset in cases and at recruitment in controls (Table 1). Within the LN-ESRD cohort, African ancestral proportions were comparable; controls with systemic lupus erythematosus (SLE) lacking nephropathy had a modest increase in percentage female relative to cases. LN-ESRD cases had a markedly earlier age of SLE onset and fewer American College of Rheumatology (ACR) SLE criteria, relative to the SLE controls without nephropathy. A power analysis for the case-control meta-analysis (2650 cases and 1656 controls) of the association with SNPs with ESRD showed that for a range of minor allele frequencies (MAF=0.1, 0.2, 0.3 and 0.4) and under an additive genetic model, the study was powered to detect odds ratios (ORs) of 1.28–1.46, and 1.25–1.41 for type 1 error rates of α=5×10−8 and α=1×10−6, respectively (Supplementary Figure S1). A parallel analysis for the SNP-by-APOL1 risk variant interaction indicates the study is powered to detect interaction effects of OR=1.36–1.60, and 1.32–1.53 for type 1 error rates of α=5×10−8 and α=1×10−6, respectively (Supplementary Figure S1).
Table 1.
Demographic and clinical characteristics of African American study cohorts†
| WF non-diabetic ESRD cases |
WF non- nephropathy controls |
P-value | LN-ESRD cases | SLE non- nephropathy controls |
P-value | |
|---|---|---|---|---|---|---|
|
|
||||||
| N | 1749* | 1136 | 901 | 520 | ||
| 0/1 copies APOL1, n | 945 | 988 | 678 | 465 | ||
| 2 copies APOL1, n | 803 | 148 | 223 | 55 | ||
| Female, n (%) | 713 (40.8%) | 563 (49.6%) | 3.38×10−6 | 789 (87.6%) | 482 (92.7%) | 0.0025 |
| Age at recruitment, years | 55.5 ± 14.3 (55) | 50.6 ± 10.9 (50) | 1.87×10−22 | 41.5 ± 11.7 (42) | 41.7 ± 11.8 (42.4) | 0.6836 |
| Age at ESRD, years | 49.4 ± 15.5 (49) | NA | NA | 33.9 ± 11.6 (33) | NA | NA |
| African ancestry, proportion | 0.84 ± 0.08 (0.86) | 0.83 ± 0.09 (0.84) | 2.48×10−7 | 0.83 ± 0.09 (0.85) | 0.82 ± 0.09 (0.84) | 0.1645 |
| Hypertension, n (%) | 780 (96.3%) | NA | NA | |||
| History of ESRD in first-degree relative | NA | NA | 59 (8.4%) | NA | NA | |
| Age at SLE onset, years | NA | NA | NA | 27.2 ± 10.8 (25) | 39.0 ± 12.3 (40) | 5.28×10−46 |
| Time from SLE onset to ESRD, years | NA | NA | NA | 7.51 ± 7.38 (5) | NA | NA |
| No. of ACR SLE criteria met (excluding renal criteria) | NA | NA | NA | 4.32 ± 1.77 (4) | 4.97 ± 1.13 (5) | 1.23×10−12 |
| Cytotoxic therapy, n (%) | NA | NA | NA | 567 (85.6%) | NA | NA |
Continuous variables reported as mean ± standard deviation (median)
One case did not have APOL1 typing, so only 1748 cases in case-only or APOL1 adjusted analyses
Abbreviations: WF Wake Forest; LN lupus nephritis; ESRD end-stage renal disease; SLE systemic lupus erythematosus; ACR American College of Rheumatology
Meta-analysis of ESRD
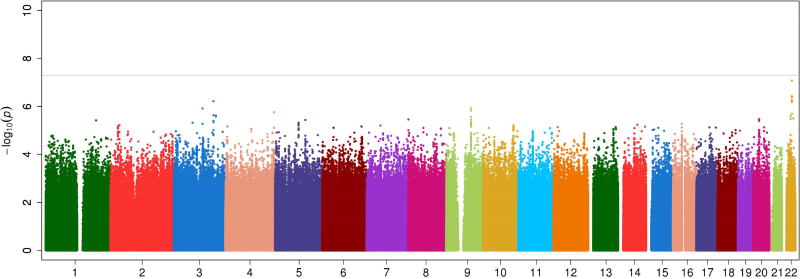
Beyond the APOL1 region, the meta-analysis across the two cohorts for SNP association with ESRD did not show evidence of association that met genome-wide significance (p<5×10−8 or a significant Benjamini-Hochberg false discovery rate adjusted p-value (PFDR) <0.05) (Supplementary Figure S2). Table 2 provides the five regions with the strongest evidence of association beyond APOL1 (i.e., smallest p-values and linkage disequilibrium [LD] support in genotyped SNPs). As expected, under a recessive genetic model the APOL1 G1/G2 risk variants were most strongly associated with ESRD (p=5.98×10−76). The OR was more than twice as large in the non-diabetic cohort (OR=7.00) compared to the SLE cohort (OR=2.72). The non-APOL1 regions exhibiting the strongest evidence of association and having at least two genotyped SNPs in LD support of the association were on chromosomes 8p23 and 14q24 (Table 2). The association at 8p23 (rs2404298, p=1.4×10−6) was observed in the WFSM non-diabetic ESRD cohort and is near disks large-associated protein 2 gene (DLGAP2), which is expressed in the brain and kidney. Although rs2404298 is within an enhancer histone mark, neither it nor a SNP in LD with it is a known expression quantitative trait locus (eQTL) for any gene and the functional link with DLGAP2 is unclear. A second association within the 8p23 region (rs4292733, p=2.1×10−6) is intronic to an eQTL for Rho guanine nucleotide exchange factor 10 gene (ARHGEF10). ARHGEF10 may play a role in neural morphogenesis but is not known to be expressed in the kidney. The association on 14q24 (rs7142086, p=2.2×10−6) is within promoter and enhancer histone marks in multiple tissues and is in proximity to DDB1- and CUL4-associated factor 4 gene (DCAF4), which has variants associated with shorting of telomere length in leukocytes. This association is potentially interesting, given the recent evidence of a link between telomere length and CKD.14;15;16
Table 2.
Genome-wide association study meta-analysis of ESRD case-control analysis
| SNP | Chr | Position | Gene Region | Minor Allele |
MAF WF non- diabetic ESRD cases (N=1749) |
MAF WF non- nephropathy controls (N=1136) |
MAF LN- ESRD cases (N=901) |
MAF SLE non- nephropathy controls (N=520) |
Meta- Analysis P-value |
OR (95% CI) |
OR Non- Diabetic ESRD |
OR LN- ESRD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1:13891452 d | 1q31.3 | 193891453 | C | 0.27 | 0.24 | - | - | 4.62×10−4 | 1.32 (1.13–1.54) | 1.32 | ||
| rs479618 i,d | 1q31.3 | 193897885 | A | 0.27 | 0.24 | 0.26 | 0.22 | 3.42 ×10−6 | 1.36 (1.19–1.55) | 1.33 | 1.39 | |
|
| ||||||||||||
| rs2404298 i | 8p23.3 | 1369019 | near DLGAP2 | A | 0.12 | 0.08 | - | - | 1.44 ×10−6 | 1.55 (1.29–1.85) | 1.55 | - |
| rs4404941 | 8p23.3 | 1372399 | near DLGAP2 | C | 0.12 | 0.08 | - | - | 4.26 ×10−6 | 1.57 (1.29–1.90) | 1.57 | - |
|
| ||||||||||||
| rs4292733 i | 8p23.3 | 1839973 | ARHGEF10 | G | 0.21 | 0.18 | 0.21 | 0.16 | 2.11 ×10−6 | 1.33 (1.18–1.50) | 1.27 | 1.42 |
| rs4242520 d | 8p23.3 | 1840697 | ARHGEF10 | G | 0.15 | 0.13 | 0.15 | 0.12 | 1.87 ×10−5 | 1.37 (1.18–1.58) | 1.37 | 1.36 |
| rs6420198 i,d | 8p23.3 | 1840952 | ARHGEF10 | T | 0.14 | 0.12 | 0.13 | 0.10 | 1.95 ×10−5 | 1.37 (1.18–1.60) | 1.38 | 1.36 |
|
| ||||||||||||
| rs112975886 | 9q22.31 | 95119255 | CENPP | G | 0.08 | 0.06 | 0.08 | 0.05 | 5.84 ×10−5 | 1.44 (1.21–1.73) | 1.41 | 1.50 |
| rs73520599 i | 9q22.31 | 95265685 | CENPP | C | 0.09 | 0.06 | 0.08 | 0.05 | 3.00 ×10−6 | 1.51 (1.27–1.79) | 1.49 | 1.54 |
|
| ||||||||||||
| rs7142086 d | 14q24.2 | 73391593 | DCAF4 | T | 0.36 | 0.40 | - | - | 2.20 ×10−6 | 0.68 (0.58–0.80) | 0.68 | - |
|
| ||||||||||||
| APOL1 G1/G2 | 22q12 | 36661784 | APOL1 | 0.46 | 0.13 | 0.25 | 0.11 | 5.98 ×10−76 | 4.74 (3.67–6.12) | 7.00 | 2.72 | |
i – imputed SNP in MegaChip cohort (all SNPs are imputed in the LN-ESRD cohort)
d or r – dominant, or recessive model; if not noted, additive model was used
Abbreviations: MAF minor allele frequency; WF Wake Forest; LN lupus nephritis; SLE systemic lupus erythematosus; OR odds ratio; CI confidence interval
Adjusting for the APOL1 G1/G2 risk genotype as a binary covariate in the logistic regression model accounted for the association within the APOL1 region but had little effect on the evidence of association across the remainder of the genome (Figure 1). The strongest evidence of association, after adjusting for the APOL1 risk genotype, was with rs13084795 at 3q25 near muscleblind-like splicing regulator 1 gene (MBNL1, p=6.08×10−7). Haploreg reports that rs13084795 is in LD (r2=0.84) with SNPs within promoter and enhancer histone marks an eQTL for RP11-788A4.1, but little is known about this gene. Thus, neither meta-analysis (adjusting and not adjusting for the APOL1 G1/G2 risk genotype) provided strong (statistically significant) evidence of a locus associated with ESRD in the two cohorts and the regions that provided evidence suggestive of association did not have clear relationships to kidney function.
Figure 1.
Genome-wide association meta-analysis for ESRD adjusting for APOL1 renal-risk genotype status (admixture proportion, age, and gender as covariates)
The ESRD association analysis was repeated stratifying by APOL1 G1/G2 risk genotype status. Again, little evidence was found for significant SNP-ESRD association within either stratum from the meta-analysis or in the individual cohorts (Supplementary Figures S3 and S4). In individuals without the APOL1 G1/G2 risk genotype, the rs2404298 association noted above was the strongest association (p=4.9×10−7, non-diabetic ESRD OR=1.76). The most intriguing association in individuals without the APOL1 risk genotype was with the intronic SNP rs17112571 on 10q23 (p=3.4×10−6, non-diabetic ESRD OR=1.45 and LN-ESRD OR=1.42) within the renalase gene (RNLS), coding for a flavoprotein that is secreted by the kidney into the blood and modulates cardiac function and systemic blood pressure (Supplementary Figure S5).17;18 This SNP is in high LD in African ancestral populations with numerous RNLS intronic SNPs that appear conserved and are within promoter and enhancer histone marks in multiple tissues and are eQTL for RNLS.19 Given the potential role of RNLS in cardiovascular disease and blood pressure control and that circulating renalase is reduced in individuals with CKD,20 this association requires further study.
The GWAS Catalog is a joint effort of the National Human Genome Research Institute (NHGRI) and the European Bioinformatics Institute (EMBL-EBI) that curates GWAS to generate a dataset of SNP associations with p-values <1.0 × 10−5. Ainsworth et al., queried the GWAS Catalog for CKD and related phenotypes (ESRD, estimated glomerular filtration rate, diabetic kidney disease, IgA nephropathy) and identified 132 SNP associations from 18 studies.21 Supplementary Table S1 reports the evidence of association with ESRD in the WFSM non-diabetic ESRD and LN-ESRD cohorts, adjusting for age, gender, admixture, and APOL1 G1/G2 risk genotype. Outside the chromosome 22q12 region that contains APOL1, none of these SNPs shows evidence of an association with ESRD. In consideration of the potential impact of the weighting strategy in GWAS, we repeated the analyses weighting by the inverse of the variance. The two GWAS methods yielded comparable results (Supplementary Table S3; Supplementary Figures S12 and S13).
GWAS for APOL1-SNP interaction
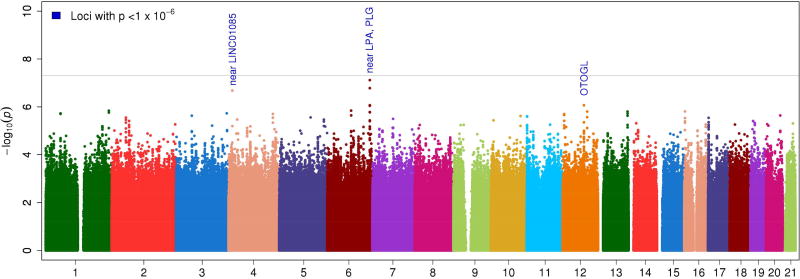
Given the magnitude of the effect of the APOL1 genotype on ESRD risk, it is possible that other variants might predispose to ESRD with effects influenced by the presence or absence of the APOL1 risk genotype. The genome-wide ESRD case-only meta-analysis for identifying SNP-by-APOL1 risk genotype interactions identified three regions with suggestive evidence of an interaction (Table 3, Figure 2). However, none met genome-wide significance (P<5×10−8) or had a significant Benjamini-Hochberg false discovery rate-adjusted p-value (PFDR) <0.05. The region with the strongest evidence of an interaction with APOL1 risk genotypes was on 6q26 (rs79741405, case-only interaction OR=2.97, p=7.7×10−8), an intergenic region 6.9kb 5’ of LPA (Supplementary Figure 6). The evidence of an interaction in ESRD cases vs. non-ESRD controls for this SNP was less significant (OR=4.44, p=0.0078). The interaction manifests as increased risk in those with APOL1 G1/G2 risk genotypes and is slightly stronger in the LN-ESRD cohort (Table 3). At ~10kb from rs79741405, rs2115868 (r2=0.95 with rs79741405) leads a group of SNPs with both promoter and enhancer histone marks in several tissues and the region appears to bind to the protein CTCF, a protein whose published effects are tied to various cancers and DNA loop structure. LD for the most strongly associated SNPs does not appear to extend to the most proximal gene, LPA, a gene that codes for a protein that comprises a substantial portion of lipoprotein(a) and has been repeatedly associated with cardiovascular disease.22 Results were suggestive, but interesting given the mixed evidence for a role of APOL1 in lipid biology and cardiovascular disease.23;24;25
Table 3.
Genome-wide association meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
| Meta-Analysis (ESRD cases only, APOL1 risk is the outcome) | Non-diabetic ESRD case- control analysis |
LN-ESRD case- SLE control Analysis |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| SNP | Chr | Position | Gene Region |
Minor Allele |
MAF WF non-diabetic ESRD cases APOL1 risk (N=803) |
MAF WF non- nephropathy controls APOL1 non- risk (N=148) |
MAF LN-ESRD cases APOL1 risk (N=223) |
MAF SLE non- nephropathy controls APOL1 non-risk (N=55) |
Meta P-value |
OR (95% CI) |
OR non- diabetic ESRD |
OR LN- ESRD |
OR APOL1 Risk |
OR APOL1 Non-Risk |
OR APOL1 Risk |
OR APOL1 Non- Risk |
| rs7660268 d | 4p15 | 14349700 | near LINC01085 | A | 0.15 | 0.10 | 0.14 | 0.12 | 2.10 ×10−7 | 1.63 (1.32–2.00) | 1.91 | 1.31 | 1.49 | 0.73 | 1.08 | 0.83 |
| rs75167652 d | 6q26 | 161091168 | near LPA, PLG | G | 0.04 | 0.02 | 0.05 | 0.02 | 1.70 ×10−6 | 2.70 (1.81–4.04) | 2.13 | 3.75 | 3.82 | 0.66 | 2.01 | 0.84 |
| rs79741405 i, d | 6q26 | 161094269 | near LPA, PLG | T | 0.05 | 0.02 | 0.05 | 0.02 | 7.70 ×10−8 | 2.97 (2.01–4.39) | 2.25 | 4.34 | 1.96 | 0.89 | 2.86 | 0.95 |
| rs1551122a | 12q21 | 80765800 | OTOGL | G | 0.19 | 0.15 | 0.21 | 0.17 | 8.61 ×10−7 | 1.49 (1.27–1.76) | 1.46 | 1.54 | 1.59 | 0.73 | 0.92 | 1.11 |
i – imputed SNP in MegaChip cohort (all SNPs are imputed in the LN-ESRD cohort)
d or r – dominant, or recessive model; if not noted, additive model was used
Abbreviations: MAF minor allele frequency; WF Wake Forest; LN lupus nephritis; SLE systemic lupus erythematosus; OR odds ratio; CI confidence interval
Figure 2.
Genome-wide ESRD case-only APOL1 × SNP interaction analysis
The second strongest evidence of an interaction was on 4p15 (rs7660268, case-only interaction OR=1.63, p=2.1×10−7). As expected, the ESRD case vs. non-ESRD control analysis provided less significant evidence of an interaction for this SNP (OR=1.64, p=0.0148). This is an intergenic region with limited evidence of histone markers or transcription factor binding sites. The region with the third strongest evidence of an interaction was on 12q21 (rs1551122, case-only OR=1.49, p=8.6×10−7; ESRD case vs. non-ESRD control analysis OR=1.61, p=0.0016). This SNP is a missense variant and intronic within the otogelin-like gene (OTOGL) which is expressed in the inner ear of vertebrates. Variants in this gene are linked to various forms of deafness. Thus, the latter two potential APOL1-SNP interaction regions provide modest evidence of an interaction and have little supporting evidence for roles in renal dysfunction or related traits.
Previously, SNPs within genomic regions near NPHS2 (SNP rs16854341), SDCCAG8 (rs2802723) and BMP4 (rs8014363) provided evidence suggestive of an interaction with APOL1.12;13 Given the overlap of these samples in the WFSM cohort, none of these SNP-by-APOL1 interactions was corroborated by the present results (rs16854341 in NPHS2 OR=0.90, p=0.0523; rs2802723 in SDCCAG8 OR=1.17, p=0.4878; rs8014363 near BMP4 OR=1.29, p=0.0197). The locus zoom plots provide evidence for APOL1-by-SNP interactions across these regions (Supplementary Figures S7, S8 and S9), highlighting a priori SNP-interactions and the most significant association within the region. None of these SNPs provide evidence of an interaction with APOL1 risk genotypes. The hemoglobin S variant was genotyped and the GSTM1 gene region amplified (to measure its product size on an agarose gel to detect the putative null allele associated with kidney disease) in cases and controls from both cohorts. No evidence of a main effect of hemoglobin S or GSTM1 (with or without adjusting for APOL1 risk) was seen for association with ESRD, nor was there an interaction between these variants and APOL1 risk variant status (Supplementary Table S2, Supplementary Figures S10 and S11). In addition, the effect of hemoglobin S alleles were assessed using LAMP (Local Ancestry in adMixed Populations)26 and results were comparable (data not shown).
Discussion
Several disorders in the APOL1-associated nephropathy spectrum have clear mediating factors; these include HIVAN and interferon-associated FSGS, collapsing variant. Others lack obvious modifying factors, such as idiopathic forms of FSGS and focal global glomerulosclerosis (FGGS). Remaining disorders in this spectrum such as severe lupus nephritis and sickle cell nephropathy currently lack known modifying second hits. It is unlikely that anti-nuclear antibodies or sickled red blood cells per se are sufficient to initiate CKD because the majority of patients with SLE and sickle cell disease with two APOL1 renal-risk variants do not develop nephropathy.7;27;28 Results of these analyses in 4 306 African Americans, including 2 649 cases with ESRD on renal replacement therapy, failed to detect genome-wide significant (or FDR-adjusted p<0.05) evidence of polymorphisms associated with ESRD beyond APOL1 (adjusting and not adjusting for the APOL1 risk genotype). Proximal genes in 8p23 and 14q24, two regions with statistical evidence suggestive of association with ESRD, were linked to renal expression, DNA looping or shortened telomere length, respectively. Telomere length has been associated with CKD.14 This study also failed to identify genome-wide significant SNP-by-APOL1 risk genotype interactions. The most intriguing SNP-by-APOL1 risk genotype interaction was on 10q23 linked to renalase, a gene whose protein has been implicated in cardiovascular disease and blood pressure regulation and whose levels are reduced in CKD. These results require further evaluation in well-powered studies.
Modifying factors causing the approximately 20% of genetically susceptible individuals to develop APOL1-associated non-diabetic kidney disease remain critical to identify. Advantages to categorizing environmental and genetic factors interacting with APOL1 to initiate CKD include the potential to develop novel treatments directed at disease modifiers and an improved ability to screen populations who possess recent African ancestry for risk of kidney disease. Individuals with the two-renal-risk APOL1 genotype have an approximate 4% lifetime risk to develop FSGS with nephrotic syndrome and 50% of HIV-infected individuals will develop HIVAN absent anti-retroviral therapy;5 approximately 20% are expected to develop sub-nephrotic FSGS or FGGS with low level proteinuria (Martin Pollak personal communication; May 2017). FSGS with low level proteinuria and FGGS are often misclassified as hypertension-attributed nephropathy, despite data that treating mild-to-moderate hypertension does not halt nephropathy progression in African Americans or members of other ethnic groups.29;30 Although other genetic loci of more modest effect size cannot be excluded and are likely to exist, the present results lead us to hypothesize that environmental modifiers are more likely clinically impactful mediators in APOL1-associated nephropathy. This hypothesis is based on existing data that reveal inflammation-driven up-regulation of the APOL1 gene, effects of interferons, and roles of interacting risk (HIV) and protective (JC polyoma) viruses in APOL1-associated nephropathy.5;8;31 In addition, the sickle variant of the hemoglobin gene did not interact with APOL1-mediated risk of nephropathy in two prior reports, suggesting the hemoglobin S allele and APOL1 variants have independent effects on CKD.27;28 In the cohorts analyzed herein, GSTM1 null alleles and hemoglobin S variants were not independent risk factors for ESRD, nor did they interact with APOL1 to modify nephropathy risk.
This study has limitations. Although it is the largest report of its type to date, the sample size was relatively modest for a GWAS meta-analysis. Here, the genome-wide case-control meta-analysis was powered to detect ORs of 1.25–1.46. Thus, true associations of more modest effect sizes, as seen in many large studies of complex genetic traits, may reside in the loci with evidence suggestive of association. A similar statement holds for the test of SNP-by-APOL1 risk genotype interactions, where the study is genome-wide powered to detect interactions ORs of 1.32 to 1.60. However, the effects of APOL1 on non-diabetic ESRD are powerful (e.g., ORs range from 3 to 89 in patients with lupus-ESRD and HIVAN, respectively) and adjusting for these effects should increase the power to detect associations.4;5;7 Imputation here is a useful tool to improve the statistical power to detect associations and interactions ultimately require genotyping of any key associations, as was done. Given this study found only suggestive evidence, independent cohorts will be required for focused assessment of the potential effects of these loci. The study was powered to detect main effects and non-APOL1 SNP-by-APOL1 interactions common to both LN-ESRD and non-diabetic ESRD. If the specific SNP association or interaction is not shared across the disorders, the study is powered only for larger effects and focused recruitment for these diseases would be required. Potential associations with the renalase gene need to be further assessed in other cohorts of African Americans with ESRD. Epigenetic factors, copy number variants, untagged indels, SNPs with small individual effects and rare SNPs remain potential second hits in APOL1-associated nephropathy. Modifying factors in the nephropathies associated with LN and sickle cell trait may differ in APOL1-associated and non-APOL1 associated forms of these diseases. This is important because APOL1 appears to be a progression factor from CKD to ESRD; hence, second hits underlying the initiation and renal histology in LN and sickle cell nephropathy may differ from those that are APOL1-associated and likely to cause progression to ESRD.
In conclusion, this relatively large GWAS failed to identify genes or genetic variants that met genome-wide (or FDR-adjusted p<0.05) significance for association with non-diabetic ESRD in African Americans, beyond the known effect of APOL1. Furthermore, no SNP met the genome-wide criteria for interaction with APOL1 risk genotype status to modify the risk of severe nephropathy. Although this finding does not exclude a role for interactive genes with weaker effects (lower ORs) and additional replication studies are required, the present results suggest that interactive SNPs are likely to individually have minor effects and will require larger sample sizes for detection. We conclude that interactive environmental factors are likely to play more clinically prominent roles in modifying nephropathy risk in individuals who are genetically susceptible to APOL1–associated nephropathy. As with use of highly active anti-retroviral therapy and rapidly falling rates of HIVAN, this interpretation provides hope that other modifiable environmental risk factors can be treated, or exposure prevented, to reduce the development of APOL1-associated kidney disease in genetically susceptible individuals.
Methods
Samples
DNA samples in this report came from non-diabetic WFSM participants who were self-described African Americans with ESRD (cases) and population-based individuals lacking nephropathy (controls).13 Causes of CKD in these non-diabetic cases included hypertension-attributed ESRD, non-specific forms of glomerulosclerosis, FSGS, HIVAN, and “unknown cause” in the absence of a kidney biopsy. Individuals with ESRD attributed to diabetes mellitus, Alport’s syndrome, renal cystic disorders, surgical nephrectomy, obstructive nephropathy, or other glomerular diseases (IgA nephropathy, membranous or membranoproliferative glomerulonephritis) were excluded.
As reported, samples collected by the LN-ESRD Consortium from 18 academic referral centers included cases with ESRD due to LN and controls with systemic lupus erythematosus (SLE) who lacked nephropathy.7 Cases met World Health Organization class III, IV or V disease in the native kidney and were documented by biopsy or physician report. Analyses in the LN-ESRD Consortium were limited to African American participants.
Genotyping
WFSM case and control DNA samples were genotyped on the Illumina Multi-Ethnic Genotyping Array (MEGA; www.illumina.com) at WFSM. LN-ESRD Consortium samples were genotyped on at least one of the following Illumina arrays as part of a large African American systemic lupus erythematosus (SLE) GWAS: Illumina HumanOmni1-Quad BeadChip, Omni1S, and Omni 2.5; a large number of samples were typed on both the Omni1 and Omni1S.
WFSM and LN-ESRD cases and controls were genotyped on the Sequenom platform for the two SNPs in the APOL1 G1 nephropathy risk locus (rs73885319 and rs60910145) and the indel for the G2 risk locus (rs71785313) using custom assays designed at WFSM. G1 and G2 calls were visually inspected for quality control. For 318 LN-ESRD Consortium samples without direct APOL1 genotyping, imputed values were used for G1 and G2 where the imputation confidence and quality was high (see below for imputation methods). Patients were determined to be in the APOL1 risk group if they possessed two APOL1 risk alleles (heterozygous for G1 and G2, or homozygous for the G1 or G2 risk alleles).
Statistical Analysis
In both cohorts, samples were excluded if their call rates were <96% across high quality SNPs (described below), had excess autosomal heterozygosity, or had self-reported sex inconsistent with genetically inferred sex. Additionally, duplicates and first- and second-degree relatives were removed retaining a case over a control, or the sample with the highest call rate in pairs with the same affection status. Any sample with missing covariate data or APOL1 G1/G2 genotyping was also removed.
Admixture estimates were computed using the program ADMIXTURE32 on a linkage disequilibrium (LD)-pruned (r2<0.2) set of autosomal, high quality SNPs. Individuals from three HapMap Phase 3 populations (CEU: Utah residents with ancestry from northern and western Europe; CHB: Han Chinese in Beijing, China; YRI: Yoruba in Ibadan, Nigeria) were included as anchoring populations. Resulting admixture proportions were used to trim out any genetic outliers.
SNPs were deemed high quality if they had call rates >95%, no evidence of differential missingness between cases and controls (p<0.05), and no evidence of departure from Hardy-Weinberg Expectation proportions (controls p<0.01, ESRD cases p<0.000001).
To facilitate a meta-analysis across cohorts and because cohorts were genotyped on different arrays, both cohorts were imputed to the 1000 Genomes Phase 1 v3 integrated reference panel using IMPUTE2 software.33 Because LN-ESRD samples were genotyped across three distinct chips, imputation was performed separately by chip. For both the WFSM and LN-ESRD sets, the imputed genotyped data for SNPs with confidence scores >0.90 and information scores >0.50 were retained. Subsequent association analyses were computed using SNPTEST, which accounts for imputation uncertainty in the statistical analysis, as outlined below.33
Validation genotyping for significant imputed variants in the case-only analysis (8 SNPs: rs7660268, rs11942293, rs2327773, rs75167652, rs4348293, rs11114411, rs10862100, rs2053894) was completed for the ESRD cases on the Sequenom platform at WFSM with an efficiency of 95.2% (94.6–95.8%). Sixty-five blind duplicates were included to ensure genotyping accuracy and were >99% concordant.
Statistical association analyses
Logistic regression models were computed to test for SNP associations with ESRD within each cohort using SNPGWA and SNPTEST adjusting for age, gender, and admixture proportions; repeating the analyses adjusting only for admixture proportions yielded comparable results. Dominant, additive and recessive genetic models were computed for SNPs conditional on at least 10 or 30 homozygotes for the minor allele for the additive and recessive models, respectively. To combine results across the two cohorts, a weighted inverse normal meta-analysis was computed by genetic model, where the weights were the cohort sample size. Benjamini-Hochberg false discovery rate (FDR) adjusted p-values were computed within the meta-analysis to adjust for the number of tests computed.
To account for the dominating effect of the APOL1 G1/G2 risk genotype, an APOL1-adjusted and an APOL1 binary risk genotype-stratified analyses were computed. Specifically, in the first analysis APOL1 G1/G2 risk genotype status was included in the model as a binary covariate with age, gender, and admixture proportions, and a weighted meta-analysis was computed as above. In the stratified analyses, the two cohorts were stratified by the binary APOL1 G1/G2 risk genotype and the case-control analysis was computed within each stratum for each cohort (Wake Forest; LN-ESRD Consortium) and combined across cohorts via the weighted inverse normal meta-analysis. As in the overall analysis, FDR-adjusted p-values were computed within each of the three analyses (i.e., APOL1 G1/G2 included as covariate, APOL1 G1/G2 non-risk stratum, and APOL1 G1/G2 risk stratum).
Given the powerful effect of APOL1 genotypes on ESRD risk it is possible that the effects of other loci are dependent on the APOL1 genotype status of an individual. To test for an interaction between an individual SNP and APOL1 risk genotype status, a case-only logistic regression analysis was computed.34 Here, APOL1 risk genotype status is the binary outcome in the logistic regression and the SNP is the independent variable of interest, adjusting for admixture, age, and gender as covariates. The case-only analysis is statistically more powerful under independence between the SNP and the APOL1 genotype34, an assumption valid on non-chromosome 22 loci due to Mendel’s Law of Independent Assortment and valid outside the chromosome 22 APOL1 region when there is no evidence of LD. In addition to the case-only analysis, the case-control analysis was computed to confirm the evidence of an interaction and to estimate the effect size (odds ratio [OR]) of the interaction.
The RegulomeDB database was used to explore what might be the most likely functional polymorphisms in LD with the top associated SNP in a region. RegulomeDB annotates SNPs with known and predicted regulatory elements (eQTLs, DNAase hypersensitivity, and binding sites of transcription factors) in the intergenic regions of the human genome.35 It includes high-throughput, experimental data sets from GEO, the ENCODE project, published literature, as well as computational predictions and manual annotations to identify putative regulatory potential and identify functional variants. HaploReg v2 is a tool for exploring annotations of the noncoding genome at variants on haplotype blocks and uses LD information from the 1,000 Genomes Project Phase 1 individuals.36 It analyzes sets of SNPs for an enrichment of cell-type specific enhancers, and includes all dbSNP build 137 SNPs, predicted chromatin state in nine cell types, conservation across mammals, motif instances from ENCODE experiments, enhancer annotations on 90 cell types from the Roadmap Epigenome Mapping Consortium and eQTLs from the GTEx eQTL browser. The query was performed using default settings, including LD calculations based on the 1,000 Genomes Phase 1 YRI individuals, and epigenome data from both the ENCODE and Roadmap Epigenome Mapping Consortium projects.
Supplementary Material
Supplementary Figure S1. Power analyses (type 1 error rate of α) for test of A) SNP association with ESRD under Case-Control (2650 ESRD cases and 1656 controls) design, and B) SNP-by-APOL1 risk variant interaction under a Case-only (1026 ESRD cases with 2 APOL1 risk alleles and 1623 ESRD cases with 0 or 1 APOL1 risk alleles) design.
Supplementary Figure S2. Genome-wide association meta-analysis of ESRD cases (non-diabetic ESRD, lupus nephritis ESRD) versus respective non-nephropathy controls
Supplementary Figure S3. Genome-wide meta-analysis of ESRD cases versus non-nephropathy controls in individuals without APOL1 renal risk genotypes
Supplementary Figure S4. Genome-wide meta-analysis of ESRD cases versus non-nephropathy controls in individuals with APOL1 renal risk genotypes
Supplementary Figure S5. Zoom plot of the renalase gene (RNLS) in the genome-wide meta-analysis of ESRD cases versus non-nephropathy controls in individuals without APOL1 renal risk genotypes
Supplementary Figure S6. Zoom plot of the LPA-PLG gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S7. Zoom plot of the NPHS2 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S8. Zoom plot of the SDCCAG8 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S9. Zoom plot of the BMP4 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S10. Zoom plot of the hemoglobin S gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S11. Zoom plot of the GSTM1 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S12. Meta-analysis p-value calculation comparison (case-only adjusted for APOL1)
Supplementary Figure S13. Meta-analysis p-value calculation comparison (case-only SNP-by-APOL1 interaction)
Acknowledgments
The authors thank all the participants in these studies and are indebted to our colleagues in the Lupus Nephritis-End-Stage Renal Disease Consortium.
Support: NIH R01 DK084149 (BIF), R01 DK070941 (BIF), and RC2 AR058951 (RPK)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Wake Forest University Health Sciences and Barry Freedman have rights to an issued United States patent related to APOL1 genetic testing. Dr. Freedman is a consultant for Ionis Pharmaceuticals. Other authors have nothing to disclose.
References
- 1.Genovese G, Friedman DJ, Pollak MR. APOL1 variants and kidney disease in people of recent African ancestry. Nat Rev Nephrol. 2013;9:240–244. doi: 10.1038/nrneph.2013.34. [DOI] [PubMed] [Google Scholar]
- 2.Freedman BI, Skorecki K. Gene-gene and gene-environment interactions in apolipoprotein L1 gene-associated nephropathy. Clin J Am Soc Nephrol. 2014;9:2006–2013. doi: 10.2215/CJN.01330214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heymann J, Winkler CA, Hoek M, Susztak K, Kopp JB. Therapeutics for APOL1 nephropathies: putting out the fire in the podocyte. Nephrol Dial Transplant. 2017;32:i65–i70. doi: 10.1093/ndt/gfw402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasembeli AN, Duarte R, Ramsay M, et al. APOL1 Risk Variants Are Strongly Associated with HIV-Associated Nephropathy in Black South Africans. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2014050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopp JB, Nelson GW, Sampath K, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen CP, Beggs ML, Saeed M, Walker PD. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol. 2013;24:722–725. doi: 10.1681/ASN.2012121180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman BI, Langefeld CD, Andringa KK, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66:390–396. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nichols B, Jog P, Lee JH, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87:332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese G, Tonna SJ, Knob AU, et al. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int. 2010;78:698–704. doi: 10.1038/ki.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skorecki KL, Lee JH, Langefeld CD, et al. A null variant in the apolipoprotei L3 gene is associated with non-diabetic nephropathy. Nephrol Dial Transplant. 2017;1–8 doi: 10.1093/ndt/gfw451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bostrom MA, Lu L, Chou J, et al. Candidate genes for non-diabetic ESRD in African Americans: a genome-wide association study using pooled DNA. Hum Genet. 2010;128:195–204. doi: 10.1007/s00439-010-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodonyi-Kovacs G, Ma JZ, Chang J, et al. Combined Effects of GSTM1 Null Allele and APOL1 Renal Risk Alleles in CKD Progression in the African American Study of Kidney Disease and Hypertension Trial. J Am Soc Nephrol. 2016;27:3140–3152. doi: 10.1681/ASN.2015050487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Divers J, Palmer ND, Lu L, et al. Gene-gene interactions in APOL1-associated nephropathy. Nephrol Dial Transplant. 2014;29:587–594. doi: 10.1093/ndt/gft423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ameh OI, Okpechi IG, Dandara C, Kengne AP. Association Between Telomere Length, Chronic Kidney Disease, and Renal Traits: A Systematic Review. OMICS. 2017;21:143–155. doi: 10.1089/omi.2016.0180. [DOI] [PubMed] [Google Scholar]
- 15.Eguchi K, Honig LS, Lee JH, Hoshide S, Kario K. Short telomere length is associated with renal impairment in Japanese subjects with cardiovascular risk. PLoS One. 2017;12:e0176138. doi: 10.1371/journal.pone.0176138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloda K, Mierzecki A, Domanski L, et al. Joint Assessment of Donor and Recipient hTERT Gene Polymorphism Provides Additional Information for Early Kidney Transplantation Outcomes. Med Sci Monit. 2017;23:1812–1818. doi: 10.12659/MSM.900406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desir GV. Regulation of blood pressure and cardiovascular function by renalase. Kidney Int. 2009;76:366–370. doi: 10.1038/ki.2009.169. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q, Fan Z, He J, et al. Renalase gene is a novel susceptibility gene for essential hypertension: a two-stage association study in northern Han Chinese population. J Mol Med (Berl) 2007;85:877–885. doi: 10.1007/s00109-006-0151-4. [DOI] [PubMed] [Google Scholar]
- 19.Stec A. Rs10887800 renalase gene polymorphism influences the level of circulating renalase in patients undergoing hemodialysis but not in healthy controls. BMC Nephrol. 2017;18:118. doi: 10.1186/s12882-017-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desir GV, Peixoto AJ. Renalase in hypertension and kidney disease. Nephrol Dial Transplant. 2014;29:22–28. doi: 10.1093/ndt/gft083. [DOI] [PubMed] [Google Scholar]
- 21.Ainsworth HC, Langefeld CD, Freedman BI. Genetic epidemiology in kidney disease. Nephrol Dial Transplant. 2017;32:ii159–ii169. doi: 10.1093/ndt/gfw270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellwig KP, Horstkotte D, van BF. Lipoprotein (a) and coronary heart disease - is there an efficient secondary prevention? Clin Res Cardiol Suppl. 2017;12:18–21. doi: 10.1007/s11789-017-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lugli EB, Pouliot M, Portela MP, Loomis MR, Raper J. Characterization of primate trypanosome lytic factors. Mol Biochem Parasitol. 2004;138:9–20. doi: 10.1016/j.molbiopara.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Weckerle A, Snipes JA, Cheng D, et al. Characterization of circulating APOL1 protein complexes in African Americans. J Lipid Res. 2016;57:120–130. doi: 10.1194/jlr.M063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean NO, Robinson TW, Freedman BI. APOL1 Gene Kidney Risk Variants and Cardiovascular Disease: Getting to the Heart of the Matter. Am J Kidney Dis. 2017 doi: 10.1053/j.ajkd.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baran Y, Pasaniuc B, Sankararaman S, et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics. 2012;28:1359–1367. doi: 10.1093/bioinformatics/bts144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hicks PJ, Langefeld CD, Lu L, et al. Sickle cell trait is not independently associated with susceptibility to end-stage renal disease in African Americans. Kidney Int. 2011;80:1339–1343. doi: 10.1038/ki.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naik RP, Irvin MR, Judd S, et al. Sickle Cell Trait and the Risk of ESRD in Blacks. J Am Soc Nephrol. 2017 doi: 10.1681/ASN.2016101086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appel LJ, Wright JT, Jr, Greene T, et al. Intensive blood-pressure control in hypertensive chronic kidney disease. N Engl J Med. 2010;363:918–929. doi: 10.1056/NEJMoa0910975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A Randomized Trial of Intensive versus Standard Blood-Pressure Control. N Engl J Med. 2015 doi: 10.1056/NEJMoa1511939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Divers J, Nunez M, High KP, et al. JC polyoma virus interacts with APOL1 in African Americans with nondiabetic nephropathy. Kidney Int. 2013;84:1207–1213. doi: 10.1038/ki.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umbach DM, Weinberg CR. Designing and analysing case-control studies to exploit independence of genotype and exposure. Stat Med. 1997;16:1731–1743. doi: 10.1002/(sici)1097-0258(19970815)16:15<1731::aid-sim595>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 35.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Power analyses (type 1 error rate of α) for test of A) SNP association with ESRD under Case-Control (2650 ESRD cases and 1656 controls) design, and B) SNP-by-APOL1 risk variant interaction under a Case-only (1026 ESRD cases with 2 APOL1 risk alleles and 1623 ESRD cases with 0 or 1 APOL1 risk alleles) design.
Supplementary Figure S2. Genome-wide association meta-analysis of ESRD cases (non-diabetic ESRD, lupus nephritis ESRD) versus respective non-nephropathy controls
Supplementary Figure S3. Genome-wide meta-analysis of ESRD cases versus non-nephropathy controls in individuals without APOL1 renal risk genotypes
Supplementary Figure S4. Genome-wide meta-analysis of ESRD cases versus non-nephropathy controls in individuals with APOL1 renal risk genotypes
Supplementary Figure S5. Zoom plot of the renalase gene (RNLS) in the genome-wide meta-analysis of ESRD cases versus non-nephropathy controls in individuals without APOL1 renal risk genotypes
Supplementary Figure S6. Zoom plot of the LPA-PLG gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S7. Zoom plot of the NPHS2 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S8. Zoom plot of the SDCCAG8 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S9. Zoom plot of the BMP4 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S10. Zoom plot of the hemoglobin S gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S11. Zoom plot of the GSTM1 gene region in the genome-wide meta-analysis of ESRD case-only APOL1 × SNP interaction analysis
Supplementary Figure S12. Meta-analysis p-value calculation comparison (case-only adjusted for APOL1)
Supplementary Figure S13. Meta-analysis p-value calculation comparison (case-only SNP-by-APOL1 interaction)