Summary
Epigenetic modifications on chromatin are most commonly thought to be involved in the transcriptional regulation of gene expression. Due to their dependency on small molecule metabolites, these modifications can relay information about cellular metabolic state to the genome for the activation or repression of particular sets of genes. In this review, we discuss emerging evidence that these modifications might also have a metabolic purpose. Due to their abundance, the histones have the capacity to store substantial amounts of useful metabolites, or to enable important metabolic transformations. Such metabolic functions for histones could help explain the widespread occurrence of particular modifications that may not always be strongly correlated with transcriptional activity.
Keywords: Epigenetics, acetyl-CoA, SAM, histone acetylation, histone methylation, acetate
Introduction
The eukaryotic genome is packaged into chromatin by histone proteins that are subject to a wide range of posttranslational modifications (PTMs) including methylation, acetylation, phosphorylation, and ubiquitylation. These histone PTMs contribute to the transcriptional regulation of gene expression by influencing chromatin states. These modifications can regulate compaction between DNA and histones, as well as the recruitment of transcriptional activators, repressors, and their associated protein complexes [1, 2]. The underlying functional roles and regulation of histone modifications in gene expression remain an active area of investigation.
In parallel to this prevailing view, increasing evidence suggests that histone modifications are significantly influenced by the metabolic states of a cell. Many chromatin-modifying enzymes are dependent on intermediary metabolites that function as their substrates or cofactors, such as S-adenosylmethionine (SAM) or acetyl-Coenzyme A (acetyl-CoA), thereby rendering them sensitive to their intracellular availability. As a result, fluctuation of these metabolites can be reflected on the histones via the abundance of such modifications, while enabling the reprogramming of gene expression in tune with cellular metabolism. Therefore, such collaborative communication between chromatin and small molecule metabolites may have a specific purpose in relaying metabolic signals to the genome during organismal growth, survival, or development.
In this review, we will focus on reciprocal relationships between the epigenome and metabolism, mainly using histone methylation and acetylation as examples, and discuss how cellular SAM and acetyl-CoA levels affect chromatin states. While histone PTMs can function in the transcriptional regulation of metabolism, what has been often overlooked is that the biochemical reactions involving the deposition and removal of histone PTMs directly influence metabolism. Because histones are so abundant, their post-translational modification can consume substantial amounts of intermediary metabolites. As such, these intermediary metabolites are transformed to facilitate metabolism or deposited as small chemical units on chromatin that can be metabolically useful. Here, we propose and discuss the concept that the histones can function as repositories that facilitate cellular metabolic functions.
SAM and acetyl-CoA, representative intermediary metabolites linking metabolic states to the epigenome
Hundreds to thousands of biochemical reactions occur within a membrane-enclosed living cell, which collectively define cellular metabolism. Among more than 8000 stable small metabolites (MW < 1500 Da), there are a select few “sentinel metabolites” that have been proposed to power major metabolic pathways [3]. These include SAM and acetyl-CoA, both of which are kinetically stable but thermodynamically activated for transfer of carbon units [3]. Pathways that generate and utilize SAM and acetyl-CoA span nearly every aspect of biological processes, including lipid synthesis and breakdown, glycolysis and the tricarboxylic acid (TCA) cycle, and protein PTMs. Therefore, the spatial and temporal regulation of SAM and acetyl-CoA abundance will be imposed by metabolism and can influence chromatin states through methylation and acetylation.
Just as SAM and acetyl-CoA serve as donors to provide methyl and acetyl units for histone modification, nicotinamide adenine dinucleotide (NAD+), flavin adenine dinucleotide (FAD), and α-ketoglutarate, among others, are co-factors required for many reversal reactions to remove these groups from histones. In principle, the dependency on each of these intermediary metabolites provides an avenue to link the placement or removal of particular marks to the metabolic state. Here, we focus discussion on emerging, fundamental links between SAM and acetyl-CoA and epigenetic modifications on chromatin, which have revealed a metabolic purpose for both histone methylation and acetylation.
SAM metabolism and histone methylation
Biochemical basis of SAM
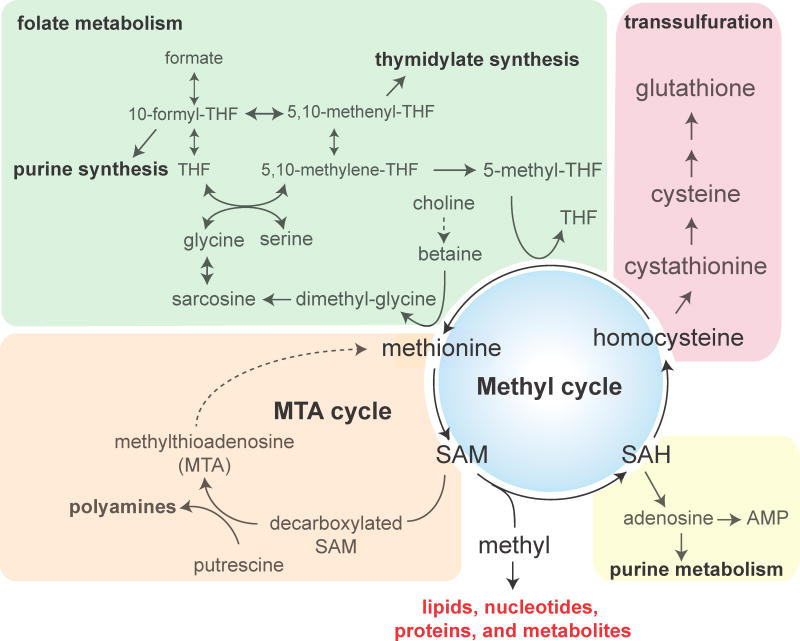
As the major methyl donor in the cell, SAM is considered the second most widely used metabolite by enzymes after ATP [4]. The estimated intracellular concentration of SAM (~10 μM) is only a small fraction of the ATP pool (1–3%) under steady state conditions, but SAM amounts can vary ~10–100 fold under physiological conditions [5–7]. Such fluctuations can potentially render SAM itself a key modulator of many cellular transmethylation reactions, such as for histone methylation. As shown in Figure 1, methionine and SAM metabolism are central to many metabolic pathways, including all SAM-dependent methylation processes, the transsulfuration reaction for cysteine and glutathione synthesis, folate metabolism, purine biosynthesis, and the methionine salvage pathway, also known as the 5′-methylthioadenosine (MTA) cycle.
Figure 1. Pathways for methionine and SAM metabolism.
SAM synthesized from methionine donates methyl groups to various methyl recipients. Alternatively, SAM can be decarboxylated for the synthesis of polyamines, and the recycling of the byproduct methylthioadenosine is known as the methionine salvage pathway. SAM-dependent methylation generates SAH, and the subsequent hydrolysis of SAH produces adenosine and homocysteine, which can feed purine metabolism and transsulfuration for cysteine and glutathione synthesis, respectively. Homocysteine can also be recycled for methionine synthesis. Folate and choline are metabolized to provide necessary methyl groups for remethylating homocysteine.
Metabolic function for SAM-dependent methylation
Approximately 1% of eukaryotic proteins encode SAM-dependent methyltransferases [8, 9]. These enzymes catalyze methyl group transfer from SAM to a variety of cellular substrates, including lipids, proteins, nucleic acids, and small metabolites. In addition to a broad range of biological functions performed by each transmethylation reaction, methylation itself is important for metabolism. This is because consumption of the methyl donor SAM is accompanied by the release of the reaction co-product, S-adenosylhomocysteine (SAH) (Figure 1). SAH is further hydrolyzed to homocysteine, the sole precursor for the biosynthesis of cysteine and glutathione in higher eukaryotes (Figure 1). Therefore, SAM turnover by methylation reactions in a cell can be essential to fuel the synthesis of cysteine, in the absence of other sources of this amino acid. In light of this, any methylation reaction that consumes substantial amounts of SAM can account for a considerable proportion of this methylation-based metabolic function. Several major SAM consumers have been postulated [Box 1], and an outstanding question pertains to how cells might employ SAM consumption as a mechanism for metabolic functions.
Box 1. Suggested major SAM consumers.
The syntheses of phosphatidylcholine by phosphatidylethanolamine (PE) methyltransferase, sarcosine by glycine N-methyltransferase (GNMT), and creatine by guanidinoacetate methyltransferase have been suggested to be three major SAM-consuming reactions. Loss of PE methyltransferase in mice resulted in a ~50% decrease in plasma homocysteine, suggesting that PE methylation accounts for a substantial portion of whole-body SAM [10]. GNMT deficiency led to very high levels of methionine and SAM in the plasma of children and in the mouse liver [11–13], underscoring a link between methionine and one-carbon metabolism by this methyltransferase. Creatine synthesis was thought to consume a significant amount of SAM-derived methyl groups in humans, which was later considered to be overestimated as dietary creatine is a major source of creatine [14, 15]. Nevertheless, given the dynamic nature of metabolism and distinct metabolic requirements, SAM-consuming capacities of methyltransferases are likely different between tissues and affected by diet. Surprisingly, histone methylation acts as a methyl sink in the absence of phospholipid methylation, which is discussed in detail in the text.
A recent study in yeast and cultured mammalian cells demonstrates that phospholipid methylation, an alternative pathway for phosphatidylcholine (PC) synthesis from methylation of phosphatidylethanolamine (PE), is the major consumer of SAM among methyltransferase enzymes [7]. Cells lacking an enzyme required for PE methylation accumulate SAM, leading to inefficient synthesis of cysteine and glutathione, sensitivity to oxidative stress, and hypermethylation of histones and a major protein phosphatase PP2A [7]. Therefore, the PE methyltransferase enzyme is entitled to two additional functions, beyond the synthesis of PC: 1) production of glutathione to defend against oxidative stress and 2) maintenance of methylation potential to sustain other critical methylation events. Indeed, it is possible that other SAM-consuming methylation reactions might be employed for a similar purpose if they become a major SAM consumer in particular settings.
SAM links metabolism to histone methylation
The Km values for SAM for various histone methyltransferases are in the low μM range [16, 17], suggesting that fluctuations of SAM concentrations can directly influence the rate of histone methylation. When dietary methionine enters cells, it is first and primarily utilized for the synthesis of SAM [4, 18]. Therefore, it is not surprising that the availability of dietary methionine can regulate histone methylation by impacting the rates of SAM synthesis [6, 7, 19–21]. SAM-consuming pathways, however, may act as a rheostat to regulate SAM levels, thereby influencing histone methylation. For example, during methionine restriction, PE methylation quickly depletes cellular SAM, which is accompanied by decreases in histone methylation [7]. In the absence of PE methylation, cellular SAM accumulates since methionine cannot be sufficiently metabolized towards the synthesis of cysteine and glutathione [7]. Therefore, a SAM-consuming pathway can affect sensitivity of histone methylation to nutritional methionine. This mechanism may be particularly important in the liver, where PE methyltransferase is most active [22]. PE methylation and one-carbon folate metabolism are coordinated to regulate SAM homeostasis and lipid biogenesis in liver cells [23, 24], and their deficiencies are often associated with conditions such as nonalcoholic fatty liver disease [18, 25]. Hereafter, we define the recipient of methyl groups during a methylation reaction (such as the phospholipid PE) as a methyl sink.
Interestingly, deficiency in nicotinamide N-methyltransferase (NNMT) in adipose tissue increases SAM amounts and methylation of H3K4 [26], suggesting nicotinamide can serve as an adipose-specific methyl sink linking SAM and NAD+ metabolism to chromatin regulation. Conversely, overexpression of NNMT in cancer cells can promote the hypomethylation of histones [27]. This further emphasizes the possibility that a methyl sink can be cell type-, disease-, or developmental stage-specific. In addition to SAM-consuming pathways, metabolic processes intersecting with SAM metabolism can also impact on histone methylation. Deficiency in folate, which provides methyl groups to re-methylate homocysteine for the synthesis of methionine, leads to aberrant histone methylation in both yeast and human cells [28] as well as in mouse liver [29]. Another example is a metabolic crosstalk between threonine and methionine in pluripotent stem cells. Metabolism of both amino acids is enhanced during acquisition of the pluripotent state [30], and threonine is catabolized and required for maintaining SAM and 5mTHF synthesis by generating glycine and acetyl-CoA [31]. Thus, depletion of threonine potently inhibits SAM synthesis, leading to a decrease in trimethylation of H3K4 [31].
Histones as a sink for methyl groups
While metabolic state and histone methylation are intrinsically linked through SAM abundance, how histone methylation is exploited for metabolic regulation remains an open question. Histone methylation occurs extensively on lysine and arginine residues throughout the histone proteins H1, H2A, H2B, H3 and H4, and more than 150 residues have been identified as methylated by mass spectrometry [32]. Among them, the most studied methylation sites are H3K4, H3K9, H3K27, H3K36, and H3K79 [Box 2]. Changes in global levels of histone methylation have been observed and linked to cancer aggressiveness [33]. Numerous genome-wide and locus-specific chromatin immunoprecipitation studies have shown both positive and negative association between histone methylation and transcription. Interestingly, there are many reported instances in which the same modification can have negligible, or even opposite effects on transcriptional activities [Box 2], leaving the role of histone methylation in gene expression obscure and complex. Considering that methylation of specific histone residues can have distinct functions depending on the gene locus, it is curious why cells might regulate histone methylation at the bulk level for modulating gene expression, if at all.
Box 2. Histone marks and transcription.
A popular view within the field of histone methylation research has been that histone methylation marks correlate with either positive or negative transcriptional states. For example, lysine methylation at H3K4, H3K36, and H3K79 and arginine methylation are often associated with transcriptional activation, while methylated H3K9, H3K27, and H4K20 are associated with transcriptional repression [2]. However, it remains difficult to predict transcriptional activity of specific gene loci based on the occupancy profiles of their methylation marks. At times, the marks associated with transcriptional activation can also be associated with transcriptional repression [50, 51]. Such examples are often seen for H3K4me3 [52–54] and H3K36me3 [55, 56].
Positively charged histone amino-terminal tails are thought to bind to DNA through electrostatic interactions or interactions between nucleosomes. Unlike methylation, acetylation neutralizes the charge of histone tails and weakens histone-DNA or nucleosome-nucleosome interactions, making the DNA of genes and their regulatory regions more accessible to nuclear protein factors, such as the transcriptional machinery [57, 58]. Therefore, histone acetylation is generally associated with active transcription.
However, histone proteins are so abundant that alterations in bulk amounts of methylated amino acid residues may impact metabolism, underlying a possible function beyond transcriptional regulation. As estimated in Table 1, each nucleosome in a mammalian cell, if fully methylated, can consume up to ~400 molecules of SAM. In theory, by this calculation, the methylation of only ~0.1% of all histone residues throughout the genome is able to deplete the entire SAM pool of the cell. Therefore, one might predict that the inhibition of histone methylation can cause SAM to accumulate, if this process does consume significant amounts of SAM. A recent study demonstrates that SAM amounts are boosted by ~100- to 200- fold by preventing single or combinatorial methylation of H3K4, H3K36, and H3K79 in cells deficient in phospholipid methylation, whose deficiency alone exhibits ~20-fold increase in SAM [7]. Underlying these marked increases in SAM in histone methyltransferase mutants is an unforeseen metabolic function for histone methylation.
Table 1.
Estimate of SAM and acetyl-CoA consumption by histones in yeast and human cells.
| Yeast (haploid) | Human | ||
|---|---|---|---|
| Genome size (bp) | 1.2×107 | 6.4×109 | |
| Nucleosome + linker DNA (bp) | 200 | 200 | |
| Nucleosome units | 6.1×104 | 3.2×107 | |
| Volume of cell (L) | 3.7×10−14 | 2.0×10−12 | |
| SAM | Estimated total methylation sites per histone octamer | 12 (K) | 96 (K) + 58 (R) |
| Molecules of SAM consumable per histone octamer | 36 | 404 | |
| Number/moles of SAM consumed per genome | 2.2×106/3.6×10−18 | 1.3×1010/2.1×10−14 | |
| Consumable SAM by histones (μM) | 98 | 11000 | |
| Cellular SAM concentration (μM) | 10 | 10 | |
| Acetyl-CoA | Estimated total acetylation sites per histone octamer | 30 | 123 |
| Number/moles of acetyl-CoA consumed per genome | 1.8×106/3.0×10−18 | 3.9×109/6.5×10−15 | |
| Consumable acetyl-CoA by histones (μM) | 82 | 3000 | |
| Cellular acetyl-CoA concentration (μM) | 20 | 20 | |
What metabolic functions might the nucleus employ the histones for? Because of the nature of the methylation reaction discussed above, histone methylation serves at least two metabolic functions: 1) Maintaining nuclear SAM homeostasis and methylation potential. In case of a high influx of SAM into the nucleus from the cytoplasm, increasing the rate of histone methylation can buffer against perturbations and minimize aberrant methylation of other substrates. In support of this idea, the absence of methylation at a particular histone residue can promote the hypermethylation of another in a compensatory-like manner [7]. Based on epistasis analysis, there appears to be a preference of particular sites on the histones for such a “sink” function: H3K36 > H3K79 > H3K4 [7]. 2) Fueling the nuclear synthesis of glutathione. All methylation reactions (including histone methylation) are accompanied by the conversion of SAM to SAH. SAH is hydrolyzed to homocysteine, the precursor for cysteine and glutathione synthesis. It is possible that histone methylation can facilitate the nuclear synthesis of glutathione, because nuclear glutathione synthesis is active [34] and a nuclear pool of related metabolic enzymes has been identified [35, 36]. It might also be necessary because reactive oxygen species that cause DNA damage are constantly generated in living cells [37], and histone demethylation reactions locally produce reactive byproducts, including formaldehyde and hydrogen peroxide (Figure 3) [38]. Intriguingly, an increase in H3K36 methylation is associated with enhanced DNA repair [39, 40] although it remains unknown whether glutathione production contributes to such a phenotype. Overall, the methylation of histones is integrated with cellular metabolism and has the capacity to significantly influence the nuclear metabolic environment.
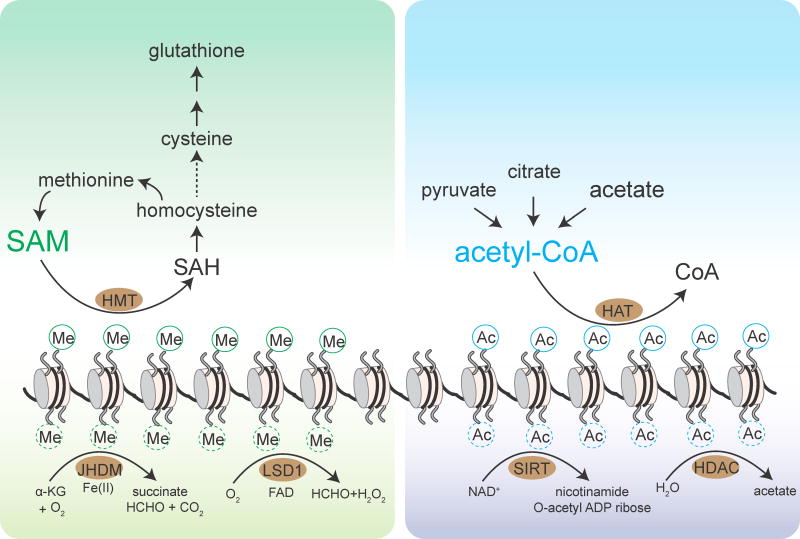
Figure 3. Dynamics of methylation and acetylation of histones.
The methylation status of histones is determined by opposing methylation and demethylation reactions. Histone methyltransferases (HMTs) catalyze the methylation of histones using SAM as the methyl donor, while histone demethylases catalyze the removal of methyl groups. Jumonji-domain containing demethylases (JHDM) consume α-KG in the demethylation reaction, while the LSD1 family of demethylases utilize FAD as a cofactor. The histones serve as a methyl sink and take methyl groups from SAM to facilitate its conversion to cysteine and glutathione through the transsulfuration pathway.
The acetylation status of histones is determined by opposing acetylation and deacetylation reactions. Histone acetyltransferases (HATs) catalyze the acetylation of histones using acetyl-CoA as the acetyl donor, while histone deacetylases catalyze the removal of acetyl groups to release free acetate (HDACs) or as O-acetyl-ADP-ribose (sirtuins). The released acetate can be converted back to acetyl-CoA to elicit metabolic or signaling functions.
Acetyl-CoA metabolism and histone acetylation
Biochemical basis of acetyl-CoA
Acetyl-CoA is composed of an acetyl moiety linked to Coenzyme A (CoA) through an energy-rich thioester bond [3], providing the chemical basis for the involvement of this metabolite in many primary anabolic processes, including fatty acid synthesis and steroidogenesis, as well as in modification of various proteins such as histones through acetyl group transfer [41–43]. The synthesis of acetyl-CoA, on the other hand, involves the central catabolic pathways that break down carbohydrates, fatty acids, and branched-chain amino acids [41–43]. Because homeostatic regulation of acetyl-CoA abundance is integral to primary metabolic processes, fluctuation of cellular acetyl-CoA levels becomes a key indicator of metabolic state for cellular physiology during growth and development. Importantly, these fluctuations have the capacity to modulate histone acetylation, resulting in epigenetic adaptation to nutrient and environmental states. Changes in histone acetylation have been implicated in many disease states, including many types of cancers [44–46].
Metabolic basis for acetyl-CoA in histone acetylation
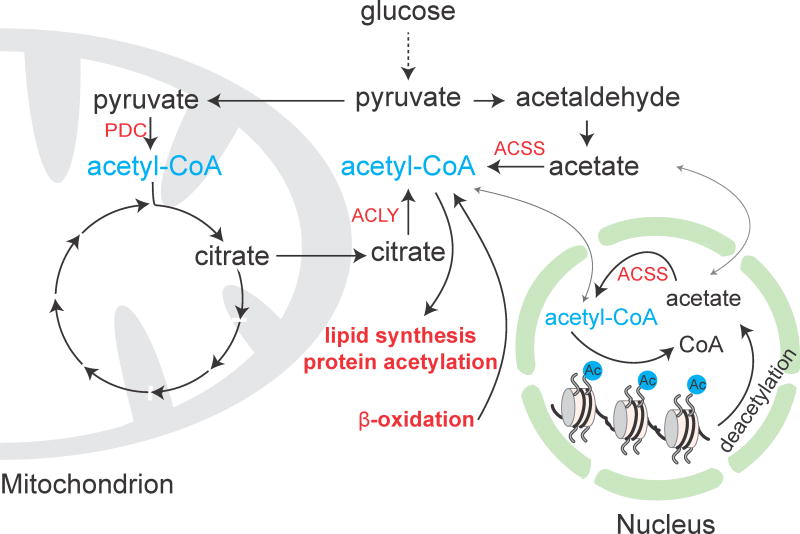
Histone acetyltransferase enzymes have a Km for acetyl-CoA in the low μM range [47, 48], which is close to estimated intracellular acetyl-CoA concentrations [49]. Just as in the case for SAM and histone methyltransferases, acetyl-CoA fluctuations can likewise modulate histone acetyltransferase activity. However, in contrast to SAM, most metabolic processes producing acetyl-CoA are central to bioenergetics that power cell growth and survival (Figure 2). During the metabolism of glucose, glycolysis breaks down glucose to pyruvate in the cytosol, which is imported into mitochondria. Mitochondrial pyruvate is then converted to acetyl-CoA by the pyruvate dehydrogenase complex. This is the canonical route through which acetyl-CoA is formed and subsequently funneled into the tricarboxylic acid (TCA) cycle. Under conditions of excess glucose, citrate generated from acetyl-CoA and oxaloacetate via the TCA cycle can be transported to the cytosol for the synthesis of cytosolic acetyl-CoA by ATP citrate lyase. This process supplies the cytosolic pool of acetyl-CoA for fatty acid biosynthesis and histone acetylation [43]. By contrast, during starvation when cells demand mitochondrial acetyl-CoA for ATP synthesis, free fatty acids are converted to acyl-CoAs in the cytosol, which are then transported into mitochondria via the carnitine shuttle and broken down to produce acetyl-CoA through β-oxidation. Parallel to these routes that are highly dependent on the fed or fasted state, acetyl-CoA can be synthesized alternatively in an ATP-dependent manner from acetate by acetyl-CoA synthetase enzymes found in the cytoplasm, nucleus, and mitochondria. Overall, acetyl-CoA metabolism is tightly orchestrated with cellular bioenergetics. This coordination necessitates compartmentalized metabolic processes to govern the spatial and temporal oscillation of acetyl-CoA levels, and a resulting nucleocytosolic acetyl-CoA pool modulates histone acetylation for genetic reconfiguration [Box 2].
Figure 2. Pathways for acetyl-CoA metabolism.
Acetyl-CoA can be synthesized via at least four routes: 1) decarboxylation of pyruvate by the pyruvate dehydrogenase complex (PDC); 2) cleavage of citrate by ATP citrate lyase (ACLY); 3) fatty acid β-oxidation; and 4) synthesis from acetate by acetyl-CoA synthetase enzymes (ACSS).
Acetyl-CoA regulates histone acetylation for the control of cell growth and survival
The role of acetyl-CoA in regulating the control of cell growth and survival is explicitly illustrated by studies of the yeast metabolic cycle (YMC) [59], during which prototrophic cells oscillate between three distinct metabolic phases [60]. Acetyl-CoA is among the most dynamic metabolites across the cycle and peaks during the oxidative phase, a growth period that is accompanied by a burst of mitochondrial respiration that culminates with entry into the cell division cycle. Bulk acetylation of histones was observed to increase periodically during this phase, perfectly coinciding with measurable increases in intracellular acetyl-CoA levels [59]. Interestingly, acetylated histones were present predominantly at growth genes during this oxidative phase [59, 61]. In addition to histones, several non-histone proteins, including components of transcriptional coactivator and chromatin remodeling complexes, are also acetylated during this phase [59], which underlies a concerted program driven by acetyl-CoA to promote expression of growth genes. Following the high acetyl-CoA oxidative phase are two reductive phases (the reductive building and reductive charging) when nucleocytosolic acetyl-CoA remains relatively low [59]. Although bulk histone acetylation is greatly reduced during both phases, acetylation of histones can be observed to increase at specific genomic loci. In this manner, particular genes associated with cell survival can be expressed. However, it remains unexplored how histones are selectively acetylated at particular DNA regions when nucleocytosolic acetyl-CoA is limiting. Consistent with the function of acetyl-CoA in determining the fate of cell growth and survival, depletion of nucleocytosolic acetyl-CoA stimulates autophagy [62, 63] and extends lifespan [62]. Mechanistically, histone hyperacetylation leads to transcriptional repression of essential autophagy genes [62]. The tight association between acetyl-CoA production and global histone acetylation has also been observed in various types of normal cells [64–66] as well as tumor cells [67, 68].
Histone acetylation is regulated by locally-produced acetyl-CoA in the nucleus
As an electrophilic molecule, acetyl-CoA is biochemically reactive enough that spontaneous acetylation of lysine residues on proteins can occur where acetyl-CoA is enriched [69–71]. This also suggests that a given molecule of acetyl-CoA may have a short half-time. As such, the local production of acetyl-CoA could be a mechanism to enable targeted protein acetylation. In particular, such spatial regulation of acetyl-CoA synthesis might be important for acetylation of histones, which can impose specific regulation of transcription at gene-specific loci. While acetyl-CoA metabolism is compartmentalized (Figure 2), many metabolic enzymes producing acetyl-CoA have been reported to be present in the nucleus, including ACSS2, ACLY, and PDC. In post-mitotic neurons, ACSS2 is recruited to chromatin and promotes expression of neuronal genes by increasing histone acetylation, which is implicated in memory formation [72]. Nuclear ACSS2 is also found in adipocytes and contributes to histone acetylation [68]. Nuclear translocation of ACSS2 has also been suggested to be regulated by AMPK in response to glucose deprivation [73]. Similar to ACSS2, ACLY is also present in both the cytosol and nucleus. Interestingly, in response to ionizing radiation-induced DNA damage, nuclear ACLY is phosphorylated at S455 that facilitates histone acetylation at double-strand break sites for DNA repair [74]. PDC, which is normally mitochondrial, can also be found in the nucleus, and the abundance of this pool is regulated by cell cycle, epidermal growth factor stimulation, and mitochondrial stress [75]. It is surprising that a variety of different metabolic enzymes can enter the nucleus to produce acetyl-CoA for histone acetylation under different physiological conditions. Nevertheless, these disparate mechanisms for generating acetyl-CoA in the nucleus underscores the importance of acetyl-CoA as means to link metabolism and chromatin regulation.
Histones as a reservoir for acetate
Because histones are abundant cellular components, a substantial number of acetyl group units can be deposited on histones by acetylation, as much as methyl groups deposited by methylation (Figure 3). As estimated in Table 1, each histone octamer for every ~146 bp of DNA can harbor up to 100 acetyl groups, which translates to 4 billion acetyl units across the human genome. Even if only ~0.1% of histone residues are active for acetylation and deacetylation, this can contribute a net of ~3 μM acetyl-CoA, compared to a total ~20 μM estimated present in the entire cell, which can again vary depending on the metabolic state [76].
There are two major differences between histone methylation and acetylation. First, acetylated histones are turned over very quickly within minutes (the half-life of acetylation is about 2–3 min [77]), whereas histone methylation displays a much slower turnover rate (estimated half-lives of methylated histone is about 0.3–4 days) [78, 79]. Second, deacetylation of histones generates acetate, which can be directly converted back to acetyl-CoA, whereas the methyl group detached from histones via demethylation is in the form of formaldehyde, which cannot be immediately utilized for SAM synthesis (Figure 1).
Due to such differences in the turnover rates and regenerative capacity between methyl and acetyl groups installed on histones, methylation forces the histones to consume SAM and thus function as a methyl sink, while acetylation enables the histones to serve as a reservoir for acetate [44, 80]. This pool of acetate can be used to regenerate acetyl-CoA, or to drive transport of protons to buffer against changes in intracellular pH [81]. Therefore, high nucleocytosolic concentrations of acetyl-CoA can promote the “bulk” acetylation of histones, which might not always be closely coupled to actual transcriptional activity. Moreover, we speculate that this reservoir of acetate can subsequently be retrieved and directed towards appropriate metabolic functions depending on the metabolic state of the cell.
Concluding Remarks
Highly sensitive protein mass spectrometry techniques have revealed that the histones are decorated by a variety of post-translational modifications, while next-generation sequencing technologies have provided a broad view of potential associations between chromatin states and gene regulation. Despite the prevalence of the “histone code” hypothesis, many of these modifications are often not well-correlated with gene transcription, causing their precise functional roles to be debated, and many unconventional functions have been proposed. Often overlooked are both the dependency of these modifications on small molecule metabolites that are indicators of the metabolic state and the direct metabolic consequences accompanying the biochemical reactions required for modifying the histones. As discussed above, we propose that due to their abundance, the histones themselves play a very underappreciated role in absorbing key intermediary metabolites for metabolic transformations, or as a readily accessible repository. In fact, an ancestral function of histone modifications could have been to enable such metabolic functions, which perhaps co-evolved with their roles in genome regulation. A proper consideration of the histones in such a role could be vital to understanding the function of these pervasive modifications in both the normal and diseased states.
Outstanding Questions Box.
What other small molecule metabolites might exhibit such intimate connections with histones and chromatin?
What information can be gleaned from bulk measurements of histone methylation or acetylation?
Could there be specialized functions for histone methylation and acetylation when SAM and acetyl-CoA are low?
How are chromatin-modifying enzymes regulated in response to metabolic cues in order to necessitate proper histone modifications?
How does this metabolism-to-histone communication impose regulation on other cellular activities, such as energy metabolism, membrane biogenesis, and inter-organelle communication?
How important are such metabolic functions of histones in human disease?
Highlights.
Histone methylation and acetylation are sensitive to metabolic states.
Histone methylation consumes SAM and functions as a methyl sink.
Histone acetylation deposits acetyl units as a source of acetate.
“Bulk” histone methylation and acetylation might reflect these metabolic functions
Acknowledgments
The authors wish to acknowledge fellowship support from the Chilton Foundation to C.Y., and funding support from NIH (R01GM094314), Welch Foundation (I-1797), and a HHMI-Simons Faculty Scholar Award to B.P.T. We apologize to those whose work could not be cited due to space and citation limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 3.Walsh CT, Tu BP, Tang Y. Eight Kinetically Stable but Thermodynamically Activated Molecules that Power Cell Metabolism. Chem Rev. 2017 doi: 10.1021/acs.chemrev.7b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61(4):503–32. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.German DC, Bloch CA, Kredich NM. Measurements of S-adenosylmethionine and L-homocysteine metabolism in cultured human lymphoid cells. J Biol Chem. 1983;258(18):10997–1003. [PubMed] [Google Scholar]
- 6.Mentch SJ, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22(5):861–73. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye C, et al. A Metabolic Function for Phospholipid and Histone Methylation. Mol Cell. 2017;66(2):180–193. e8. doi: 10.1016/j.molcel.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke S. Protein carboxyl methyltransferases: two distinct classes of enzymes. Annu Rev Biochem. 1985;54:479–506. doi: 10.1146/annurev.bi.54.070185.002403. [DOI] [PubMed] [Google Scholar]
- 10.Noga AA, et al. Plasma homocysteine is regulated by phospholipid methylation. J Biol Chem. 2003;278(8):5952–5. doi: 10.1074/jbc.M212194200. [DOI] [PubMed] [Google Scholar]
- 11.Mudd SH, et al. Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninaemia. J Inherit Metab Dis. 2001;24(4):448–64. doi: 10.1023/a:1010577512912. [DOI] [PubMed] [Google Scholar]
- 12.Luka Z, et al. A glycine N-methyltransferase knockout mouse model for humans with deficiency of this enzyme. Transgenic Res. 2006;15(3):393–7. doi: 10.1007/s11248-006-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luka Z, Mudd SH, Wagner C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J Biol Chem. 2009;284(34):22507–11. doi: 10.1074/jbc.R109.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mudd SH, et al. Methyl balance and transmethylation fluxes in humans. Am J Clin Nutr. 2007;85(1):19–25. doi: 10.1093/ajcn/85.1.19. [DOI] [PubMed] [Google Scholar]
- 15.Stead LM, et al. Is it time to reevaluate methyl balance in humans? Am J Clin Nutr. 2006;83(1):5–10. doi: 10.1093/ajcn/83.1.5. [DOI] [PubMed] [Google Scholar]
- 16.Horiuchi KY, et al. Assay development for histone methyltransferases. Assay Drug Dev Technol. 2013;11(4):227–36. doi: 10.1089/adt.2012.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patnaik D, et al. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J Biol Chem. 2004;279(51):53248–58. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- 18.Mato JM, Martinez-Chantar ML, Lu SC. Methionine metabolism and liver disease. Annu Rev Nutr. 2008;28:273–93. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- 19.Shiraki N, et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19(5):780–94. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Dobosy JR, et al. A methyl-deficient diet modifies histone methylation and alters Igf2 and H19 repression in the prostate. Prostate. 2008;68(11):1187–95. doi: 10.1002/pros.20782. [DOI] [PubMed] [Google Scholar]
- 21.Tang S, et al. Methionine metabolism is essential for SIRT1-regulated mouse embryonic stem cell maintenance and embryonic development. Embo j. 2017;36(21):3175–3193. doi: 10.15252/embj.201796708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vance DE. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochim Biophys Acta. 2014;1838(6):1477–87. doi: 10.1016/j.bbamem.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Walker AK. 1-Carbon Cycle Metabolites Methylate Their Way to Fatty Liver. Trends Endocrinol Metab. 2017;28(1):63–72. doi: 10.1016/j.tem.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker AK, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147(4):840–52. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song J, et al. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD) FASEB J. 2005;19(10):1266–71. doi: 10.1096/fj.04-3580com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus D, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508(7495):258–62. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9(5):300–6. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadhu MJ, et al. Nutritional control of epigenetic processes in yeast and human cells. Genetics. 2013;195(3):831–44. doi: 10.1534/genetics.113.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia BA, et al. Folate deficiency affects histone methylation. Med Hypotheses. 2016;88:63–7. doi: 10.1016/j.mehy.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325(5939):435–9. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–6. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H, et al. SnapShot: histone modifications. Cell. 2014;159(2):458–458.e1. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? Am J Cancer Res. 2012;2(5):589–97. [PMC free article] [PubMed] [Google Scholar]
- 34.Markovic J, et al. Role of glutathione in cell nucleus. Free Radic Res. 2010;44(7):721–33. doi: 10.3109/10715762.2010.485989. [DOI] [PubMed] [Google Scholar]
- 35.Boukouris AE, Zervopoulos SD, Michelakis ED. Metabolic Enzymes Moonlighting in the Nucleus: Metabolic Regulation of Gene Transcription. Trends Biochem Sci. 2016;41(8):712–730. doi: 10.1016/j.tibs.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Li S, et al. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol Cell. 2015;60(3):408–21. doi: 10.1016/j.molcel.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 37.Barzilai A, Yamamoto K. DNA damage responses to oxidative stress. DNA Repair (Amst) 2004;3(8–9):1109–15. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–76. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fnu S, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108(2):540–5. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pai CC, et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun. 2014;5:4091. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125–31. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietrocola F, et al. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21(6):805–21. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Sivanand S, Viney I, Wellen KE. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem Sci. 2018;43(1):61–74. doi: 10.1016/j.tibs.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Comerford SA, et al. Acetate dependence of tumors. Cell. 2014;159(7):1591–602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mashimo T, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159(7):1603–14. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schug ZT, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27(1):57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langer MR, et al. Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. J Biol Chem. 2002;277(30):27337–44. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- 48.Berndsen CE, et al. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46(3):623–9. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Z, Cai L, Tu BP. Dietary control of chromatin. Curr Opin Cell Biol. 2015;34:69–74. doi: 10.1016/j.ceb.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–26. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442(7098):96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 54.Dai Z, et al. Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat Commun. 2018;9(1):1955. doi: 10.1038/s41467-018-04426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furuhashi H, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3(1):15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6(9):e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64(2):435–59. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 59.Cai L, et al. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42(4):426–37. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tu BP, et al. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310(5751):1152–8. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 61.Kuang Z, et al. High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nat Struct Mol Biol. 2014;21(10):854–63. doi: 10.1038/nsmb.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenberg T, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19(3):431–44. doi: 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marino G, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53(5):710–25. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 64.Wong BW, et al. The role of fatty acid beta-oxidation in lymphangiogenesis. Nature. 2017;542(7639):49–54. doi: 10.1038/nature21028. [DOI] [PubMed] [Google Scholar]
- 65.Covarrubias AJ, et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife. 2016:5. doi: 10.7554/eLife.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carrer A, et al. Impact of a High-fat Diet on Tissue Acyl-CoA and Histone Acetylation Levels. J Biol Chem. 2017;292(8):3312–3322. doi: 10.1074/jbc.M116.750620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JV, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20(2):306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai L, Tu BP. On acetyl-CoA as a gauge of cellular metabolic state. Cold Spring Harb Symp Quant Biol. 2011;76:195–202. doi: 10.1101/sqb.2011.76.010769. [DOI] [PubMed] [Google Scholar]
- 70.Paik WK, et al. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213(2):513–22. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- 71.Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288(40):29036–45. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mews P, et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature. 2017;546(7658):381–386. doi: 10.1038/nature22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li X, et al. Nucleus-Translocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Mol Cell. 2017;66(5):684–697. e9. doi: 10.1016/j.molcel.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sivanand S, et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol Cell. 2017;67(2):252–265.e6. doi: 10.1016/j.molcel.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sutendra G, et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158(1):84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 76.Shurubor YI, et al. Determination of Coenzyme A and Acetyl-Coenzyme A in Biological Samples Using HPLC with UV Detection. Molecules. 2017;22(9) doi: 10.3390/molecules22091388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jackson V, et al. Studies on highly metabolically active acetylation and phosphorylation of histones. J Biol Chem. 1975;250(13):4856–63. [PubMed] [Google Scholar]
- 78.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12(12):917–30. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 79.Zee BM, et al. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285(5):3341–50. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurdistani SK. Chromatin: a capacitor of acetate for integrated regulation of gene expression and cell physiology. Curr Opin Genet Dev. 2014;26:53–8. doi: 10.1016/j.gde.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McBrian MA, et al. Histone acetylation regulates intracellular pH. Mol Cell. 2013;49(2):310–21. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]