Abstract
Campylobacter jejuni is considered as the leading cause of gastroenteritis all over the world. This bacterium has the CRISPR–cas9 system, which is used as a gene editing technique in different organisms. However, its role in bacterial virulence has just been discovered; that discovery, however, is just the tip of the iceberg. The purpose of this study is to find out the relationship between cas9 and virulence both phenotypically and genotypically in C. jejuni NCTC11168. Understanding both aspects of this relationship allows for a much deeper understanding of the mechanism of bacterial pathogenesis. The present study determined virulence in wild and mutant strains by observing biofilm formation, motility, adhesion and invasion, intracellular survivability, and cytotoxin production, followed by the transcriptomic analysis of both strains. The comparative gene expression profile of wild and mutant strains was determined on the basis of De-Seq transcriptomic analysis, which showed that the cas9 gene is involved in enhancing virulence. Differential gene expression analysis revealed that multiple pathways were involved in virulence, regulated by the CRISPR-cas9 system. Our findings help in understanding the potential role of cas9 in regulating the other virulence associated genes in C. jejuni NCTC11168. The findings of this study provide critical information about cas9's potential involvement in enhancing the virulence of C. jejuni, which is a major public health threat.
Keywords: Campylobacter jejuni, cas9, virulence, transcriptome analysis, differential gene expression
Introduction
Campylobacteriosis is one of the most common intestinal diseases (WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR)., 2011), mainly caused by C. jejuni in humans (Tauxe, 2001). This bacteria infects humans by contaminating poultry products, milk and water (Manavathu et al., 1988; Chai et al., 2009; Pitkänen, 2013; Huang et al., 2015). Outbreaks of C. jejuni infection in humans are sporadic and relatively rare. However, serious possible consequences of Campylobacter infection include an autoimmune mediated disease known as Guillain-Barre Syndrome (GBS) and the closely related Miller Fisher Syndrome (Han et al., 2007). The pathogenesis of C. jejuni is achieved by numerous factors interacting together, including motility, adhesion, invasion, and toxin production (Bolton, 2015).
Motility is the key for bacteria to against various chemotactic conditions encountered in the gastrointestinal tract (Bolton, 2015). In the presence of viscous substances, Campylobacter shows unusual motility (Ferrero and Lee, 1988). C. jejuni motility factors include flaA (Lertsethtakarn et al., 2011), flab, and flgI (Sommerlad and Hendrixson, 2007), fliA and rpoN (Hendrixson, 2006). C. jejuni adherence to the host intestinal epithelial cells is also important for colonization, which is facilitated by various adhesions present on bacterial surfaces (Jin et al., 2001). These adhesion factors are cadF (Konkel et al., 1997), capA, and jlpA (Jin et al., 2001), and virB11 (Dasti et al., 2010).
Along with motility, flagella also function as an export apparatus (T3SS) during the host invasion (Poly and Guerry, 2008). In various studies, it was revealed that the flagellar apparatus functions as a type III secretion system (T3SS), transporting C. jejuni invasion antigens (Cia) into the host cell (Ziprin et al., 2001; Konkel et al., 2004; Biswas et al., 2007). The invasion antigen (CiaI) is secreted by C. jejuni via T3SS and consists of amino-terminal type III secretion sequences (Konkel et al., 2004). It enables the C. jejuni to survive within epithelial cells (Buelow et al., 2011). Campylobacter invasion factors include flaC, flhA, flhB, fliO, flip, and fliR (Carrillo et al., 2004), ciaB (Konkel et al., 2004), ciaC (Christensen et al., 2009), ciaI (Buelow et al., 2011), iamA (Carvalho et al., 2001), htrA (Bæk et al., 2011) and virK (Poly et al., 2007). C. jejuni produces a number of cytotoxins (McFarland and Neill, 1992); however, detailed study has been conducted only on cytolethal distending toxin (CDT; Pickett and Whitehouse, 1999). Nucleotide sequence analysis revealed that the cluster of CDT genes consists of cdtA, cdtB, and cdtC in C. jejuni (Pickett et al., 1996; Purdy et al., 2000; Lara-Tejero and Galán, 2001; Dasti et al., 2010). Along with CDT, C. jejuni also have cgtB and wlaN toxin factors (Gilbert et al., 2000; Linton et al., 2000).
Clustered regularly interspaced short palindromic repeats (CRISPRs) are short sequences of repeats that have been found in different bacterial genomes (Price et al., 2007). The term CRISPR was first used in 2002, and refers to the specific structure of loci. The CRISPR system is divided into three types on the basis of specific proteins present in each system. CRISPR system type I has cas3, type II has cas9, and type III has cas10 protein. The CRISPR type II system present in C. jejuni and cas9 plays an important role in the attachment and invasion of these bacteria in the colorectal epithelial cell model (Shabbir et al., 2016). Adherence to the host epithelial cells play an important role in bacterial infection (Soto and Hultgren, 1999). In 2013, scientists observed that cas9 of Francisella novicida uses CRISPR cas-associated RNA (scaRNA) for the repression of an endogenous transcription, which encodes a bacterial lipoprotein. This helps in the intracellular survivability of the pathogen within the host, as TLR2 activation is dampened (Sampson et al., 2014). Scientists in 2013 also reported that C. jejuni (cst-II-positive) strains almost lost their virulence after the deletion of the cas9 gene, and they also observed that GBS patients' serum was highly visible in the human immune system (Louwen et al., 2013).
The mechanism underlying the role of cas9 as a virulence factor in C. jejuni is not completely understood. Therefore, the purpose of the current study was to observe how the cas9 gene enhances virulence by regulating the virulence of associated genes. Therefore, transcriptomic analysis was performed after the deletion of the cas9 gene in C. jejuni and the wild type was compared with Δcas9 mutant strain to find out the role of cas9 as a virulence factor in C. jejuni. Along with transcriptomic profiling, we also checked and compared the virulence of C. jejuni wild and Δcas9 mutant strains by phenotypic methods, such as determination of cytotoxin assay, biofilm assay, adhesion, invasion, and intra-cellular survivability assay, along with motility. Our results showed that the cas9 gene plays a critical role in enhancing virulence. The findings of the present study shed light on the importance of the cas9 gene in the virulence of C. jejuni, which is a major public health concern.
Materials and methods
Bacterial strain and growth condition
The C. jejuni NCTC11168 strain used in this study was provided by China's center for disease control and prevention. C. jejuni was grown on Skirrow agar supplemented with 5% sheep blood and Skirrow addition agent at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) for 24–48 hours (h).
Construction of Cas9 isogenic mutant
The cas9 (Gene ID: 905809) gene was found by an NCBI blast in the genome of C. jejuni NCTC11168. For the amplification of the cas9 gene, adjacent upstream flanking sequences (about 1300bp) and downstream flanking sequences (about 1300bp) were used. The kanamycin resistance (Kan) cassette (pmdT- TKarmA- TKarmB -Kan- ef1a- BFP-WPRE-polyA) was amplified by PCR, which was donated by Huazhong agricultural university. These three fragments were ligated together by adopting the method of overlapping PCR. This overlapping PCR product was then ligated to the pGem-Teasy vector (Promega; Hao et al., 2016). The resulting constructs were sequenced and electroporated into the C. jejuni NCTC11168 strain and recombinants were selected on blood agar plates containing 5% sheep blood (Becton Dickinson) and 50 μg/mL kanamycin (Sigma-Aldrich). Junction PCR was used to confirm the double crossover events and the orientation of the Kan resistance cassette was assessed, with primers up and downstream of the area involved in the homologous recombination and primers (see S1 Table).
Electro-transformation of the C. jejuni isolate
For the electro-transformation of C. jejuni, 5% sheep blood agar plates (with and without antibiotic) were put into a 37°C incubator, to pre-warm. C. jejuni was harvested from the inoculated Skirrow blood agar plate (Becton Dickinson) using Luria broth (LB; Becton Dickinson) supplemented with 1 × nonessential amino acids (NEAA; Invitrogen). C. jejuni was pelleted by centrifugation for 5 min at 8,000 rpm and re-suspended with 1.5 mL of LB containing 1 × NEAA (Invitrogen), which was repeated one time. For resuspension, a transformation buffer (0.5 mL) without 1 × NEAA (Invitrogen) was used and aliquoted in 1.5 mL centrifuge tubes having 100 μL samples of the competent cells. To make Δcas9 mutants, plasmid DNA (2 μg) was transformed into the 100 μL sample of competent cells. The mixture was transferred to an electroporation cuvette and pulsed at 2.5 kV, 200 Ω, and 25 μF. Then 1 mL of LB supplemented with 1 × NEAA (Invitrogen) was added to the cuvette and re-suspended by pipetting thoroughly. This mixture (200 μL) from the cuvette was plated and spread onto the surface of pre-warmed Columbian blood agar plates (Becton Dickinson). Plates were incubated for 5 h at 42°C under micro-aerophilic conditions. The cells were recovered from these plates using 2 mL of LB supplemented with 1 × NEAA (Invitrogen). After centrifugation, suspension was spread on the surface of a new pre-warmed blood agar plate containing 50 μg/mL kanamycin (Sigma-Aldrich) for the selection of knock-out mutants, and ampicillin (Sigma-Aldrich) was used with a concentration of 50 μg/mL to screen for supplemented Δcas9 clones. Plates were incubated at 42°C under micro-aerophilic conditions for 2–5 days, until colonies were visible. Colonies resistant to ampicillin or kanamycin were passaged five times on new plates to generate stable clones. From these stable clones, a PCR assay was used to test for the correctness of the supplementation assay. For supplementation, we used the primer pairs presented in S2 Table to confirm that the supplemented strains had the gene inserted in the same sense orientation (Louwen et al., 2013).
Construction of complemented mutant strain
Complemented cas9 mutant strain was constructed according to the previously described method (Howard et al., 2009). Briefly, target gene Cj1523c (cas9, Gene ID: 905809) was amplified from extracted genomic DNA of C. jejuni NCTC11168. The amplified gene product was subsequently cloned into the pGEM-Teasy vector. Sequencing was performed to confirm the absence of mutation in the target gene. The target gene was electroporated into the cas9 mutant strain and transformants were selected on skirrow's agar plates. Positive colonies were confirmed by PCR using the targeted gene primers (S2 Table).
Determination of standard growth curve
The standard growth curves of C. jejuni NCTC11168, complemented mutant and Δcas9 mutant strains were determined as per method described previously (Davis and DiRita, 2008). Biphasic MH (50% MH broth and MH agar) medium was used to perform growth curves. The inoculum of three strains (1 × 106 CFU/mL) was incubated separately under micro-aerobic conditions for 16 to 18 h. Cultures of wild, complemented and mutant strains were suspended to an optical density of 0.4 at 600 nm. Suspensions were diluted (1:10) and inoculated (80 μL) into 20 mL of biphasic medium. Biphasic medium of each strain was incubated under both static and shaking micro-aerobic conditions. Orbital shaker was used to determine the growth curves under shaking condition according to the previously described method (Ghaffar et al., 2015). Viable colonies of three strains were determined at 0 h to 40 h with 4 h interval.
Biofilm assays
To measure the biofilm formation of the C. jejuni and its Δcas9 mutant strain, a microtiter plate biofilm test was used as previously described for C. jejuni and other bacteria (Stepanović et al., 2000) with some modifications. Briefly, 200 uL of C. jejuni and Δcas9 mutant strain fresh culture (OD570 = 0.05) were added to 96-well microtiter plates (flat bottom) and incubated at 42°C under microaerobic conditions for 24, 48, and 72 h. After every mentioned time interval, wells were washed three times with PBS (phosphate buffered saline) to remove the planktonic cells. Methanol (200 uL) was added in all wells and incubated for 15 min, followed by drying at room temperature and subsequently, 200 uL of 0.1% Hucker crystal violet solution were added into the plates and incubated for 5 min at room temperature. PBS was used to wash out the unbound crystal violet, and the plates were dried at 60°C. Glacial acetic acid (30%) was used to dissolve the bounded crystal violet. A plate reader(BioTek, uQuant microplate reader, USA)was used to determine absorbance (570 nm). Sterile media in wells were used as a blank control. To report biofilm production in C. jejuni and Δcas9 mutant strains, their blank corrected absorbance values were used. Based on bacterial films (ODs), both strains were categorized into non-adherent (0), weakly (*), moderately (**), and strongly (***) adherent. For the microtitration test, three standard deviations of negative control were considered as the cut-off OD.
Assays were repeated three times with three technical replicates and the results were averaged.
Adhesion and invasion assay
The adhesion and invasion of C. jejuni and Δcas9 mutant strains were determined as previously described for C. jejuni and other bacteria (Hao et al., 2016). Briefly, 5.0 × 107 CFU/mL of the C. jejuni and Δcas9 mutant was used to inoculate the monolayers of macrophages RAW264.7 at multiplicity of infection (MOI) of 100. For adhesion and invasion to occur, the infected monolayers were incubated for 3 h. In order to determine the total number of adherent and internalized bacteria, Dulbecco's Modified Eagle's Medium (DMEM) without antibiotic was used for the washing (three times) of monolayers to get rid of the extracellular unbound bacteria. The intracellular bacteria were released by the lysis of monolayers. Invading bacteria were determined by washing the monolayers with aspirated DMEM medium and 100 ug/mL gentamicin was added for 2 h to kill the bound as well as extracellular bacteria. The intracellular bacteria were released after washing and lysis of monolayers. The adherent bacteria number was calculated by subtracting the number of internalized bacteria from the total number of adherent and internalized bacteria (Iqbal et al., 2016).
Intracellular survivability assay
To test the intracellular survivability of C. jejuni and Δcas9 mutant strains within the macrophages RAW264.7, the post infection invasion period of both strains was extended to 3, 6, 12, 18, 24, 48, and 72 h. Same procedure was applied for washing and lysis of cells as described in the adhesion and invasion assay section. The intracellularly survived bacteria were counted by serial dilutions on Skirrow blood agar (Iqbal et al., 2016).
Cytotoxin assay
RAW264.7 macrophages were used to determine the cytotoxic effects of the C. jejuni NCTC 11168 and Δcas9 mutant strains. C. jejuni can produce a unique cytotoxin (Guerrant et al., 1987). A cytotoxin assay was performed according to the previously described method (Guerrant et al., 2001). Briefly, C. jejuni and Δcas9 mutant strains were treated by 2000 μg/mL polymyxin B and incubated at 37°C for 1 h (Ashkenazi and Cleary, 1990); subsequently, centrifuged at 8,000 rpm for 20 min. The supernatant was filtered with a 0.22 μm filter. 0.3% TritonX-100-PBS was used as a positive control, and polymyxin B-treated microbial and polymyxin B-PBS were used as negative control. Suspensions of macrophages RAW264.7 (100 μL:5 × 105 cells) were placed into the microtitration plates for 1 to 3 h to allow adherence. The prepared serial twofold dilutions of C. jejuni and Δcas9 mutant strain filtrates were used for tissue culture monolayers in 100 μL volumes, and incubated at 37°C in 5% CO2 for 24 h. The monolayers were examined for the percentage of cells of round cells by phase-contrast microscopy. A Trypan Blue Dye uptake (A540 value) was used to determine the cell death and after that a staining technique known as “Giemsa staining” was used to correlate with normal cell morphology. The index of cytotoxin effect was calculated by the formula (Hao et al., 2016) 100 × {1–[A540 (test) A540 (positive)] ÷[A540(negative)–A540 (positive)]}.
Motility test
The two strains (C. jejuni NCTC11168 and Δcas9 mutant strains) used to test mobility, a fresh culture of C. jejuni and Δcas9 mutant strains, were inoculated into Mueller Hinton (MH) broth and cultured to their logarithmic growth phase. Then 3 uL of bacteria were stabbed into 0.4 % MH agar and plates were incubated at 42°C under microaerobic conditions for 24 h. From each plate, motility was scored by measuring growth diameter (mm). Assays were repeated at least three times with three technical replicates and the results were averaged (Kim et al., 2015).
Frozen transmission electron microscopy
The flagellar length of C. jejuni NCTC11168 and Δcas9 mutant strains were examined using frozen transmission electron microscopy according to a previously described method (Hao et al., 2017). Briefly, plates were washed with 2 mL PBS to get bacterial suspensions and spotted on carbon-coated copper grids. Bacterial cells were stained with 2% phosphotungstic acid (pH 6.7) for 1 min. Samples were observed employing a transmission electron microscope (H-7650, Japan).
Transcriptome analysis by RNA-Seq
After the phenotypic determination of C. jejuni NCTC 11168 and Δcas9 mutant strains, RNA-Seq was performed as described previously (Iqbal et al., 2016). C. jejuni and Δcas9 mutant strains (two each sample) were harvested at log phase. By following the manufacturer's instructions, total RNAs were isolated from the samples using TRIzol (Invitrogen Inc., California, USA). RNase free DNase I (Ambion Inc., Texas, USA) was used to remove DNA and the quality of RNA was checked with the help of Agilent 2100 system with RIN (RNA integrity number) over 7. From total RNA, rRNA was removed by using the Ribozero kit and strand specific RNA-Seq protocol was followed on the illumine DE-Seq platform (Paired end sequencing; 100 bp fragment) at Shanghai personal biotechnology. Briefly, synthesis of cDNA first strand was carried out using the Super-ScriptII (Invitrogen, Carlsbad, CA) in the presence of random hexamer primers and synthesis of cDNA second strand was performed before end repair and dA tailing. Ligation of the DNA fragment was performed by using a truSeq adapter and amplified with truSeq PCR primers for sequencing. Reads more than 35 nucleotide (nt) were selected. Furthermore, paired reads were removed that were matched to the sliva database (http://www.arb-silva.de/download/arb-files/). With the help of the DE-Seq package, all genes' expression in different samples were transformed to count per gene (CPG) by using blind and fit-only parameters. From the respective repeats of C. jejuni, NCTC 11168 and Δcas9 mutant mean CPG gene expression were determined and comparison was made between the differentially expressed genes (DEGs) of both strains. The DEGs having P-value ≤ 0.05 with fold change ≥1.5 were regarded as up-regulated genes, while DEGs having fold change ≤ 1.5 were considered as down-regulated genes. For the functional gene classification, gene ontology (GO) from an internationally established system was followed. This system not only comprehensively describes properties of genes but also provides their products in any organism. Further analyses of DEGs were performed on the basis of biological process, molecular function and cellular components. The Kyoto Encyclopedia of genes and genomes (KEGG) database was used to find out the role of DEGs in various pathways. The data has been submitted to Gene expression omnibus (GEO) under the accession number SUB2870584.
Validation by RT-qPCR
RNA sequencing results were verified by RT-qPCR. For that, seven genes were selected on the basis of their role in virulence and differential expression in RNA-Seq results. These included Cj0762c (aspartate aminotransferase), Cj1523c (CRISPR-associated protein), Cj1222c (two-component sensor histidine kinase), Cj0996 (GTP cyclohydrolase II), Cj0725c (molybdenum cofactor biosynthesis), Cj0454c (membrane protein), and Cj0330c (50S ribosomal protein L32). For RT-qPCR, a 16SrRNA gene was used as a housekeeping gene. Primers used in RT-qPCR are given in the S3 Table.
Statistical analysis
For statistical analyses, SPSS version 22.0 (IBM corp., Armonk, NY, USA) was used to determine the mean ± standard deviation (MSD) and significance level by applying a two-tailed t-test comparing C. jejuni NCTC11168, and its Δcas9 mutant strain for in vitro experiments, such as biofilm formation, adhesion, and invasion, and intracellular survivability. A Pearson's correlation coefficient (r) was computed between RNA-Seq and RT-qPCR results with a significance cutoff of P ≤ 0.05.
Results
Cas9 gene role on growth
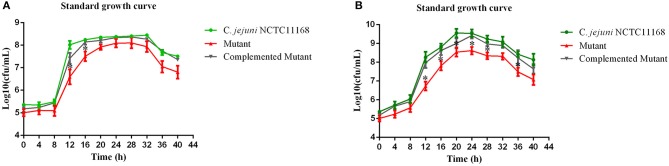
Campylobacter jejuni NCTC11168, complemented mutant and Δcas9 mutant strains were grown separately in biphasic media of MH for the determination of standard growth curve. This experiment was performed to determine growth characteristics of three strains. Compared to growth of C. jejuni NCTC11168, complemented and Δcas9 mutant under static condition (Figure 1A), we found significant difference in growth between three strains, being grown under shaking conditions (Figure 1B).
Figure 1.
(A,B) Standard growth curve of mutant with its parent strain under static and shaking conditions. (A) represents the bacterial growth curve under static conditions, while (B) shows the bacterial growth under shaking conditions. The Y axis shows the growth of C. jejuni NCTC11168, complemented and Δcas9 mutant strains in Log10 (cfu/mL) in static condition. One asterisk (*) represent statistically significant differences at P ≤ 0.05, in comparisons of C. jejuni NCTC11168, complemented and the Δcas9 mutant.
Biofilm formation of the C. jejuni NCTC11168 and mutant
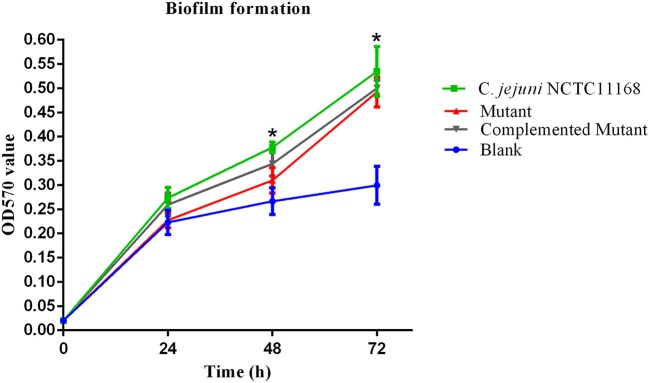
It has been suggested that biofilm formation helps the C. jejuni to survive in the environment against various stressors. In other words, biofilm formation aids the virulence of bacteria. Therefore, we determined the biofilm of C. jejuni NCTC11168, complemented and Δcas9 mutant strains in vitro at different time intervals. The highest level of biofilm formation occurred after 72 h of incubation. It seemed that the level of biofilm formation had a positive correlation with the incubation time. Compared to the Δcas9 mutant strain, a significant difference in biofilm formation was observed in the C. jejuni NCTC11168 strain after each incubation time, indicating that the C. jejuni NCTC11168 strain may have stronger ability to form biofilm (Figure 2).
Figure 2.
Biofilm formation of C. jejuni NCTC11168 and Δcas9 mutant at different time points. The Y axis is the OD570 value of crystal violet in biofilm. The results were obtained from three independent repeats. The asterisk (*) indicates statistical significance at P ≤ 0.05 for the comparison of C. jejuni NCTC11168, complemented, and the Δcas9 mutant.
Adhesion and invasion of C. jejuni NCTC11168 and mutant in macrophages RAW264.7
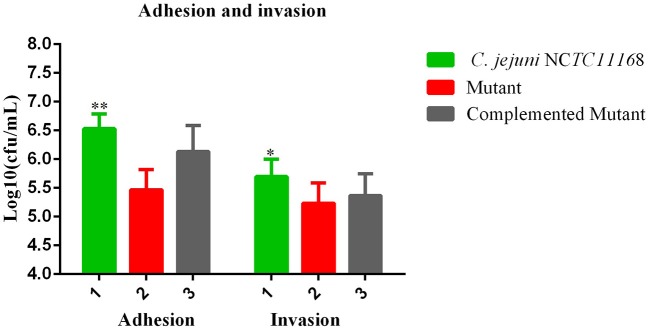
Most of the bacteria having a CRISPR-cas type II system are considered pathogenic bacteria. Therefore, adhesion and invasion assay was performed to determine the role of the cas9 gene in virulence. We observed that C. jejuni NCTC11168 exhibited remarkably higher adhesion and invasion of macrophage RAW264.7 cells than the Δcas9 mutant strain (Figure 3). A significant change of adhesion to macrophage cells (P < 0.05) was observed in C. jejuni NCTC1168 compared to its Δcas9 mutant strain. While, almost similar trend of complemented strain with C. jejuni NCTC11168 in terms of adhesion and invasion was observed.
Figure 3.
Adhesion and invasion of C. jejuni NCTC11168 and Δcas9 mutant on macrophage RAW 264.7 cells. The Y axis is the mean of log10 CFU/mL of each strain in the RAW264.7 cell. The results were obtained from three independent repeats. One asterisk (*) and two asterisks (**) represent statistically significant differences at P ≤ 0.05and P ≤ 0.01, respectively, in comparisons of C. jejuni NCTC11168, complemented, and the Δcas9 mutant.
Intracellular survivability of C. jejuni NCTC11168 in macrophages RAW264.7
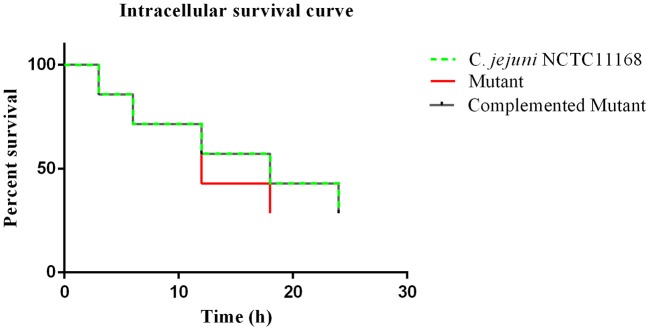
An intracellular survivability test was performed to determine whether the cas9 gene has a role in the survivability of C. jejuni intracellularly or not. It was observed in the present study that the intracellular survivability of C. jejuni NCTC11168 and complemented strains are greater than that of the Δcas9 mutant strain (Figure 4). The Δcas9 mutant strain had a shorter survival time (18 h) than the C. jejuni NCTC11168 strain, which was able to survive for 24 h. However, a considerable decrease in the number of internalized bacteria was observed at all stages. At each time point, the post-infection number of surviving C. jejuni NCTC11168 was higher than the Δcas9 mutant, suggesting that the C. jejuni NCTC11168 strain had greater survivability.
Figure 4.
Intracellular survivability of C. jejuni NCTC11168 and Δcas9 mutant in macrophage RAW 264.7 cells. The Y axis shows the percent survival of C. jejuni NCTC11168, complemented and Δcas9 mutant strain in the RAW264.7 cell. The analysis was performed by using the Kaplan-Meier estimate method. The results showed that C. jejuni NCTC11168 and complemented strains had more survivability than Δcas9 mutant strain.
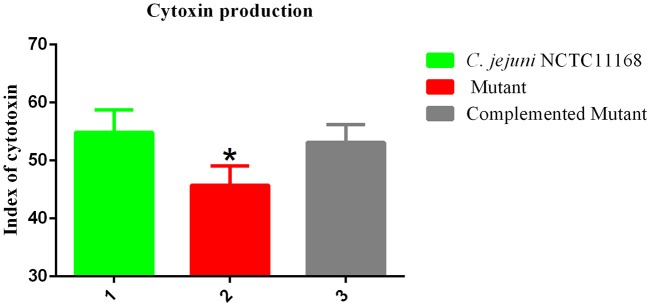
Cytotoxin of the C. jejuni NCTC11168 and Δcas9 mutant in macrophage cells
Campylobacter jejuni NCTC11168 produces toxins, which is another factor in virulence. Therefore, the cytotoxin production ability of C. jejuni NCTC11168, complemented and Δcas9 mutant strain was determined. The present study showed the killing effect (index) of cytotoxin released by C. jejuni NCTC11168, complemented and Δcas9 mutant strains in murine macrophage RAW264.7 cells (Figure 5). It was observed that C. jejuni NCTC11168 and complemented strains had the ability to produce more toxin than the Δcas9 mutant strain.
Figure 5.
Cytotoxic production of the C. jejuni NCTC11168 and Δcas9 mutant in murine macrophage RAW264.7 cells. The Y axis is the index of cytotoxic production of each strain. The results were obtained from three independent repeats. One asterisk (*) indicates a statistically significant difference at P ≤ 0.05 between C. jejuni NCTC11168, complemented, and the Δcas9 mutant.
Motility of the C. jejuni NCTC11168 and Δcas9 mutant
Flagella were also considered a virulence factor that helps in the motility and colonization of C. jejuni in the gastrointestinal tract of host. Therefore, motility of C. jejuni NCTC11168 and Δcas9 mutant strains were determined. The present study showed that there was an apparent difference between diameter growth rings of the C. jejuni NCTC11168 and Δcas9 mutant strains on a 0.4% MH agar plate (Figure 6). It was observed that Δcas9 mutant strains have less motility than the C. jejuni NCTC11168 strain.
Figure 6.
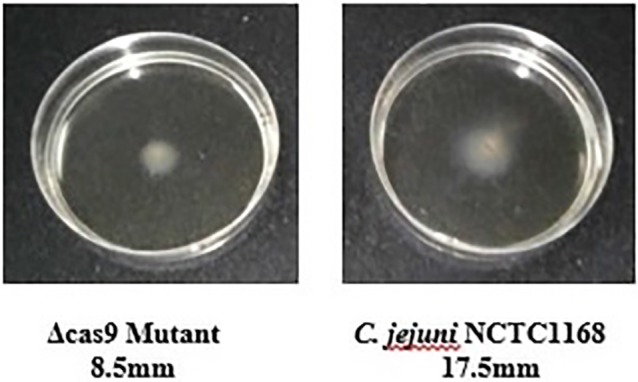
Motility of the C. jejuni NCTC 11168 and Δcas9 mutant strains on 0.4% MH agar plates. Motility is positively correlated to the diameter of the growth ring of each strain.
Frozen transmission electron microscopy
C. jejuni NCTC11168 have polar flagella, which are necessary for motility. In present study, flagellar morphology was observed by frozen transmission electron microscopy in two strains. Apparently, no significant morphological change was found in the flagella of both C. jejuni NCTC 11168 and Δcas9 mutant strains (Figure 7).
Figure 7.
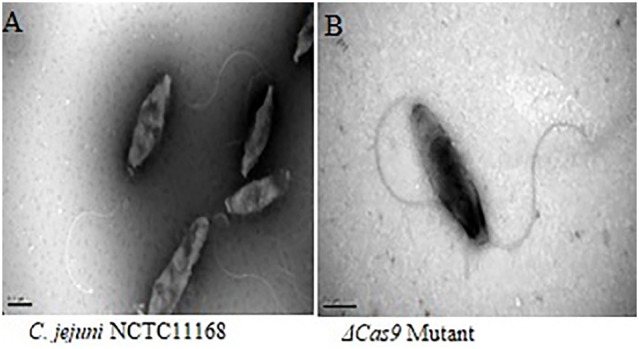
Frozen transmission electron microscopy of the C. jejuni NCTC 11168 and Δcas9 mutant strains. C. jejuni NCTC 11168 (A), Δcas9 mutant (B). The magnification used for TEM images in the capionis is 1μm.
Transcriptome analysis on the basis of RNA-Seq
In order to further understand the different regulators involved in virulence, we investigated the gene expression profile of the Δcas9 mutant strain and compared the transcriptome profile with the C. jejuni NCTC11168 strain. By comparing the transcriptome profile of both strains, a total of 2.89 M reads with clean ratio 99.8% were obtained for C. jejuni NCTC11168. A total of 2.79 M reads with clean ratio 99.75% were obtained for the Δcas9 mutant strain.
Upon differential expression analysis with the C. jejuni NCTC11168, we found a number of important genes related to our interest were down-regulated in Δcas9 mutant strain (Table 1). A number of membrane encoded genes, such as Cj1053c, Cj1203c, Cj0844c, Cj0884, and Cj0660c are involved in maintaining the integrity and permeability of bacterial cell membrane. Six lipoprotein encoding genes (Cj0983, Cj0113, Cj0950c, Cj1029c, Cj0978c, and Cj0842) were also found to be down-regulated. Flagella related genes such as Cj0547 and Cj0440c involved in the biosynthesis and assembly of flagella were also found to be down-regulated. Cj0923c (CheR) and Cj0924c (CheB) genes involved in chemotaxis were also found to be down-regulated. Several genes, such as Cj0535, Cj0441, and Cj1299 that play a role in energy metabolism were also found to be down-regulated. Five genes (Cj0330c, Cj0664c, Cj0471, Cj1708c, and Cj1694c) encoding ribosomal proteins were also found to be down-regulated.
Table 1.
Important down-regulated genes in Δcas9 mutant during differential expression analysis.
| Gene ID | Gene product | Fold change | P-value | q-value |
|---|---|---|---|---|
| Cj1053c | Integral membrane protein | 1.28 | 0.0037 | 0.164 |
| Cj0844c | Integral membrane protein | 1.14 | 0.0167 | 0.381 |
| Cj0884 | Integral membrane protein | 1.19 | 0.0224 | 0.430 |
| Cj0660c | Transmembrane protein | 1.20 | 0.0485 | 0.623 |
| Cj1203c | Integral membrane protein | 1.29 | 0.0061 | 0.218 |
| Cj0983 | Lipoprotein | 1.17 | 0.0497 | 0.623 |
| Cj0113 | Peptidoglycan associated lipoprotein | 1.10 | 0.0313 | 0.496 |
| Cj0950c | Lipoprotein | 1.15 | 0.0266 | 0.460 |
| Cj1029c | Lipoprotein (mapA) | 1.16 | 0.0222 | 0.430 |
| Cj0978c | Lipoprotein | 1.37 | 0.0124 | 0.330 |
| Cj0842 | Lipoprotein | 1.30 | 0.0060 | 0.218 |
| Cj1331 | Acylneuraminate cytidylyltransferase (ptmB) | 1.21 | 0.0406 | 0.553 |
| Cj0547 | Flagellar protein FlaG possibly involved in flagella export | 1.20 | 0.0117 | 0.327 |
| Cj0440c | Transcriptional regulator | 1.35 | 0.0039 | 0.164 |
| Cj0535 | 2-oxoglutarate-acceptor oxidoreductase subunit (OorD) | 1.28 | 0.0069 | 0.223 |
| Cj0441 | Acyl carrier protein carries the fatty acid chain in fatty acid biosynthesis (acpP) | 1.22 | 0.0024 | 0.134 |
| Cj1299 | Acyl carrier protein (acpP2) | 1.11 | 0.0205 | 0.429 |
| Cj0330c | 50S ribosomal protein L32 (rpmF) | 1.13 | 0.0261 | 0.460 |
| Cj0664c | 50S ribosomal protein (rplI) | 1.10 | 0.0209 | 0.429 |
| Cj0471 | 50S ribosomal protein (rpmG) | 1.15 | 0.0143 | 0.350 |
| Cj1708c | 30S ribosomal protein S10 NusE (rpsJ) | 1.20 | 0.0122 | 0.330 |
| Cj1694c | 30S ribosomal protein (rpsN) | 1.28 | 0.0024 | 0.134 |
| Cj0923c | MCP protein methyltransferase (CheR) | 1.14 | 0.0144 | 0.350 |
| Cj0924c | MCP protein-glutamate methylesterase (cheB) | 1.15 | 0.0189 | 0.406 |
| Cj0304c | Biotin synthesis protein (bioC) | 1.13 | 0.0267 | 0.460 |
Apart from down-regulated genes, several up-regulated genes were also observed in the present study. Cj0551, responsible for encoding elongation factor P, Cj0725c encode molybdenum cofactor biosynthesis protein, Cj0836 encode methylated DNA protein systeine methyltransferase, Cj0996 encode GTP cyclohydrolase II and Cj1587c encode putative ABC transporter proteins (Table 2). Furthermore, detail of additional DEGs were given in the S4 Table.
Table 2.
Up-regulated genes in Δcas9 mutant during differential expression analysis.
| Gene ID | Gene product | Fold change | P-value | q-value |
|---|---|---|---|---|
| Cj0551 | Elongation factor P | 2.09 | 2.67E−04 | 0.0195 |
| Cj0725c | Molybdenum cofactor biosynthesis protein | 2.45 | 3.71E−06 | 5.01E−04 |
| Cj0836 | Methylated DNA protein cysteine methyltransferase | 2.38 | 4.25E−15 | 1.49E−12 |
| Cj0996 | GTP cyclohydrolase II | 3.08 | 1.07E−04 | 0.0093 |
| Cj1587c | Putative ABC transporter | 2.70 | 0.039 | 0.544 |
Verification by RT-qPCR
Seven genes (Cj1523c, Cj0762c, Cj1222c, Cj0996, Cj0725c, Cj0454c, Cj0330c) encoding proteins related to the virulence mechanism of C. jejuni NCTC1168 were selected for validation of RNA-Seq results. Among the seven selected genes only Cj1523c was down-regulated in the Δcas9 mutant strain. Other genes such as Cj0762c, Cj1587c, Cj1222c, Cj0996, Cj0725c, Cj0454c, and Cj0330c were up-regulated in the Δcas9 mutant strain. For these seven genes, a trend of expression level by RT-qPCR was similar to that of the RNA-Seq with a correlation coefficient of r = 0.938, which indicated a strong correlation between the two techniques (S1 Figure).
Discussion
Over the past decade, C. jejuni has been reported as the leading cause of food poisoning in Europe (Bronnec et al., 2016). Due to its public health or zoonotic significance, it is important to understand the pathogenic mechanisms adopted by this pathogen, because the pathogenicity of this bacterium is largely unknown (Young et al., 2007). The CRISPR-cas system, which is an adaptive immune system of bacteria, also plays its part in controlling endogenous transcription as well as regulating bacterial pathogenicity. Similarly, C. jejuni has a type II CRISPR-cas system, and the functional protein of this system is cas9; therefore, this system has the ability to regulate bacterial pathogenesis (Shabbir et al., 2016). Therefore, in the present study we determined the pathogenesis of both C. jejuni NCTC1168 and its Δcas9 mutant strains.
Campylobacter jejuni NCTC 11168 has the ability to form biofilm, which is an important determinant of virulence. Biofilm develops in response to the aggregation of bacterial cells, which are surrounded by a self-producing matrix layer and adhere to the host surface. Moreover, biofilm protects the bacteria from different environmental stresses and provides resistance against the response of the host immune system (Bronnec et al., 2016). We observed in our study that biofilm formation was time dependent in C. jejuni NCTC11168 and this strain has a higher ability to form biofilm as compared to the Δcas9 mutant strain. A recent study also supports our findings that the C. jejuni wild strain has more capability to form biofilm as compared to mutant strains (Teh et al., 2017). This showed that the cas9 gene might have a role in biofilm formation or enhancing virulence.
Adherence and invasion of C. jejuni in the host epithelial cells is considered as the main feature of its pathogenesis. There are several proteins present on the surface of bacterial cells that help the bacteria in adherence to the host cell, such as Cj1478c (cadF; Jin et al., 2001), and bacterial invasion into the epithelial cells is achieved by a number of secretory proteins (Konkel et al., 2004). Scientists in 2011 reported that the C. jejuni strain has a higher adhesion and invasion rate than its mutant strain (Almofti et al., 2011). In our in vitro study, we also got the same findings in C. jejuni NCTC11168 and its Δcas9 mutant strain. Based on this finding, someone can guess that the cas9 gene might play its part in the regulation of these genes.
In light of several studies, it was observed that C. jejuni has the ability to multiply and survive within the macrophage cells (Day et al., 2000; Hickey et al., 2005; Iovine et al., 2008). In the present study, intracellular survivability of C. jejuni NCTC11168 and its Δcas9 mutant strain were also determined. We observed that intracellular survivability of the Δcas9 mutant was less than its wild strain. Our finding was in line with a previous study that also found that intracellular survivability of C. jejuni is greater than that of its mutant strain (Almofti et al., 2011).
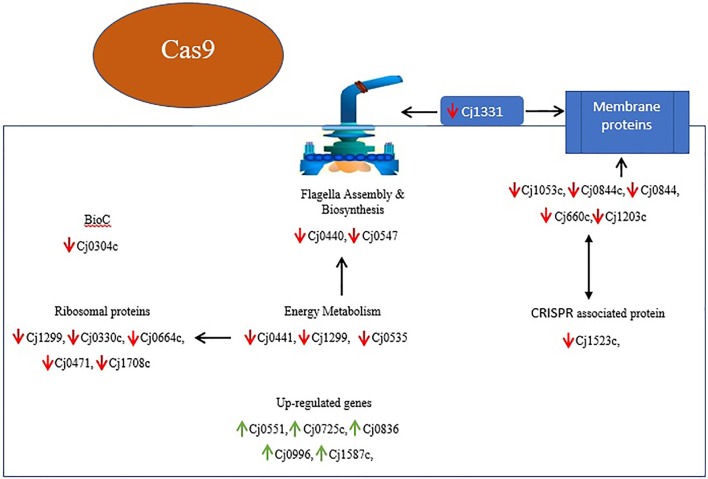
Upon the transcriptome analysis of C. jejuni NCTC11168 and its Δcas9 mutant strains, we observed a number of down-regulated genes that play a role in enhancing bacterial virulence directly or indirectly. Of which the important gene is Cj1331 (PtmB) involved in the glycosylation (McNally et al., 2007). Glycosylation helps in the modification of outer membrane proteins (OMPs) as well as in the assembly of flagella. Furthermore, it was also reported that PtmB mutagenesis does effect the motility of bacteria (Howard et al., 2009). Proteins related to outer membrane play role in maintaining the integrity and permeability of bacterial membrane (Gotoh et al., 1989). In the present study, several putative membrane associated genes (Cj0884, Cj0884c, Cj1203c, Cj1053c, Cj0842, Cj0978c) were found down-regulated that are additional candidates for virulence. As it was reported that membranous proteins essential for adhesion and invasion (Ye et al., 2015). Flagella formation is a key in the pathogenesis of campylobacter in terms of motility, biofilm formation, and adherence to and invasion of the host cell (Guerry, 2007). In the current study, we found repressed expression of genes involved in the biosynthesis and assembly of flagella such as Cj0440c and Cj0547 (FlaG) which leads to reduced flagellar motility. Our findings are in line with those of previous study in which they reported that Cj0440c involved in the biosynthesis of flagella (Hao et al., 2017). According to our findings, it can be speculated that cas9 might play a role in regulating the genes responsible for membrane integrity and adherence to the host cells as well as genes involved in the assembly and biosynthesis of flagella. This might be one reason we found less virulence in the Δcas9 mutant strain as compared to the wild strain.
Chemotaxis is a process in which bacteria move toward or away from chemical stimuli such as chemoeffectors or ligands that can either attractants or repellents present in the environment. By using this process, C. jejuni enable themselves in term of adaptation against disparate niches. In present study, we found two enzymes such as methylesterase CheB and methyltransferase CheR with repressed expression. These two enzymes involved in chemotaxis system of Campylobacter spp. (Chandrashekhar et al., 2017). Down-regulation of these two important enzymes of chemotaxis system suggest that cas9 gene also play its part in the enhancement of virulence by regulating this system.
Acyl carrier protein (ACP) plays a key role in the biosynthesis of bacterial fatty acids (Martinez et al., 2010). Fatty acids play a multifaceted role in maintaining the viability and virulence of bacteria (Ma et al., 2017). In the present study, two ACP genes (Cj0441, and Cj1299) involved in fatty acid biosynthesis were found down-regulated, which explained altered pathogenic characteristics in the Δcas9 mutant strain compared to the wild strain. Moreover, Cj0304c (bioC) encoding biotin synthesis protein was also found to be down-regulated in present study. Biotin plays an important role in the transfer of CO2 in key metabolic processes such as carboxylation, decarboxylation or transcarboxylation reactions (Bi et al., 2016). BioC enzyme that methylates ω-carboxyl group of malonyl-ACP to form malonyl-ACP methyl ester, this molecule is accepted by the fatty acid synthesis pathway (Bi et al., 2016). Additionally, Cj0535 (oorD) gene which play role in the generation of Succinyl-CoA via 2-oxoglutarate: acceptor oxidoreductase during citrate cycle (TCA cycle) was found down-regulated in current study. It was reported that oorD as well as porD (pyruvate:flavodoxin oxidoreductase) seems to be essential for bacterial survival (Su et al., 2006). Our result also supported by previous study in which they also found that oorD necessary for H. pylori survival (Su et al., 2006).
In light of several studies, it is assumed that ribosomal proteins involved in various processes which ultimately helps in the survival of bacteria. They reported that increased expression of ribosomal proteins facilitates the multiplication and survivability of C. jejuni (Cecchini et al., 2007; Flint et al., 2010). However, in current study down-regulated expression of ribosomal proteins such as Cj1694c (rpsN), Cj1708c (rpsJ), Cj0471 (rpmG), Cj0664c (rplI), and Cj0330c (rpmF) was observed. This suggest that cas9 have role in the regulation of ribosomal proteins or in translation of several proteins which are essential for the survivability of bacteria (Figure 8).
Figure 8.
Overview of DEGs and their relationship in Δcas9 mutant strain on the basis of KEGG and STRING protein network analysis. Genes with red arrows indicate down-regulation, while genes with green arrows indicate up-regulation. Black double sided arrows indicate possible gene interaction.
Conclusion
In conclusion, it can be speculated based on the phenotypic determination of virulence characteristics along with transcriptomic profiling that CRISPR-cas9 is involved in the regulation of numerous virulence associated genes and enhanced the virulence of C. jejuni, which has zoonotic importance. Our study elucidates the genetic picture of C. jejuni in term of their up or down-regulation after the deletion of cas9. We can say in other words without any doubt that the CRISPR-cas9 system can change the pathophysiology of C. jejuni0. However, there is further need to explore the potential by which cas9 enhances virulence in C. jejuni.
Author contributions
YT, HH, and ZY conceived and designed the experiments. ZX and ML performed the experiments. YT and MS analyzed the data. GC, MD, XW, ZL, HH, and ZY contributed reagents, materials, analysis tools. YT and MS wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Key Research and Development program (2016YFD0501302/2017YFD0501406), the National Natural Science Foundation of China (31772791), the Fundamental Research Funds for the Central Universities (2662018JC001), and the National Program for Risk Assessment of Quality and Safety of Livestock and Poultry Products (GJFP2016008). The funders had no participation in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00285/full#supplementary-material
References
- Almofti Y. A., Dai M., Sun Y., Haihong H., Yuan Z. (2011). Impact of erythromycin resistance on the virulence properties and fitness of Campylobacter jejuni. Microb. Pathog. 50, 336–342. 10.1016/j.micpath.2011.02.009 [DOI] [PubMed] [Google Scholar]
- Ashkenazi S., Cleary T. G. (1990). A method for detecting Shiga toxin and Shiga-like toxin-l in pure and mixed culture. J. Med. Microbiol. 32, 255–261. 10.1099/00222615-32-4-255 [DOI] [PubMed] [Google Scholar]
- Bæk K. T., Vegge C. S., Brøndsted L. (2011). HtrA chaperone activity contributes to host cell binding in Campylobacter jejuni. Gut Pathog. 3:13. 10.1186/1757-4749-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H., Zhu L., Jia J., Cronan J. E. (2016). A biotin biosynthesis gene restricted to helicobacter. Sci. Rep. 6, 1–13. 10.1038/srep21162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Fernando U. M., Reiman C. D., Willson P. J., Townsend H. G. G., Potter A. A., et al. (2007). Correlation between in vitro secretion of virulence-associated proteins of Campylobacter jejuni and colonization of chickens. Curr. Microbiol. 54, 207–212. 10.1007/s00284-006-0295-z [DOI] [PubMed] [Google Scholar]
- Bolton D. J. (2015). Campylobacter virulence and survival factors. Food Microbiol. 48, 99–108. 10.1016/j.fm.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Bronnec V., Turonová H., Bouju A., Cruveiller S., Rodrigues R., Demnerova K., et al. (2016). Adhesion, biofilm formation, and genomic features of Campylobacter jejuni Bf, an atypical strain able to grow under aerobic conditions. Front. Microbiol. 7:1002. 10.3389/fmicb.2016.01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelow D. R., Christensen J. E., Neal-Mckinney J. M., Konkel M. E. (2011). Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol. Microbiol. 80, 1296–1312. 10.1111/j.1365-2958.2011.07645.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo C. D., Taboada E., Nash J. H. E., Lanthier P., Kelly J., Lau P. C., et al. (2004). Genome-wide expression analyses of campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and we examined two variants of the Genome-sequenced. J Biol Chem. 279, 20327–20338. 10.1074/jbc.M401134200 [DOI] [PubMed] [Google Scholar]
- Carvalho A. C. T., Ruiz-palacios G. M., Ramos-cervantes P., Cervantes L., Jiang X. I., Pickering L. K. (2001). Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates. J Clin. Microbiol. 39, 1353–1359. 10.1128/JCM.39.4.1353-1359.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini K. R., Gorton T. S., Geary S. J. (2007). Transcriptional responses of Mycoplasma gallisepticum strain R in association with eukaryotic cells. J. Bacteriol. 189, 5803–5807. 10.1128/JB.00667-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai L. C., Ghazali F. M., Bakar F. A., Lee H. Y., Suhaimi L. R. A., Talib S. A., et al. (2009). Occurrence of thermophilic Campylobacter spp. contamination on vegetable farms in Malaysia. J. Microbiol. Biotechnol. 19, 1415–1420. 10.4014/jmb.0901.0002 [DOI] [PubMed] [Google Scholar]
- Chandrashekhar K., Kassem I. I., Rajashekara G. (2017). Campylobacter jejuni transducer like proteins: Chemotaxis and beyond. Gut Microbes 8, 323–334. 10.1080/19490976.2017.1279380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J. E., Pacheco S. A., Konkel M. E. (2009). Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol. Microbiol. 73, 650–662. 10.1111/j.1365-2958.2009.06797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasti J. I., Tareen A. M., Lugert R., Zautner A. E., Groß U. (2010). Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 300, 205–211. 10.1016/j.ijmm.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Davis L., DiRita V. (2008). Growth and laboratory maintenance of Campylobacter jejuni. Curr. Protoc. Microbiol. 10.1002/9780471729259.mc08a01s10 [DOI] [PubMed] [Google Scholar]
- Day W. A., Sajecki J. L., Pitts T. M., Joens L. A. (2000). Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68, 6337–6345. 10.1128/IAI.68.11.6337-6345.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero R. L., Lee A. (1988). Motility of Campylobacter jejuni in a viscous environment: comparison with conventional rod-shaped bacteria. Microbiology 134, 53–59. 10.1099/00221287-134-1-53 [DOI] [PubMed] [Google Scholar]
- Flint A., Butcher J., Clarke C., Marlow D., Stintzi A. (2010). Use of a rabbit soft tissue chamber model to investigate Campylobacter jejuni-host interactions. Front. Microbiol. 1:126. 10.3389/fmicb.2010.00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffar N. M., Connerton P. L., Connerton I. F. (2015). Filamentation of campylobacter in broth cultures. Front. Microbiol. 6:657. 10.3389/fmicb.2015.00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M., Brisson J., Karwaski M., Michniewicz J., Cunningham A., Wu Y., et al. (2000). Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. J Biol Chem. 275, 3896–3906. 10.1074/jbc.275.6.3896 [DOI] [PubMed] [Google Scholar]
- Gotoh N., Wakebe H., Yoshihara E., Nakae T., Nishino T. (1989). Role of protein F in maintaining structural integrity of the Pseudomonas aeruginosa outer membrane. J. Bacteriol. 171, 983–990. 10.1128/jb.171.2.983-990.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrant R. L., Van Gilder T., Steiner T. S., Thielman N. M., Slutsker L., Tauxe R. V., et al. (2001). Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32, 331–351. 10.1086/318514 [DOI] [PubMed] [Google Scholar]
- Guerrant R. L., Wanke C. A., Pennie R. A., Barrett L. J., Lima A. A., O'Brien A. D. (1987). Production of a unique cytotoxin by Campylobacter jejuni. Infect. Immun. 55, 2526–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P. (2007). Campylobacter flagella: not just for motility. Trends Microbiol. 15, 456–461. 10.1016/j.tim.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Han K., Jang S. S., Choo E., Heu S., Ryu S. (2007). Prevalence, genetic diversity, and antibiotic resistance patterns of Campylobacter jejuni from retail raw chickens in Korea. Int. J. Food Microbiol. 114, 50–59. 10.1016/j.ijfoodmicro.2006.10.042 [DOI] [PubMed] [Google Scholar]
- Hao H., Fang X., Han J., Foley S. L., Wang Y., Cheng G., et al. (2017). Cj0440c affects flagella formation and in vivo colonization of erythromycin-susceptible and-resistant Campylobacter jejuni. Front. Microbiol. 8:729. 10.3389/fmicb.2017.00729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Ren N., Han J., Foley S. L., Iqbal Z., Cheng G., et al. (2016). Virulence and genomic feature of multidrug resistant Campylobacter jejuni isolated from broiler chicken. Front. Microbiol. 7:1605. 10.3389/fmicb.2016.01605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson D. R. (2006). A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61, 1646–1659. 10.1111/j.1365-2958.2006.05336.x [DOI] [PubMed] [Google Scholar]
- Hickey T. E., Majam G., Guerry P. (2005). Intracellular survival of Campylobacter jejuni in human monocytic cells and induction of apoptotic death by cytholethal distending toxin. Infect. Immun. 73, 5194–5197. 10.1128/IAI.73.8.5194-5197.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S. L., Jagannathan A., Soo E. C., Hui J. P. M., Aubry A. J., Ahmed I., et al. (2009). Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect. Immun. 77, 2544–2556. 10.1128/IAI.01425-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Brooks B. W., Lowman R., Carrillo C. D. (2015). Campylobacter species in animal, food, and environmental sources, and relevant testing programs in Canada. Can. J. Microbiol. 61, 701–721. 10.1139/cjm-2014-0770 [DOI] [PubMed] [Google Scholar]
- Iovine N. M., Pursnani S., Voldman A., Wasserman G., Blaser M. J., Weinrauch Y. (2008). Reactive nitrogen species contribute to innate host defense against Campylobacter jejuni. Infect. Immun. 76, 986–993. 10.1128/IAI.01063-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z., Seleem M. N., Hussain H. I., Huang L., Hao H., Yuan Z. (2016). Comparative virulence studies and transcriptome analysis of Staphylococcus aureus strains isolated from animals. Sci. Rep. 6:35442. 10.1038/srep35442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Joe A., Lynett J., Hani E. K., Sherman P., Chan V. L. (2001). JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 39, 1225–1236. 10.1111/j.1365-2958.2001.02294.x [DOI] [PubMed] [Google Scholar]
- Kim G., Lee S. E., Yang H., Park H. R., Son G. W., Park C., et al. (2015). β2 integrins (CD11/18) are essential for the chemosensory adhesion and migration of polymorphonuclear leukocytes on bacterial cellulose. J. Biomed. Mater. Res. Part A 103, 1809–1817. 10.1002/jbm.a.35316 [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Garvis S. G., Tipton S. L., Anderson D. E., Jr., Cieplak W., Jr. (1997). Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24, 953–963. 10.1046/j.1365-2958.1997.4031771.x [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Klena J. D., Rivera-amill V., Monteville M. R., Biswas D., Raphael B., et al. (2004). Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export Apparatus 186, 3296–3303. 10.1128/JB.186.11.3296-3303.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M., Galán J. E. (2001). CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect. Immun. 69, 4358–4365. 10.1128/IAI.69.7.4358-4365.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P., Ottemann K. M., Hendrixson D. R. (2011). Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65, 389–410. 10.1146/annurev-micro-090110-102908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D., Gilbert M., Hitchen P. G., Dell A., Morris H. R., Wakarchuk W. W., et al. (2000). Phase variation of a β-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37, 501–514. 10.1046/j.1365-2958.2000.02020.x [DOI] [PubMed] [Google Scholar]
- Louwen R., Horst-Kreft D., De Boer A. G., Van Der Graaf L., de Knegt G., Hamersma M., et al. (2013). A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain–Barré syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 32, 207–226. 10.1007/s10096-012-1733-4 [DOI] [PubMed] [Google Scholar]
- Ma J., Wu Y., Cao D., Zhang W., Wang H., Wang H. (2017). Only acyl carrier protein 1 ( AcpP1 ) functions in Pseudomonas aeruginosa fatty acid synthesis. Front. Microbiol. 8:2186. 10.3389/fmicb.2017.02186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathu E. K., Hiratsuka K., Taylor D. E. (1988). Nucleotide sequence analysis and expression of a tetracycline-resistance gene from Campylobacter jejuni. Gene 62, 17–26. 10.1016/0378-1119(88)90576-8 [DOI] [PubMed] [Google Scholar]
- Martinez M. A., de Mendoza D., Schujman G. E. (2010). Transcriptional and functional characterization of the gene encoding acyl carrier protein in Bacillus subtilis. Microbiology 156, 484–495. 10.1099/mic.0.033316-0 [DOI] [PubMed] [Google Scholar]
- McFarland B. A., Neill S. D. (1992). Profiles of toxin production by thermophilic Campylobacter of animal origin. Vet. Microbiol. 30, 257–266. 10.1016/0378-1135(92)90119-E [DOI] [PubMed] [Google Scholar]
- McNally D. J., Aubry A. J., Hui J. P. M., Khieu N. H., Whitfield D., Ewing C. P., et al. (2007). Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem. 282, 14463–14475. 10.1074/jbc.M611027200 [DOI] [PubMed] [Google Scholar]
- Pickett C. L., Pesci E. C., Cottle D. L., Russell G., Erdem A. N., Zeytin H. (1996). Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect. Immun. 64, 2070–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett C. L., Whitehouse C. A. (1999). The cytolethal distending toxin family. Trends Microbiol. 7, 292–297. 10.1016/S0966-842X(99)01537-1 [DOI] [PubMed] [Google Scholar]
- Pitkänen T. (2013). Review of Campylobacter spp. in drinking and environmental waters. J. Microbiol. Methods 95, 39–47. 10.1016/j.mimet.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Poly F., Ewing C., Goon S., Hickey T. E., Rockabrand D., Majam G., et al. (2007). Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect. Immun. 75, 3859–3867. 10.1128/IAI.00159-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poly F., Guerry P. (2008). Pathogenesis of campylobacter. Curr. Opin. Gastroenterol. 24, 27–31. 10.1097/MOG.0b013e3282f1dcb1 [DOI] [PubMed] [Google Scholar]
- Price E. P., Smith H., Huygens F., Giffard P. M., Al P. E. T. (2007). High-resolution DNA melt curve analysis of the clustered, regularly interspaced short-palindromic-repeat Locus of Campylobacter jejuni ? †. Appl. Environ. Microbiol. 73, 3431–3436. 10.1128/AEM.02702-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy D., Buswell C. M., Hodgson A. E., McAlpine K., Henderson I., Leach S. A. (2000). Characterisation of cytolethal distending toxin (CDT) mutants of Campylobacter jejuni. J. Med. Microbiol. 49, 473–479. 10.1099/0022-1317-49-5-473 [DOI] [PubMed] [Google Scholar]
- Sampson T. R., Napier B. A., Schroeder M. R., Louwen R., Zhao J., Chin C.-Y., et al. (2014). A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc. Natl. Acad. Sci. U.S.A. 111, 11163–11168. 10.1073/pnas.1323025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir M. A. B., Hao H., Shabbir M. Z., Hussain H. I., Iqbal Z., Ahmed S., et al. (2016). Survival and evolution of CRiSPR–Cas system in prokaryotes and its applications. Front. Immunol. 7:375. 10.3389/fimmu.2016.00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerlad S. M., Hendrixson D. R. (2007). Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189, 179–186. 10.1128/JB.01199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G. E., Hultgren S. J. (1999). Bacterial adhesins : common themes and variations in architecture and assembly MINIREVIEW bacterial adhesins : common themes and variations in architecture and assembly. 181, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanović S., Vuković D., Dakić I., Savić B., Švabić-Vlahović M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179. 10.1016/S0167-7012(00)00122-6 [DOI] [PubMed] [Google Scholar]
- Su Z., Xu H., Zhang C., Shao S., Li L., Wang H., et al. (2006). Mutations in Helicobacter pylori porD and oorD genes may contribute to furazolidone resistance. Croat. Med. J. 47, 410–415. [PMC free article] [PubMed] [Google Scholar]
- Tauxe R. V. (2001). “Incidence, Trends and Sources of Campylobacteriosis in Developed Countries: An Overview. increasing Incid. Campylobacteriosis Humans,” in Rep. Proc. a WHO Consult. Expert. Geneva: World Heal. Organ. [Google Scholar]
- Teh A. H. T., Lee S. M., Dykes G. A. (2017). Identification of potential Campylobacter jejuni genes involved in biofilm formation by EZ-Tn5 Transposome mutagenesis. BMC Res. Notes 10:182. 10.1186/s13104-017-2504-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) (2011). Critically Important Antimicrobials for Human Medicine - 3rd Revision 2011. World Heal. Organ., 1–38. [Google Scholar]
- Ye Y., Gao J., Jiao R., Li H., Wu Q., Zhang J., et al. (2015). The membrane proteins involved in virulence of cronobacter sakazakii virulent G362 and attenuated L3101 isolates. Front. Microbiol. 6:1238. 10.3389/fmicb.2015.01238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. T., Davis L. M., DiRita V. J. (2007). Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5, 665–669. 10.1038/nrmicro1718 [DOI] [PubMed] [Google Scholar]
- Ziprin R. L., Young C. R., Byrd J. A., Stanker L. H., Hume M. E., Gray S. A., et al. (2001). Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis. 549–557. 10.2307/1592894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.