Abstract
Myosin VI is a molecular motor involved in intracellular vesicle and organelle transport. To carry out its cellular functions myosin VI moves toward the pointed end of actin, backward in relation to all other characterized myosins. Myosin V, a motor that moves toward the barbed end of actin, is processive, undergoing multiple catalytic cycles and mechanical advances before it releases from actin. Here we show that myosin VI is also processive by using single molecule motility and optical trapping experiments. Remarkably, myosin VI takes much larger steps than expected, based on a simple lever-arm mechanism, for a myosin with only one light chain in the lever-arm domain. Unlike other characterized myosins, myosin VI stepping is highly irregular with a broad distribution of step sizes.
Myosin VI is a molecular motor that is ubiquitously expressed across organisms and tissue types and is involved in a variety of functions (1–3). It is the motor that is defective in Snell's waltzer mice, characterized by deafness and coordination problems, suggesting that myosin VI is involved in stereocilia function in cochlear hair cells (4). Additionally, myosin VI is believed to be a vesicle transporter in other cell types. Immunocytochemistry has shown that myosin VI is directly associated with vesicles in Drosophila embryos (5), and imaging of green fluorescent protein (GFP) fusions indicates that myosin VI is broadly localized in the trans-Golgi network and in protrusions in the plasma membrane (6). Unique among characterized myosin motors, myosin VI moves toward the pointed ends of actin filaments (7). Actin is typically oriented with the barbed end toward the plasma membrane and the pointed end toward the cell interior. This fact, coupled with the observation that myosin VI colocalizes with clathrin-coated pits (8), suggests that myosin VI is involved in endocytosis. Kinetic characterization of single-headed constructs shows that myosin VI is a high-duty-ratio motor, meaning that it spends much of its ATPase cycle strongly bound to actin. Furthermore, myosin VI is kinetically processive, meaning that after a diffusional encounter with actin, it hydrolyzes multiple ATPs before completely releasing again (9). Together, these kinetic and functional characteristics led to the expectation that myosin VI may be capable of transporting cargo at the single molecule level. In this study, we show that myosin VI is a processive motor with an unusually large step size.
Methods
Protein Constructs and Expression.
To create the double-headed myosin VI/GFP construct, the porcine myosin VI cDNA was truncated at Arg-994 to include 20 native heptad repeats of predicted coiled-coil and was followed by a leucine zipper (GCN4) to ensure dimerization (10). This was then followed by the cDNA for enchanced GFP (EGFP; CLONTECH), and then a Flag tag (encoding GDYKDDDDK) at the C terminus to facilitate purification (11). This cDNA was used to generate a recombinant baculovirus that was used for coexpression of the myosin VI/GFP with chicken calmodulin. To create the double-headed myosin V/GFP construct, the chicken myosin V cDNA was truncated at Glu-1099 and was followed by a leucine zipper (GCN4) to ensure dimerization (10). This was then followed by the cDNA for EGFP, and then a Flag tag (encoding GDYKDDDDK) at the C terminus to facilitate purification (11). This cDNA was used to generate a recombinant baculovirus that was used for coexpression of the myosin V/GFP with separate recombinant baculoviruses coding for calmodulin and two essential light chains, LC1-sa and LC23. The generation of recombinant baculovirus, expression in SF9 cells, and protein purification followed published procedures (9, 11). Human fascin protein and cDNA were the generous gift of Steve Almo (Albert Einstein College of Medicine, Bronx, NY). Bacterial expression and purification of fascin followed published procedures (12).
Motility Assays.
In all assays described, assay buffers included 25 mM imidazole HCl (pH 7.4), 25 mM KCl, 5 μM calmodulin, 1 mM EGTA, 10 mM DTT, 4 mM MgCl2, 2 mM ATP, and an oxygen scavenging system to retard photobleaching (25 μg⋅ml−1 glucose oxidase, 45 μg⋅ml−1 catalase, and 1% glucose). Assays were performed at 30°C, with the exception of the optical trapping assays, which were performed at 23°C. Actin bundles were made by polymerizing G-actin in the presence of a 10-fold molar excess of purified fascin. Loose bundles appeared after >1 week at 4°C. Single-molecule fluorescence assays were performed on an evanescent field microscope patterned after the design of Yanagida and coworkers (13) (R.S.R. and J.A.S., unpublished work). Only spots that moved >300 nm and for >1 s were tabulated. Landing and continuous movement assays were performed as previously described (14). Flow cells were pretreated with anti-GFP monoclonal antibodies (Quantum, Durham, NC, 0.05 mg⋅ml−1), followed by 1 mg⋅ml−1 BSA in assay buffers, before addition of the GFP-tagged myosin VI. All surface myosin VI densities given assume that every molecule entered the flow cell, none were denatured, and half were adsorbed to each surface of the flow cell. Fits to the continuous movement assay were described by
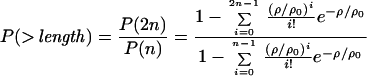 |
where n is the number of motor units required for motility, ρ is the surface density, and ρ0 is a fit parameter. All errors are SD unless otherwise indicated.
Optical Trap.
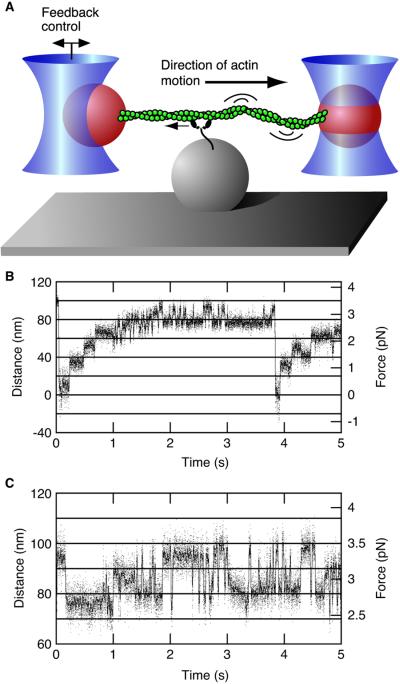
A single actin filament attached at both ends to optically trapped 1-μm diameter polystyrene beads was stretched to tension, then relaxed slightly. This actin dumbbell was moved near surface-bound silica spheres that served as platforms, which were decorated sparsely with myosin VI molecules. Both polystyrene beads were tracked with nm and ms resolution. Force feedback was performed as described (15), with feedback control on one of the two trapped beads. The actin filament was arranged so that the motor protein would pull the bead under feedback control out of its trap. Because the actin filament between the bound motor and the second bead was slack (see Fig. 2A), the second bead did not move over the entire feedback range. As a result, the second trap did not contribute to the optical load experienced by myosin VI, and constant load could be maintained through feedback control of a single trap. Steps in the optical trapping records were tabulated manually, and all steps within the feedback range were tabulated.
Figure 2.
Processive stepping by myosin VI observed by the dual-beam optical trap. (A) Dual-bead optical trap scheme. An actin filament is held between two 1-μm polystyrene beads. Bead positions are recorded with single nm resolution over 10 kHz. For fixed trap measurements (B and C) the optical load increases with displacement. (B) Sample trace of stepping behavior at 2 mM ATP with a fixed trap. Dwell periods at low load (<1.75 pN) fit single exponential statistics, with a mean stepping rate of 9.1 ± 0.6 s−1 (n = 234). This rate is similar to the 8- to 9-s−1 steady-state ATPase rate for single-headed constructs (9). (C) Oscillatory behavior observed at 2 mM ATP and high load (>2.5 pN).
Results and Discussion
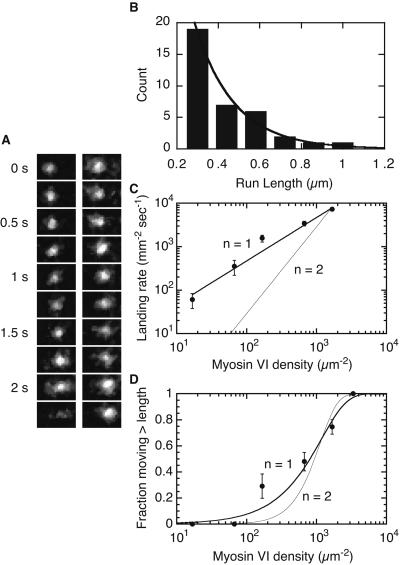
Processive movement of myosin VI was observed by in vitro motility assays using total internal reflection fluorescence microscopy to track fluorescently labeled molecules (baculovirus expressed truncated myosin VI with GFP engineered at the C terminus; ref. 16). Although no processive motility was seen on single actin filaments attached to the glass surface, myosin VI did move processively on loosely bundled, unidirectional actin filaments created by crosslinking with fascin (Fig. 1A) (12). The actin bundles were attached to the glass surface. Myosin VI molecules in solution were observed to move as single fluorescent spots along filaments that were frayed out of the bundles at their ends, perhaps tracking around the actin helix as they moved. The velocity of moving myosin VI spots in these assays was 291 ± 77 nm⋅s−1 (n = 36) similar to that observed in sliding filament assays (312 ± 17 nm⋅s−1, n = 212). Moving fluorescent spots disappeared in at most two events, as expected for the sequential photobleaching of each of the two GFP dyes fused to a single two-headed myosin VI. The average processive run length of myosin VI in these assays is 226 nm (n = 36, r2 = 0.96, single exponential, Fig. 1B). This run length may be an underestimate because of steric hindrance from the fascin or interaction of filaments with the glass coverslip.
Figure 1.
Processivity assays. (A) Motility of GFP-tagged myosin VI on fascin-actin bundles as seen by total internal reflection fluorescence. Two separate runs are shown. Frame width, 1.5 μm. (B) Run-length distribution for single, fluorescently labeled myosin VI molecules. Exponential curves were fit to the data by using only runs that lasted longer than 1 s, all of which were ≥0.3 μm. n = 36 measurements, histogram bin widths are 0.14 μm. (C) Actin filament landing rates as a function of myosin VI density. First-power (reduced χ2 = 2.45) and second-power (reduced χ2 = 17.4) fits are shown. (D) Fraction of filaments that moved more than their length before dissociating, as a function of motor density. Fits describing single-molecule motility (reduced χ2 = 2.2) and double molecule motility (reduced χ2 = 8.9) are shown. Error bars are SE obtained from counting statistics.
Independent evidence for processivity came from myosin VI motility observed at a variety of surface densities. At high density (1,600 μm−2), myosin VI exhibits smooth and continuous actin filament movement. Motility was observed at low densities (16 μm−2) as well, where actin appeared to thread through and swivel about isolated surface attachments, as observed for other processive cytoskeletal motors (17–20). The actin velocity did not fall as the myosin VI surface density was decreased, consistent with a high duty ratio. Such continuous motility may be caused by the coincident colocalization of several nonprocessive motors on the surface. To exclude this possibility, actin filament landing rates and distances moved were measured over a range of surface densities. There was a first-power dependence of the landing rate on the surface density, as well as a gradual transition (from 0 to 1) in the fraction of filaments that moved greater than their length before detaching (Fig. 1 C and D). Both of these results are expected for processive motors, where encounters with functional motor units are processes that are first-order in motor density and differ markedly from the behavior of nonprocessive motors (21).
Dual bead optical trapping (Fig. 2A) was used to examine the stepping process of myosin VI in detail (22, 23). Of 177 platforms tested, 14 (8%) exhibited processive binding events with multiple steps as in Fig. 2B, whereas only 4 platforms (2%) yielded single-step binding events such as seen for myosin II. Based on the observed incidence of binding (10% of platforms tested), 95% of the binding events detected should have been produced by single molecules (24). The small fraction of platforms that give rise to single-step binding events are likely due to motors that have been damaged by surface adsorption.
In a typical processive event, the actin filament and its attached bead were pulled for 3–5 steps before reaching a stall point, detaching from the surface, and then returning to the baseline position (Fig. 2B). Myosin VI stepped against the increasing load from the optical trap until it reached a stall force of 2.8 ± 0.3 pN (n = 34), comparable to the 3 pN seen for myosin V (18). Unlike myosin V, which slows its stepping rate and occasionally steps backward under high load, myosin VI exhibited remarkable oscillatory behavior at high load with rapid transitions between two bound positions, as previously observed for RNA-folding transitions (25). Either forward steps or detachment events followed all backsteps. Because the rate of forward (and reverse) stepping in such oscillations against high load exceeds the ATPase rate of unloaded myosin VI, these oscillations reflect a reversible or off-pathway process in the chemomechanical cycle of myosin VI.
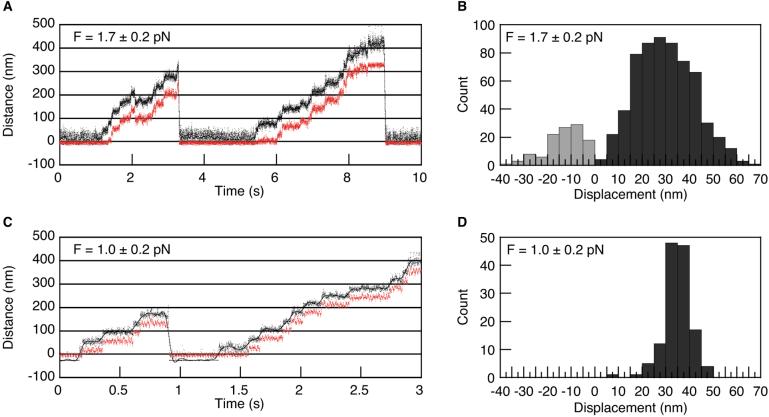
To enable an accurate measurement of myosin VI step size, a force-feedback system was used (Fig. 2A) (15). This system maintains a constant load on myosin VI by moving one of the optical traps to follow the trapped bead. This system enables myosin VI molecules to take a large number of steps (≈10) and eliminates errors in step size measurements because of compliance of the protein-bead linkages in the system. Large forward steps were interrupted with less frequent, smaller backward steps (Fig. 3A). Application of a moderate load (1.7 ± 0.2 pN, below the level that produced the oscillatory behavior in Fig. 2 B and C) yielded a broadly distributed step-size histogram (Fig. 3B). Two clear lobes were observed, one from forward steps (30 ± 12 nm, modal value 27 nm, n = 615) and a smaller one from backsteps (−13 ± 8 nm, modal value −11 nm, n = 114). In each of these lobes, the spread of the step size distribution greatly exceeds the 1.4-nm measurement precision. Therefore, it appears that myosin VI binds actin in a promiscuous manner, attaching to within ± ≈3 sites of the preferred binding site along the helical actin filament. This spread is significantly greater than the ±6 nm observed for myosin V in both single-bead (15) and dual-bead trap geometries (Fig. 3 C and D). Myosin V has been shown by electron microscopy to predominantly bind within one site of the preferred binding site 36 nm away (26) (Fig. 4 Lower, green monomers).
Figure 3.
Force feedback measurements. Constant separation between the bead center and the trap center was maintained by a feedback loop for the left-hand bead (see Fig. 2A). Forward steps indicate positive displacements of the left-hand bead out of its trap, and do not reflect the polarity of the actin filament. (A) Sample force-feedback trace of myosin VI stepping under 1.7 pN of load, and 2 mM ATP. Black trace, bead position; red trace, trap position. (B) Histogram of measured myosin VI step sizes. Forward steps were 30 ± 12 nm (n = 615), and backsteps were −13 ± 8 nm (n = 114). Both the |mean| and variances of the forward and backward steps differed significantly (t probability and F probability < 0.0001). Therefore the backsteps observed here arise from a different process than the oscillations observed at high load in Fig. 2 B and C (where backward and forward transitions are of equal magnitude). (C) Sample force-feedback trace of myosin V stepping under 1 pN of load and 2 mM ATP, using the same dual-bead trapping setup described for myosin VI (see text). (D) Step size histogram of myosin V. The narrow distribution of myosin V step sizes near 36 nm (35 ± 6 nm, n = 131) is in agreement with published results (15). This indicates that the broad distribution of myosin VI steps around 30 nm is not due to the geometry used, and the myosin VI step size is significantly less than 36 nm (t probability < 0.0001).
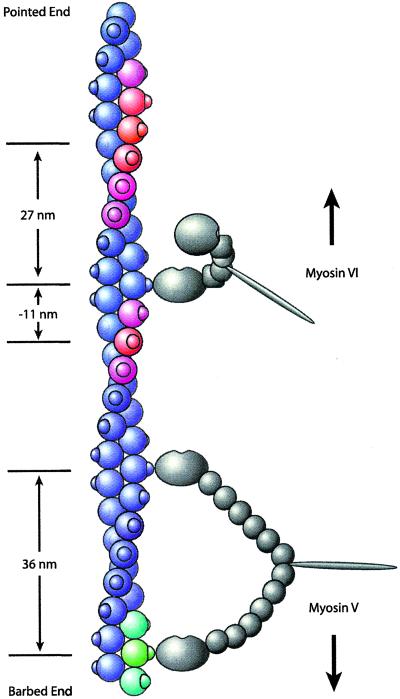
Figure 4.
Model of myosin VI stepping. Myosin V and VI molecules are shown in gray, and the actin filament is shown in color. Knobs on the actin filament indicate stereospecific myosin-binding sites (although the actual binding site is located between two actin monomers, binding is shown on only one monomer for simplicity). Both motors are bound to actin subunits facing directly right. Myosin V (Lower), having six light chains, spans the 36-nm actin pseudorepeat to within ± 1 binding site (green and blue-green). Myosin VI (Upper), having only one light chain and the unique insert (square), cannot. Instead, the bound myosin VI head swings the unbound head to the left side of the actin filament as suggested by the transition from the ADP-bound state to the rigor state in electron microscopy structures (7). The actin subunits shown in red indicate the preferred binding sites of the unbound myosin VI head toward both the pointed end (≈27 nm) and the barbed end (≈11 nm) of actin, corresponding to the modal values seen in the optical trapping experiments. The spread of the color from red to blue reflects the step size histogram. Therefore, nearby actin subunits shown in purple are somewhat less accessible, but steps to these subunits can occur. Subunits shown in blue are relatively inaccessible because their myosin VI-binding sites are on either the right side or the underside of the actin filament. The measured step sizes of myosin VI toward both the pointed end (modal value, 27 nm) and the barbed end (modal value, −11 nm) of actin are a direct result of the accessibility of binding sites on the left side of the actin filament to the unbound myosin VI head.
Myosin VI has only a single calmodulin light chain in the lever-arm domain, which, according to a simple lever-arm model, implies a maximum working stroke of ≈5 nm. However, there is a unique 53-residue insert of unknown function between the converter domain and the light chain-binding domain (7, 27). This insert (and perhaps adjacent structures) may adopt an extended conformation to allow myosin VI to take the observed 30-nm steps. Myosin VI likely operates by the combination of a short powerstroke in the strongly bound head, coupled with a significant conformational change that allows the free head to extend to the next available binding site. It is formally possible that the 30-nm step of myosin VI involves a conformational change of the actin filament, or that the motor takes two rapid, tightly coupled 15-nm steps in succession to create an apparent 30-nm forward step. However, no motor has been shown to move by either of these two mechanisms. If myosin VI is indeed taking single 30-nm steps without altering the actin track, it is not operating by a canonical lever-arm mechanism. Alternate stepping models that do not involve a rotation of the lever arm have been proposed for other myosin classes (28, 29).
To explain the step size histogram in Fig. 3B, a mechanism that biases the step 20–30 nm toward the pointed end of actin is required. A model for the stepping of myosin VI is outlined in Fig. 4. The actin filament shown has two right-handed long-pitch helices with a pseudorepeat of 36 nm, and the pointed end is at the top of the figure. In this model (in which a state with one bound head and one free head is shown), the bound head undergoes a powerstroke parallel to the long axis of actin, toward the pointed end, but in addition has a perpendicular component that involves a left-handed rotation around the actin filament. This rotation positions the free head to preferably bind the red monomers shown in Fig. 4. Modal values for steps are indicated (27 nm and −11 nm, respectively). Binding sites within 1 SD of these modal values are located ≈5–15 actin subunits (14–41 nm) toward the pointed end and ≈2–6 actin subunits (5–17 nm) toward the barbed end of the filament, and are shown in red and purple. In this manner, the step size is determined largely by the periodicity of the actin filament. This model is supported by two observations: (i) electron microscopy studies of single headed myosin VI have shown a left-handed rotation of the lever arm domain in the transition from the ADP state to the rigor state (7); and (ii) the distance between the two lobes of the step size distribution is ≈38 nm, or roughly one pseudohelical repeat of actin.
Myosin VI has evolved features that make it unique among motor proteins. Not only is it processive, it is also backward directed (7), making myosin the first family of motor proteins shown to move processively in both directions. This property of myosins greatly facilitates steady-state cellular trafficking, because separate tracks for trafficking in each direction need not be assembled. The processive nature of myosin VI demonstrates that a long light chain-binding region like that of myosin V is not required for myosin processivity (18, 30). The stepping behavior of myosin VI is also quite revealing. Unlike all other characterized motors, myosin VI rapidly oscillates at its stall load and has different forward and backward step sizes at lower loads. It is possible that the insert between the converter and light chain-binding domains, along with adjacent structures, undergoes a major conformational change to extend the reach of myosin VI, and that the geometry of the actin filament determines the myosin-VI binding sites.
Acknowledgments
We thank A. Mehta, J. Dawson, A. Sääf, and D. Robinson for their comments on the manuscript and E. Landahl for stimulating discussions. R.S.R. is a Helen Hay Whitney postdoctoral fellow. S.E.R. is a supported by the Tumor Biology Training Program, awarded by the National Cancer Institute. J.A.S. and H.L.S. are supported by grants from the National Institutes of Health.
Abbreviation
- GFP
green fluorescent protein
References
- 1.Kellerman K A, Miller K G. J Cell Biol. 1992;119:823–834. doi: 10.1083/jcb.119.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Titus M A. Curr Biol. 2000;10:R294–R297. doi: 10.1016/s0960-9822(00)00433-4. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez O C, Cheney R E. Trends Cell Biol. 2000;10:307–311. doi: 10.1016/s0962-8924(00)01797-9. [DOI] [PubMed] [Google Scholar]
- 4.Avraham K B, Hasson T, Steel K P, Kingsley D M, Russell L B, Mooseker M S, Copeland N G, Jenkins N A. Nat Genet. 1995;11:369–375. doi: 10.1038/ng1295-369. [DOI] [PubMed] [Google Scholar]
- 5.Mermall V, McNally J G, Miller K G. Nature (London) 1994;369:560–562. doi: 10.1038/369560a0. [DOI] [PubMed] [Google Scholar]
- 6.Buss F, Kendrick-Jones J, Lionne C, Knight A E, Cote G P, Paul Luzio J. J Cell Biol. 1998;143:1535–1545. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells A L, Lin A W, Chen L Q, Safer D, Cain S M, Hasson T, Carragher B O, Milligan R A, Sweeney H L. Nature (London) 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 8.Buss F, Arden S D, Lindsay M, Luzio J P, Kendrick-Jones J. EMBO J. 2001;20:3676–3684. doi: 10.1093/emboj/20.14.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De La Cruz E M, Ostap E M, Sweeney H L. J Biol Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 10.Trybus K M, Freyzon Y, Faust L Z, Sweeney H L. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney H L, Rosenfeld S S, Brown F, Faust L, Smith J, Xing J, Stein L A, Sellers J R. J Biol Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- 12.Tseng Y, Fedorov E, McCaffery J M, Almo S C, Wirtz D. J Mol Biol. 2001;310:351–366. doi: 10.1006/jmbi.2001.4716. [DOI] [PubMed] [Google Scholar]
- 13.Tokunaga M, Kitamura K, Saito K, Iwane A H, Yanagida T. Biochem Biophys Res Commun. 1997;235:47–53. doi: 10.1006/bbrc.1997.6732. [DOI] [PubMed] [Google Scholar]
- 14.Rock R S, Rief M, Mehta A D, Spudich J A. Methods. 2000;22:373–381. doi: 10.1006/meth.2000.1089. [DOI] [PubMed] [Google Scholar]
- 15.Rief M, Rock R S, Mehta A D, Mooseker M S, Cheney R E, Spudich J A. Proc Natl Acad Sci USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto T, Amitani I, Yokota E, Ando T. Biochem Biophys Res Commun. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 17.Howard J, Hudspeth A J, Vale R D. Nature (London) 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 18.Mehta A D, Rock R S, Rief M, Spudich J A, Mooseker M S, Cheney R E. Nature (London) 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 19.Sakakibara H, Kojima H, Sakai Y, Katayama E, Oiwa K. Nature (London) 1999;400:586–590. doi: 10.1038/23066. [DOI] [PubMed] [Google Scholar]
- 20.Mehta A. J Cell Sci. 2001;114:1981–1998. doi: 10.1242/jcs.114.11.1981. [DOI] [PubMed] [Google Scholar]
- 21.Hancock W O, Howard J. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finer J T, Simmons R M, Spudich J A. Nature (London) 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 23.Mehta A D, Finer J T, Spudich J A. Methods Enzymol. 1998;298:436–459. doi: 10.1016/s0076-6879(98)98039-9. [DOI] [PubMed] [Google Scholar]
- 24.Block S M, Goldstein L S, Schnapp B J. Nature (London) 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 25.Liphardt J, Onoa B, Smith S B, Tinoco I J, Bustamante C. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 26.Walker M L, Burgess S A, Sellers J R, Wang F, Hammer J A, 3rd, Trinick J, Knight P J. Nature (London) 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 27.Homma K, Yoshimura M, Saito J, Ikebe R, Ikebe M. Nature (London) 2001;412:831–834. doi: 10.1038/35090597. [DOI] [PubMed] [Google Scholar]
- 28.Kitamura K, Tokunaga M, Iwane A H, Yanagida T. Nature (London) 1999;397:129–134. doi: 10.1038/16403. [DOI] [PubMed] [Google Scholar]
- 29.Yanagida T, Iwane A H. Proc Natl Acad Sci USA. 2000;97:9357–9359. doi: 10.1073/pnas.97.17.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard J. Nature (London) 1997;389:561–567. doi: 10.1038/39247. [DOI] [PubMed] [Google Scholar]