Abstract
Previous work has shown that ductile/intrinsically disordered proteins (IDPs) and residues (IDRs) are found in all unicellular and multicellular organisms, wherein they are essential for basic cellular functions and complement the function of rigid proteins. In addition, computational studies of diverse phylogenetic lineages have revealed: (1) that protein ductility increases in concert with organismic complexity, and (2) that distributions of IDPs and IDRs along the chromosomes of plant species are non-random and correlate with variations in the rates of the genetic recombination and chromosomal rearrangement. Here, we show that approximately 50% of aligned residues in paralogs across a spectrum of algae, bryophytes, monocots, and eudicots are IDRs and that a high proportion (ca. 60%) are in disordered segments greater than 30 residues. When three types of IDRs are distinguished (i.e., identical, similar and variable IDRs) we find that species with large numbers of chromosome and endoduplicated genes exhibit paralogous sequences with a higher frequency of identical IDRs, whereas species with small chromosomes numbers exhibit paralogous sequences with a higher frequency of similar and variable IDRs. These results are interpreted to indicate that genome duplication events influence the distribution of IDRs along protein sequences and likely favor the presence of identical IDRs (compared to similar IDRs or variable IDRs). We discuss the evolutionary implications of gene duplication events in the context of ductile/disordered residues and segments, their conservation, and their effects on functionality.
Keywords: IDPs, polyploidy, protein ductility, protein disorder, paralogs, genome duplication, plants
Introduction
There is wide consensus that spontaneous whole genome duplications (WGD, autopolyploidy) and interspecific hybridization (allopolyploidy), followed by post-polyploid diploidization (PPD) have contributed significantly to the evolution of the land plants and to the angiosperms in particular (Wendel, 2000; Ramsey and Schemske, 2002; Soltis and Soltis, 2009; Jackson and Chen, 2010; Mandáková and Lysak, 2018; Ren et al., 2018). In general, new species emerging from either type of polyploidy tend to exhibit improved growth vigor and adaptive resilience to adverse environments thereby conferring significant evolutionary advantages (Song and Chen, 2015). Although the reasons remain unclear, plant genomes tend to have larger numbers of duplicated genes compared with the genomes of non-photosynthetic eukaryotes, although recent reports suggest that WGD events have also been frequent in insects (Li et al., 2018). Among the angiosperms, there is evidence that major clade-wide WGD events have occurred multiple times over the past 200 Mya (Lyons et al., 2008; Renny-Byfield and Wendel, 2014; del Pozo and Ramírez-Parra, 2015; Landis et al., 2018; Ren et al., 2018) in contrast to duplication events within major vertebrate lineages (Panopoulou et al., 2003; Dehal and Boore, 2005). In addition, a whole genome triplication events (triploidization, WGT, or hexaploidization) occurred in the ancestor of the core eudicots (approximately 125 Mya) and another more recent event (between 23 and 47 Mya) occurred in Brassica species, which affected the evolution of many agriculturally important species (Zheng et al., 2013; Kagale et al., 2014; Parkin et al., 2014). Thus, ancient and recent autopolyploidy have profoundly influenced the evolution of the flowering plants and have contributed to improved important agronomic traits, such as grain quality, fruit shape, and flowering time (Kellogg and Bennetzen, 2004; Dubcovsky and Dvorak, 2007; Leitch and Leitch, 2008; Jackson and Chen, 2010; Panchy et al., 2016). Given the fact that PPD events are recurrent over the course of angiosperm evolution, many extant diploid genomes contain a record of ancient WGD events that can be inferred by the analysis of duplicated genes with conserved co-linearity across genomic segments (Proost et al., 2012; Ren et al., 2018).
Despite their overarching importance, the consequences of polyploidy remain poorly understood. Studies have documented rapid and dynamic changes in genomic structure and gene expression in plant polyploids, which reflect genomic and functional plasticity of duplicated genes (Dubcovsky and Dvorak, 2007; Leitch and Leitch, 2008; Jackson and Chen, 2010). However, it is uncertain as to whether individual genes or WGD have contributed equally to the evolution and functional plasticity of plant genomes (see Dehal and Boore, 2005; De Storme and Mason, 2014). This ambiguity results in part because a direct causal link between an adaptive phenotype and a specific gene duplication event are difficult to ascertain because they usually occur at different times (Lawton-Rauth, 2003).
Studies during the past two decades have provided valuable information about intrinsically disordered/ductile proteins (IDPs) and disordered regions (Xie et al., 2007; Oldfield and Dunker, 2014; van der Lee et al., 2014; Wright and Dyson, 2015; Yan et al., 2016). IDPs do not fold into well-defined three-dimensional (3D) structures and can be either entirely disordered or partially disordered, with regions spanning just a few contiguous disordered residues (<10 aa) or containing long segments (≥30 aa) of contiguously disordered residues. Numerous researchers have developed algorithms that use amino acid sequences as inputs to predict the probability to be structured or disordered for each residue as outputs (He et al., 2009; Meng et al., 2017). By applying such disorder predictors to sequences of proteins with known functions, the biological activities of IDPs can be inferred from large collections of proteins (Ward et al., 2004; Xie et al., 2007). From these and other studies it has been concluded that 25–50% of all eukaryotic proteins contain at least one long IDP region and that 33–50% of eukaryotic proteomes have IDPs regions.
At the molecular level, it is uncertain how disordered/ductile proteins evolve in the scenario of WGD and PPD events. Nevertheless, there is ample evidence that disordered residues (IDRs) confer flexibility to proteomes (Tompa, 2002; Schad et al., 2011; Yruela and Contreras-Moreira, 2012; Yruela et al., 2017). Moreover, IDPs are known (1) to have played a significant role in the evolution of multicellularity and/or cell type specification (Niklas et al., 2014, 2018; Dunker et al., 2015; Niklas and Dunker, 2016; Yruela et al., 2017), (2) to contribute to organismic plasticity by facilitating protein multifunctionalities and nucleic acid interactions through complex gene regulatory network dynamics (Dyson and Wright, 2002, 2005; Xie et al., 2007; Habchi et al., 2014; van der Lee et al., 2014; O’Shea et al., 2015; Wright and Dyson, 2015; Yruela, 2015; Covarrubias et al., 2017), and (3) to be associated with proteins involved in signaling, cellular regulation, nuclear localization, chaperone activity, and RNA, DNA, and protein binding among many other functions (Xie et al., 2007; Kovacs et al., 2008; Babu et al., 2011; Buljan et al., 2013; Pazos et al., 2013; Oldfield and Dunker, 2014; Skupien-Rabian et al., 2016; Olsen et al., 2017). Moreover, IDPs and IDRs collaborate with alternative splicing (AS) and post-transcriptional modifications (PTMs) to markedly enhance the complexity of signaling networks (Niklas et al., 2015). By means of these collaborations, the same gene products bring about alternative signaling outcomes that depend on the use by IDPs or IDRs of shape-shifts to bind to multiple, different partners and that depend on the further alteration of partner binding by AS and/or PTMs (Dunker et al., 2008, 2015; Rautureau et al., 2010; Hsu et al., 2013; Niklas et al., 2015; Zhou et al., 2018). This collaboration of IDPs or IDRs with AS and/or PTMs appears to have contributed significantly to the evolution of multicellularity in all major eukaryotic lineages (Niklas et al., 2018).
The goal of this paper is to evaluate the hypothesis that WGD (or WTG) events have disproportionately increased IDRs in plant proteomes thereby contributing to plant “evolvability.” To investigate this hypothesis, we determined the fraction of IDRs in co-linear paralogs of several model and economically important plant species including green algae, bryophytes, monocots, and eudicots.
Materials and Methods
Protein Sequences
Protein sequences of co-linear paralogs of one chlorophyte (Chlamydomonas reinhardtii, n = 32), one bryophyte (Physcomitrella patens, n = 3,716), four monocots (Zea mays, n = 14,062; Sorghum bicolor, n = 5,336; Oryza sativa, n = 6,503; Brachypodium distachyon, n = 4,670), and thirteen eudicots (Glycine max, n = 74,584; Populus trichocarpa, n = 27,976; Brassica oleracea, n = 41,318; Manihot esculenta, n = 18,954; Vitis vinifera, n = 8,836; Gossypium raimondi, n = 25,880; Capsicum annuun, n = 996; Solanum lycopersicum, n = 6,796; Arabidopsis thaliana, n = 7,894; Prunus persica, n = 4,784; Beta vulgaris, n = 774; Medicago truncatula, n = 6,664; Cucumis sativus, n = 2,198) were retrieved from PLAZA 4.0,1 (van Bel et al., 2017). These species were selected because (1) some are model experimental systems (e.g., C. reinhardtii P. patens, A. thaliana, B. distachyon), (2) others are economically extremely important (e.g., Z. mays, G. max, V. vinifera), and (3) all have full representative genome and chromosome assemblies. Co-linear regions within genomes are annotated in PLAZA 4.0 by application of the i-ADHoRe algorithm (Proost et al., 2012). Co-linear paralogs are encoded by genes from the same gene family and are located in genomic segments that share the same gene content in the same order.
WGD and WGT in Plants
Ancient large-scale duplication (WGD) and triplication (WGT) events, or more recent duplications have been reported in the literature for the following model plant systems: P. patens (1 recent duplication), Z. mays (6 duplications), S. bicolor (5 duplications), O. sativa (5 duplications), B. distachyon (5 duplications), G. max (4 duplications and 1 triplication), P. trichocarpa (3 duplications and 1 triplication), V. vinifera (2 duplications and 1 triplication), B. oleracea (2 duplications), M. esculenta (2 duplications and 1 triplication), G. raimondi (2 duplications and 1 undefined event), S. lycopersicum (2 duplications and 2 triplications), A. thaliana (5 duplications), P. persica (2 duplications and 1 triplication), M. truncatula (3 duplications and 1 triplication) (Panchy et al., 2016; for more details about WGD and WGT history see Gaut et al., 2000; Blanc and Wolfe, 2004; Yu et al., 2005; Schmutz et al., 2010; Song et al., 2012; Tiley et al., 2016).
Mean reported Ks values, which represent WGDs and the divergence of duplicate gene pairs in plant families, are as follows: Angiosperms (Ks > 3), Solanaceae (Ks = 0.60), Fabaceae (Ks = 0.60), Poaceae (Ks = 0.90), Brassicaceae (Ks = 0.80) (Schranz et al., 2012).
Prediction of Disordered Residues
More than 60 predictors of disorder have been developed (He et al., 2009). A comparison of predictors and their variants across 1,765 proteomes reveals considerable variation in their ability to identify IDRs (Oates et al., 2013), indicating that the reliability of the predictor used in this study had to be evaluated critically before detailed analyses were under taken. In a detailed comparison of 16 commonly used predictors, PONDR VSL2b (Peng et al., 2006) had the best overall accuracy for long disordered regions (Peng and Kurgan, 2012). Nevertheless, we also explored DISOPRED v3.1 (Jones and Cozzetto, 2015) using a selected group of model monocot and eudicot species. Our analyses showed that overall DISOPRED v3.1 provided consistent results with the predictions of PONDR VSL2b (r2 = 0.60 and r2 = 0.96 for predicted IDRs and disordered regions with L > 30 aa, respectively) (Supplementary Figures S1–S3).
Data Analysis
Bar-plots and statistical analysis were performed with Origin Pro8.6. The coefficient of determination, r2, of standard linear regression protocols were calculated as:
where RSS is the residual sum of square and TSS is the total sum of square (Anderson-Sprecher, 1994).
Sequence Alignment
Pairwise alignments of co-linear paralogous sequences were determined using Clustal Omega (Sievers et al., 2011). For each aligned pair, the aligned disorder predictions were compared in order to calculate three types of IDRs: (1) identical disordered residues, where both the amino acid sequence and disorder predictions were identical (denoted hereafter as “identical IDRs”), (2) similar disordered residues, where the disorder predictions matched but the amino acid sequence varied (“similar IDRs”), and (3) variable residues where disorder predictions were not conserved (“variable IDRs”) (Yruela et al., 2017). The same three IDRs types were also computed for the subset of residues that were predicted to be disordered within long segments of at least 30 contiguous disordered residues (L > 30 aa). In all cases the fraction of IDRs were computed by dividing the number of aligned IDRs with the total aligned residues.
The IDR categories described previously (Yruela et al., 2017) and used here were inspired by work of Bellay et al. (2011) but differ in some details reported previously by others. Bellay et al. (2011) focused on orthologs and did not consider insertions and deletions, only sequences that could be aligned. In contrast, here we are studying paralogs. The three categories of IDRs described above for our work provided useful categories for the IDRs found in these proteins. For such proteins, we were particularly interested in examples in which disordered/ductile regions were present and absent in a given paralogous pair of proteins, and as noted above, Bellay et al. (2011), did not consider insertions or deletions at all.
Gene Ontology (GO) Enrichment Analysis
Gene Ontology annotations for the complete proteomes analyzed were retrieved from PLAZA 4.0 and genome GO term (expected) frequencies computed for each species. In addition, the subset of GO annotations corresponding to pairs of paralogs harboring long disordered segments (L > 30 aa) were used to compute (1) observed GO term frequencies for co-linear genes and (2) observed GO term frequencies for co-linear genes harboring at least two thirds of identical IDRs. Enrichment was computed by applying Fisher’s exact test with Bonferroni correction2 to compare the observed and expected GO term frequencies. When possible, plant-specific GO-slim terms were assigned to enriched terms by parsing file3.
Results and Discussion
We focused on the paralogs of 19 important plant species across a broad spectrum of the Chlorobionta (i.e., green algae to eudicots) whose proteome size, basic haploid chromosome number and number of co-linear paralogous pairs differ significantly (Table 1). From 37 to 52% total aligned paralogous sequences were identified by PONDR VSL2b as having an IDR signature. These results are consistent with previous whole proteome analyses (Yruela and Contreras-Moreira, 2012). The highest percentages of aligned IDRs were found in monocot species (50–52%); the lowest percentage was found in the green alga C. reinhardtii (37%). The range of total aligned IDRs observed for the 19 species examined in this study accords reasonably well with the evolutionary origins of these taxa (i.e., total aligned IDRs tend to increase with more recent descent). It is worth nothing that these values were on average much lower than those predicted using DisoPred v3.1 (Supplementary Table S1) and also much lower than those previously reported (Yruela and Contreras-Moreira, 2012) using DisoPred v2.42 (Ward et al., 2004). The differences observed using both versions of DisoPred are attributed to different sensitivities to IDRs longer than 20 amino acids (Jones and Cozzetto, 2015).
Table 1.
Characteristics of the nineteen plant species examined in this study.
| Plant species | Proteome size | Haploid chromosome number (n) | Pair of co-linear paralogs | Fraction of total aligned IDRs PONDR VSL2 |
Fraction of aligned IDRs in segments L > 30 aa PONDR VSL2b |
|---|---|---|---|---|---|
| Chlorophyte (n) | |||||
| Chlamydomonas reinhardtii | 14,488 | 17 | 16 | 0.37 | 0.55 |
| Bryophyte | |||||
| Physcomitrella patens | 48,022 | 27 | 1,858 | 0.50 | 0.62 |
| Monocots (2n = 2×) | |||||
| Zea mays | 58,615 | 10 | 7,031 | 0.52 | 0.66 |
| Sorghum bicolor | 39,248 | 10 | 2,668 | 0.52 | 0.65 |
| Oryza sativa | 40,881 | 12 | 3,251 | 0.50 | 0.62 |
| Brachypodium distachyon | 33,844 | 5 | 2,335 | 0.50 | 0.63 |
| Eudicots (2n = 2×) | |||||
| Glycine max | 71,525 | 20 | 37,292 | 0.45 | 0.56 |
| Populus trichocarpa | 73,110 | 19 | 13,988 | 0.46 | 0.55 |
| Vitis vinífera | 41,208 | 19 | 1,916 | 0.44 | 0.53 |
| Brassica oleracea | 56,687 | 9 | 20,660 | 0.48 | 0.57 |
| Manihot esculenta | 43,286 | 18 | 9,477 | 0.47 | 0.58 |
| Gossypium raimondi | 59,057 | 13 | 12,941 | 0.47 | 0.58 |
| Capsicum annuum | 40,627 | 12 | 498 | 0.42 | 0.48 |
| Solanum lycopersicum | 36,010 | 12 | 3,399 | 0.46 | 0.55 |
| Arabidopsis thaliana | 48,148 | 5 | 3,947 | 0.47 | 0.59 |
| Prunus persica | 32,595 | 8 | 2,392 | 0.47 | 0.58 |
| Beta vulgaris | 32,874 | 9 | 387 | 0.47 | 0.57 |
| Medicago truncatula | 57,661 | 8 | 3,322 | 0.47 | 0.57 |
| Cucumis sativus | 25,668 | 7 | 1,099 | 0.49 | 0.59 |
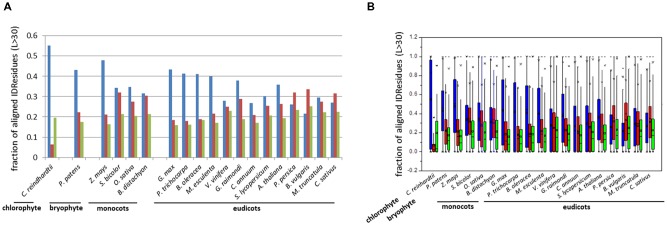
The fraction of aligned residues in long disordered segments (L > 30 aa) was also calculated using PONDR VSL2b, which revealed high proportions of IDRs located in such segments L > 30 aa (ca. 60%) (Table 1). Similar results were obtained using DisoPred v3.1 (Supplementary Table S1). Thus, the results consistently indicated that a high proportion of IDRs reside in long ductile segments (L > 30 aa) in paralogous pairs.
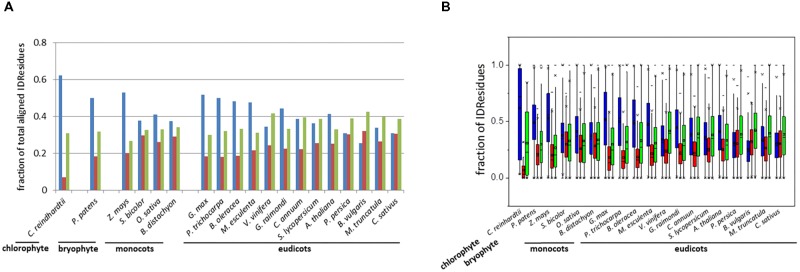
The distribution of the three types of IDRs (i.e., identical, similar, and variable IDRs) was analyzed for the paralogous pairs across all 19 species (Figure 1). Here we use the terms “identical IDRs” for those that are conserved with respect to sequence, length and location from one paralog to the next, “similar IDRs” for those that show substantial sequence variations but are conserved with respect to length and location from one paralog to the next and “variable IDRs” for those that are observed in some paralogs but absent in others.
FIGURE 1.
Bar-plot of the fraction (A) and box-plot distribution (B) of total aligned IDResidues. Identical IDRs (blue), similar IDRs (red), and variable IDRs (green) based on PONDR VSL2b predictions.
Analyses indicated that the percentage of aligned identical IDRs in paralogous sequences predicted by PONDR VSL2b ranged between 30 and 60%. It was highest in the green algae C. reindhardtii and lowest in the eudicot B. vulgaris (Figure 1). These data are in agreement with previous results (Yruela and Contreras-Moreira, 2013). The predicted fractions of similar IDRs and variable IDRs were highest in B. vulgaris, M. truncatula, P. persica, and C. sativus (Figures 1, 2). We speculate that the differences observed among the three different kinds of predicted IDRs reflect the history of genome duplication/polyploidy events (i.e., both chromosome number and the number of paralogs) in the species investigated in this study (Table 1). It is worth noting that the basic haploid chromosome number of B. vulgaris, M. truncatula, P. persica, and C. sativus are much reduced (n = 7 – 9) compared with those of the green alga C. reinhardtii (n = 17), other monocots such as O. sativa (n = 12) and Z. mays (n = 10), and eudicots such as G. max (n = 20) and P. trichocarpa (n = 19) (Table 1). Furthermore, the combination of multiple ancestral WGD and more recent polyploidy events promoting high rates of duplicated gene retentions (e.g., P. trichocarpa, G. max, B. oleoracea) (Parkin et al., 2014; Panchy et al., 2016) likely also favored the increase of identical IDRs.
FIGURE 2.
Bar-plot of the fraction (A) and box-plot distribution (B) of aligned residues in ductile regions (L > 30 aa). Identical IDRs (blue), similar IDRs (red), and variable IDRs (green) based on PONDR VSL2b predictions.
With the exception of the green alga C. reinhardtii and the bryophyte P. patens, a statistically significant and positive correlation (r2 = 0.45, P = 8 × 10-4) was observed between the number of co-linear paralogous pairs and the haploid number of chromosomes across the 17 vascular plant species (Supplementary Figure S2A). The proteome size and the number of paralogs were also significantly correlated with one another (r2 = 0.53, P = 5 × 10-3) (Supplementary Figure S2B).
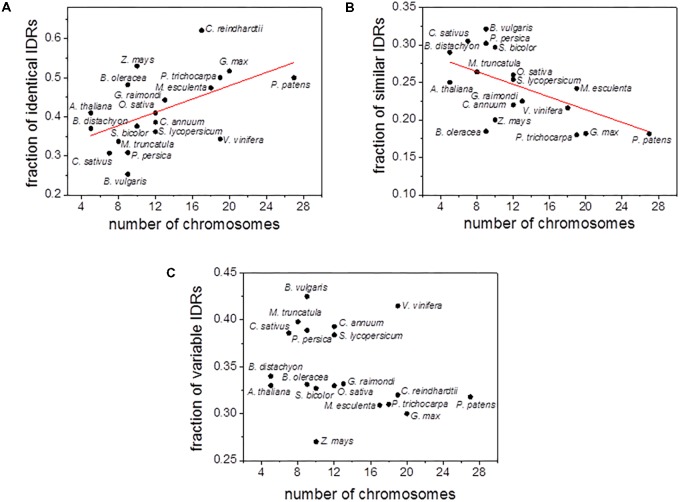
In order to further explore the relationship between polyploidy and IDRs content, we analyzed the correlation between the number of chromosomes and the fraction of the three types of IDRs (i.e., identical, similar and variable IDRs). A statistically positive and significant correlation (r2 = 0.42, P = 5 × 10-3) was observed between the number of chromosomes and the fraction of identical IDRs (Figure 3A and Supplementary Figure S3A). In contrast, a statistically significant negative correlation (r2 = 0.42, P = 5 × 10-3) was observed for the fraction of similar IDRs (Figure 3B and Supplementary Figure S3B). Little or no correlation was observed between the number of chromosomes and the fraction of variable IDRs (Figure 3C and Supplementary Figure S3C).
FIGURE 3.
Scatter plots of chromosome numbers versus the fraction of aligned identical IDRs (A), similar IDRs (B), variable IDRs (C) for C. reindhartii (n = 17), P. patens (n = 27), Z. mays (n = 10), S. bicolor (n = 10), O. sativa (n = 12), B. distachyon (n = 5), G. max (n = 20), P. trichocarpa (n = 19), B. oleracea (n = 9), M. esculenta (n = 18), V. vinifera (n = 19), G. raimondi (n = 13), C. annuun (n = 12), S. lycopersicum (n = 12), A. thaliana (n = 5), P. persica (n = 9), B. vulgaris (n = 9), M. truncatula (n = 8), C. sativus (n = 7). Disordered predictions are based on PONDR VSL2b.
It has been reported that most of the retained duplicated genes in angiosperms are enriched in Gene Ontology (GO) categories associated with protein targeting, synthesis, and post-translational modification (Ren et al., 2018). In order to put in perspective our results and get additional insights, we investigated the GO annotations function of (1) co-linear paralogous proteins in all 19 plant species studied, and (2) the group of co-linear paralogous harboring a majority of identical IDRs. The analysis revealed that on average paralogs are enriched in biological processes (P) (50–60%), molecular functions (F) (20–30%) and cellular components (C) (15–30%) GO categories with corrected p-values < 10E-6. Similar trends were found in the group of paralogs enriched in identical IDRs (p-values < 10E-5). Regarding biological processes, we found that paralogs with identical IDRs are mainly associated with terms such as “catalytic activity,” “metabolic process,” “biosynthetic process,” “development,” “cell differentiation” and “cell proliferation” (p-values < 10E-6). The most significant association among specific molecular functions was with “molecular binding” and “transport” terms (p-values < 10E-6). Regarding cellular components we found that paralogs with identical IDRs are associated with “plasma membrane” and “thylakoid” terms (p-values < 10E-6).
Differences in the distribution of the fraction of IDRs across the co-linear paralogous sequences could be the result of differences in the locations of paralogous genes along chromosomes. This attribution is based on a positive correlation between genetic recombination rates and protein disorder frequency observation, and on the fact that ductile segments are more conserved between paralogs located in regions close to (as opposed to distant from) centromeres (Yruela and Contreras-Moreira, 2013). It is clear from previous analyses and the results presented here that significant evolutionary differences exist in proteomes and in the “dynamics” of IDRs protein sequence sorting during polyploidy events, i.e., our data indicate that polyploidy incurs a disproportionate increase in highly conserved flexibility/ductility compared with less conserved and random disordered/ductile protein regions.
Differences in the distribution of the three types of IDRs have been also observed in a set of transcription factor orthologs involved in key developmental processes such as cellular differentiation, cell division, cell cycle, and cell proliferation (Yruela et al., 2017). Analyses indicated that the fraction of predicted aligned identical IDRs is higher in the green algae (chlorophytes) and non-vascular land plants (bryophytes) compared to vascular plants and animals, whereas the fraction of less conserved IDRs (similar and variable IDRs) is lower in the green algae (chlorophytes) and the non-vascular plants in comparison to vascular plants and animals (Yruela et al., 2017).
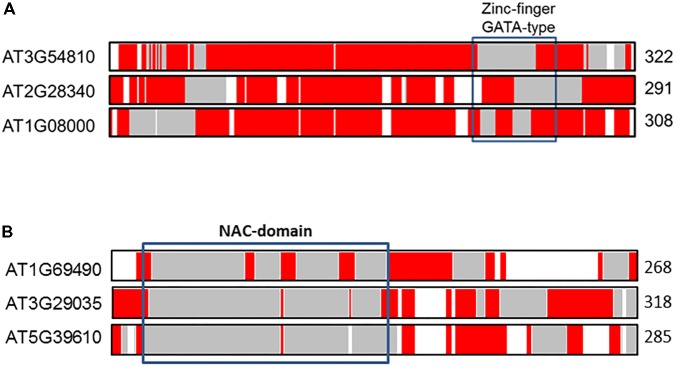
To illustrate differences of IDRs in paralogs compared with orthologs we selected two transcription factors of A. thaliana previously examined by Yruela et al. (2017). Figure 4 shows the distribution of predicted aligned IDRs along the sequences of the GATA10 and NAC92 transcription factor paralogs, which belong to zinc finger and NAC families, respectively. Inspection of Figure 4A shows that co-linear paralogs of GATA10 (AT1G08000), located on chromosome 2 (AT2G28340) and chromosome 3 (AT3G54810), have important differences in the distribution of IDRs, as indicated in the marked zinc-finger GATA-type binding domain. Although the three transcription factors have a high proportion of IDRs (ca. 90%), analysis indicates that between 13 and 30% of the aligned residues correspond to identical IDRs. The percentage of similar and variable IDRs ranges from between 30 and 46%. It has been speculated that the three paralogs are involved in cell differentiation, and that they might be involved in the regulation of some light-responsive genes. We speculate further that variations observed in the distribution of IDRs around the DNA-binding motif might result in different paralog functionalities. Such differences contrast with those observed in GATA orthologs (Yruela et al., 2017). In particular the distribution of IDRs in the zinc-finger GATA-type binding domain is more conserved and manifests a progressive gain of IDRs from green algae to vascular plants, which increases flexibility/ductility in the functional domain.
FIGURE 4.
Alignments of the co-linear GATA10 (A) and NAC92 (B) paralogs in A. thaliana. The protein length is given to the right of each sequence. The color coded bars in each sequence depict predicted disorder. Disordered residues are shown in red, ordered residues are shown in gray, and alignment gaps are shown in white. Typical DNA-binding domains are shown as black boxes.
The alignment of NAC92 (AT5G39610) paralogs on chromosome 3 (AT3G29035 or NAC59) and chromosome 1 (AT1G69490 or NAC29) also reveals notable differences in IDRs distributions. The percentage of total aligned IDRs is ca. 40%, and that of identical IDRs is ca. 10–20%. Such differences likely contributed to functional divergences. NAC92 and NAC59 are involved in senescence, salt stress responses, and lateral root development (Balazadeh et al., 2010), whereas NAC29 is involved in heat stress responses (Zhao et al., 2018).
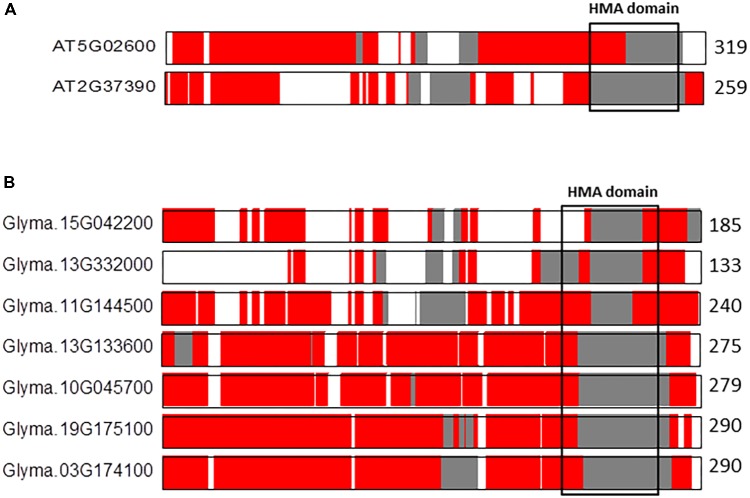
An additional interesting example is the comparison between the co-linear NAKR paralogs in A. thaliana (n = 5) and G. max (n = 20) (Figure 5). The pair of co-linear NAKR1 (AT5G02600) and NAKR2 (AT2G37390) paralogs located on A. thaliana chromosomes 5 and 3, respectively, show notable differences in the IDRs distribution along sequences (Figure 5A). The percentage of total aligned IDRs is ca. 82% in contrast to the percentage of identical IDRs, which is only ca. 24%. The percentage of similar and variable IDRs is 52 and 22%, respectively. The alignment reveals once again differences in the distribution of IDRs, particularly in the functional HMA domain. NAKR1 (Sodium Potassium Root Defective1) is a heavy metal-binding protein expressed in phloem. It interacts with the FLOWERING LOCUS T (FT) transcription factor and regulates flowering through both the transcriptional regulation and transport of FT, especially in response to potassium availability (Negishi et al., 2018). The precise function of NAKR2 is still unclear. In contrast, in G. max the differences in the IDRs composition of the HMA domain across the seven co-linear NAKR1 paralogs are smaller, in particular among four of them, Glyma13G133600, Glyma10G045700, Glyma19G175100, and Glyma03G174100 located on G. max chromosomes 13, 10, 19, and 3, respectively (Figure 5B). The proportion of aligned IDRs on average ranges from 53 to 81%. The fraction of identical IDRs is ca. 87%, and those of similar IDRs and variable IDRs are ca. 1 and 0.6%, respectively. These observations once again support our hyphothesis that polyploidy likely favors increases in highly conserved flexibility/ductility. This fact might have preserved essential functionalities during the course of angiosperm evolution.
FIGURE 5.
Alignments of the co-linear NAKR1 paralogs in A. thaliana (A) and G. max (B). The protein length is given to the right of each sequence. Sequences are represented by color coded bars representing predicted disorder. Disordered residues are shown in red), ordered residues are shown in gray, and alignment gaps are shown in white. Typical DNA-binding domains are shown as black boxes.
Conclusion
In summary, the results reported here indicate: (1) a positive correlation between chromosome number and the fraction of paralogous sequence that are identified as identical IDRs, and (2) a negative correlation between chromosome number and the fraction of paralogous sequences that are identified as similar IDRs. We interpret these findings to indicate (1) retention of paralogs with identical IDRs after WGD (or WTG) could be favored by selection because identical IDRs (as opposed to similar/variable IDRs) facilitated essential functions involved in development, and (2) the retention of genes with high proportions of similar/variable IDRs after WGD (or WTG) could be less likely and therefore tended to be lost in one of paralogs. We argue that the patterns observed for similar/variable IDRs pattern are simply a byproduct of recent WGD (or WTG) events. Thus, ancient WGD (or WGT) events in species such as Z. mays, G. max, and P. trichocarpa have disproportionately favored an increase in aligned identical IDRs across paralogs, thereby contributing to the stability of functions such as the catalytic activity of proteins, metabolic and transport processes, and molecular binding. Based on these characteristics, it is not unreasonable to speculate that, over evolutionary time, duplication events have stabilized proteome adaptive functionalities.
Author Contributions
IY and AKD conceived the study. IY analyzed the data, wrote the original draft of the manuscript, and reviewed and edited the manuscript. BC-M did data analyses, and reviewed and edited the manuscript. AKD and KN contributed to discussion, and wrote, reviewed, and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by Gobierno de Aragón (DGA-GC E35_17R and A08_17R). These grants were partially financed by the EU FEDER Program. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01216/full#supplementary-material
Average fraction (A,B) and box-plot distribution (C,D) of total aligned IDRs (A,B) and aligned residues in ductile regions (L > 30 aa). Identical IDRs (blue), similar IDRs (red), and variable IDRs (green). The data represent the average of paralogs in the proteomes of three monocots (Z. mays, O. sativa, B. distachyon) and two eudicots (P. trichocarpa, A. thaliana). Disordered predictions are based on DisoPred v3.1.
Scatter plots of (A) number of chromosomes versus number of paralogous pairs and (B) proteome size versus number of paralogous pairs in monocots (Z. mays, O. sativa, B. distachyon) and eudicots (P. trichocarpa, A. thaliana). Disordered predictions are based on DisoPred v3.1.
Scatter plots of number of chromosomes versus the fraction of aligned identical IDRs (A), similar IDRs (B), and variable IDRs (C) in monocots Z. mays (n = 10), O. sativa (n = 12), B. distachyon (n = 5), and eudicots P. trichocarpa (n = 19) and A. thaliana (n = 5). Disordered predictions are based on DisoPred v3.1.
Characteristics of the plant species examined with DisoPred v3.1.
References
- Anderson-Sprecher R. (1994). Model comparisons and R2. Am. Stat. 48 113–117. [Google Scholar]
- Babu M. M., van der Lee R., de Groot N. S., Gsponer J. (2011). Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 21 432–440. 10.1016/j.sbi.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Siddiqui H., Allu A. D., Matallana-Ramirez L. P., Caldana C., Mehrnia M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62 250–264. 10.1111/j.1365-313X.2010.04151.x [DOI] [PubMed] [Google Scholar]
- Bellay J., Han S., Michaut M., Kim T., Costanzo M., Andrews B. J., et al. (2011). Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 12:R14. 10.1186/gb-2011-12-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K. H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16 1667–1678. 10.1105/tpc.021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buljan M., Chalancon G., Dunker A. K., Bateman A., Balaji S., Fuxreiter M., et al. (2013). Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr. Opin. Struct. Biol. 23 443–450. 10.1016/j.sbi.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Covarrubias A. A., Cuevas-Velazquez C. L., Romero-Pérez P. S., Rendón-Luna D. F., Chater C. C. C. (2017). Structural disorder in plant proteins: where plasticity meets sessility. Cell Mol. Life Sci. 74 3119–3147. 10.1007/s00018-017-2557-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N., Mason A. (2014). Plant speciation through chromosome instability and ploidy change: cellular mechanisms, molecular factors and evolutionary relevance. Curr. Plant Biol. 1 10–33. 10.1016/j.cpb.2014.09.002 [DOI] [Google Scholar]
- Dehal P., Boore J. L. (2005). Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3:e314. 10.1371/journal.pbio.0030314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J. C., Ramírez-Parra E. (2015). Whole genome duplications in plants: an overview from Arabidopsis. J. Exp. Bot. 66 6991–7003. 10.1093/jxb/erv432 [DOI] [PubMed] [Google Scholar]
- Dubcovsky J., Dvorak J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316 1862–1866. 10.1126/science.1143986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A. K., Bondos S. E., Huang F., Oldfield C. J. (2015). Intrinsically disordered proteins and multicellular organisms. Semin. Cell Dev. Biol. 37 44–55. 10.1016/j.semcdb.2014.09.025 [DOI] [PubMed] [Google Scholar]
- Dunker A. K., Silman I., Uversky V. N., Sussman J. L. (2008). Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18 756–764. 10.1016/j.sbi.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Wright P. E. (2002). Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12 54–60. 10.1016/S0959-440X(02)00289-0 [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Wright P. E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6 197–208. 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- Gaut B. S., Le Thierry d’Ennequin M., Peek A. S., Sawkins M. C. (2000). Maize as a model for the evolution of plant nuclear genomes. Proc. Natl. Acad. Sci. U.S.A. 97 7008–7015. 10.1073/pnas.97.13.7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habchi J., Tompa P., Longhi S., Uversky V. N. (2014). Introducing protein intrinsic disorder. Chem. Rev. 114 6561–6588. 10.1021/cr400514h [DOI] [PubMed] [Google Scholar]
- He B., Wang K., Liu Y., Xue B., Uversky V. N., Dunker A. K. (2009). Predicting intrinsic disorder in proteins: an overview. Cell Res. 19 929–949. 10.1038/cr.2009.87 [DOI] [PubMed] [Google Scholar]
- Hsu W. L., Oldfield C. J., Xue B., Meng J., Huang F., Romero P., et al. (2013). Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 22 258–273. 10.1002/pro.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S., Chen Z. J. (2010). Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 13 153–159. 10.1016/j.pbi.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Cozzetto D. (2015). DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31 857–863. 10.1093/bioinformatics/btu744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Robinson S. J., Nixon J., Xiao R., Huebert T., Condie J., et al. (2014). Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26 2777–2791. 10.1105/tpc.114.126391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E. A., Bennetzen J. L. (2004). The evolution of nuclear genome structure in seed plants 1. Am. J. Bot. 91 1709–1725. 10.3732/ajb.91.10.1709 [DOI] [PubMed] [Google Scholar]
- Kovacs D., Kalmar E., Torok Z., Tompa P. (2008). Chaperone activity of ERD10 and ERD 14, two disordered stress-related plant proteins. Plant Physiol. 147 381–390. 10.1104/pp.108.118208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis J. B., Soltis D. E., Li Z., Marx H. E., Barker M. S., Tank D. C., et al. (2018). Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2018:2. 10.1002/ajb2.1060 [DOI] [PubMed] [Google Scholar]
- Lawton-Rauth A. (2003). Evolutionary dynamics of duplicate genes in plants. Mol. Phylogenet. Evol. 29 396–409. 10.1016/j.ympev.2003.07.004 [DOI] [PubMed] [Google Scholar]
- Leitch A. R., Leitch I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320 481–483. 10.1126/science.1153585 [DOI] [PubMed] [Google Scholar]
- Li Z., Tiley G. P., Galuska S. R., Reardon C. R., Kidder T. I., Rundell R. J., et al. (2018). Multiple large-scale gene and genome duplications during the evolution of hexapods. Proc. Natl. Acad. Sci. U.S.A. 115 4713–4718. 10.1073/pnas.1710791115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Alam M., Ming R., Tang H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar and grape: CoGe with rosids. Plant Physiol. 148 1772–1781. 10.1104/pp.108.124867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Lysak M. A. (2018). Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol. 42 55–65. 10.1016/j.pbi.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Meng F., Uversky V., Kurgan L. (2017). Computational prediction of intrinsic disorder in proteins. Curr. Protoc. Protein Sci. 88 2.16.1–2.16.14. 10.1002/cpps.28 [DOI] [PubMed] [Google Scholar]
- Negishi K., Endo M., Abe M., Araki T. (2018). SODIUM POTASSIUM ROOT DEFECTIVE1 regulates FLOWERING LOCUS T expression via the microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module in response to potassium conditions. Plant Cell Physiol. 59 404–413. 10.1093/pcp/pcx199 [DOI] [PubMed] [Google Scholar]
- Niklas K. J., Bondos S. E., Dunker A. K., Newman S. A. (2015). Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Front. Cell Dev. Biol. 3:8. 10.3389/fcell.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas K. J., Cobb E. D., Dunker A. K. (2014). The number of cell types, information content, and the evolution of complex multicellularity. Acta Soc. Bot. Pol. 83 337–347. 10.5586/asbp.2014.034 [DOI] [Google Scholar]
- Niklas K. J., Dunker A. K. (2016). “Alternative splicing, intrinsically disordered proteins, calmodulin, and the evolution of multicellularity,” in Multicellularity: Origins and Evolution, eds Niklas K. J., Newman S. A. (Cambridge, MA: MIT Press; ), 17–40. [Google Scholar]
- Niklas K. J., Dunker A. K., Yruela I. (2018). The evolutionary origins of cell type diversification and the role of intrinsically disordered proteins. J. Exp. Bot. 69 1437–1446. 10.1093/jxb/erx493 [DOI] [PubMed] [Google Scholar]
- Oates M. E., Romero P., Ishida T., Ghalwash M., Mizianty M. J., Xue B., et al. (2013). D2P2: database of disordered protein predictions. Nucleic Acids Res. 41 D508–D516. 10.1093/nar/gks1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield C. J., Dunker A. K. (2014). Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83 553–584. 10.1146/annurev-biochem-072711-164947 [DOI] [PubMed] [Google Scholar]
- Olsen J. G., Teilum K., Kragelund B. B. (2017). Behaviour of intrinsically disordered proteins in protein-protein complexes with an emphasis on fuzziness. Cell Mol. Life Sci. 74 3175–3183. 10.1007/s00018-017-2560-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea C., Jensen M. K., Stender E. G. P., Kragelund B. B., Willemoës M., Skriver K. (2015). Protein intrinsic disorder in Arabidopsis NAC transcription factors: transcriptional activation by ANAC013 and ANAC046 and their interactions with RCD1. Biochem. J. 465 281–294. 10.1042/BJ20141045 [DOI] [PubMed] [Google Scholar]
- Panchy N., Lehti-Shiu M., Shiu S.-H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171 2294–2316. 10.1104/pp.16.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulou G., Hennig S., Groth D., Krause A., Poustka A. J., Herwig R., et al. (2003). New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res. 13 1056–1066. 10.1101/gr.874803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin I. A., Koh C., Tang H., Robinson S. J., Kagale S., Clarke W. E., et al. (2014). Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 15:R77. 10.1186/gb-2014-15-6-r77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F., Pietrosemoli N., García-Martín J. A., Solano R. (2013). Protein intrinsic disorder in plants. Front. Plant Sci. 4:363. 10.3389/fpls.2013.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Radivojac P., Vucetic S., Dunker A. K., Obradovic Z. (2006). Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 7:208. 10.1186/1471-2105-7-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z.-L., Kurgan L. (2012). Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 13 6–18. 10.2174/138920312799277938 [DOI] [PubMed] [Google Scholar]
- Proost S., Fostier J., Witte D., Dhoedt B., Demeester P., Van de Peer Y., et al. (2012). i-ADHoRe 3.0—fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 40:e11. 10.1093/nar/gkr955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J., Schemske D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33 589–639. 10.1146/annurev.ecolsys.33.010802.150437 [DOI] [Google Scholar]
- Rautureau G. J., Day C. L., Hinds M. G. (2010). Intrinsically disordered proteins in bcl-2 regulated apoptosis. Int. J. Mol. Sci. 11 1808–1824. 10.3390/ijms11041808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R., Wang H., Guo C., Zhang N., Zeng L., Chen Y., et al. (2018). Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol Plant. 11 414–428. 10.1016/j.molp.2018.01.002 [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S., Wendel J. F. (2014). Doubling down on genomes: polyploidy and crop plants. Am. J. Bot. 101 1711–1725. 10.3732/ajb.1400119 [DOI] [PubMed] [Google Scholar]
- Schad E., Tompa P., Hegyi H. (2011). The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 12:R120. 10.1186/gb-2011-12-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Schranz M. E., Mohammadin S., Edger P. P. (2012). Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 15 147–153. 10.1016/j.pbi.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D. G., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skupien-Rabian B., Jankowska U., Swiderska B., Lukasiewicz S., Ryszawy D., Dziedzicka-Wasylewska M., et al. (2016). Proteomic and bioinformatics analysis of a nuclear intrinsically disordered proteome. J. Proteomics 130 76–84. 10.1016/j.jprot.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Soltis P. S., Soltis D. E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60 561–588. 10.1146/annurev.arplant.043008.092039 [DOI] [PubMed] [Google Scholar]
- Song C., Guo J., Sun W., Wang Y. (2012). Whole genome duplication of intra- and inter-chromosomes in the tomato genome. J. Genet. Genomics 39 361–368. 10.1016/j.jgg.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Song Q., Chen Z. J. (2015). Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 24 101–109. 10.1016/j.pbi.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiley G. P., Ané C., Burleigh J. G. (2016). Evaluating and characterizing ancient whole-genome duplications in plants with gene count data. Genome Biol. Evol. 8 1023–1037. 10.1093/gbe/evw058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27 527–533. 10.1016/S0968-0004(02)02169-2 [DOI] [PubMed] [Google Scholar]
- van Bel M., Diels T., Vancaester E., Kreft L., Botzki A., van de Peer Y., et al. (2017). PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46 D1190–D1196. 10.1093/nar/gkx1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R., Buljan M., Lang B., Weatheritt R. J., Daughdrill G. W., Dunker A. K., et al. (2014). Classification of intrinsically disordered regions and proteins. Chem. Rev. 114 6589–6631. 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. (2004). The DISOPRED server for the prediction of protein disorder. Bioinformatics 20 2138–2139. 10.1093/bioinformatics/bth195 [DOI] [PubMed] [Google Scholar]
- Wendel J. F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. 10.1023/A:1006392424384 [DOI] [PubMed] [Google Scholar]
- Wright P. E., Dyson H. J. (2015). Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16 18–29. 10.1038/nrm3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Vucetic S., Iakoucheva L. M., Oldfield C. J., Dunker A. K., Uversky V. N., et al. (2007). Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 6 1882–1898. 10.1021/pr060392u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Dunker A. K., Uversky V. N., Kurgan L. (2016). Molecular recognition features (MoRFs) in three domains of life. Mol. Biosyst. 12 697–710. 10.1039/c5mb00640f [DOI] [PubMed] [Google Scholar]
- Yruela I. (2015). Plant development regulation: overview and perspectives. J. Plant Physiol. 182 62–78. 10.1016/j.jplph.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Yruela I., Contreras-Moreira B. (2012). Protein disorder in plants: a view from the chloroplast. BMC Plant Biol. 12:165. 10.1186/1471-2229-12-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yruela I., Contreras-Moreira B. (2013). Genetic recombination is associated with intrinsic disorder in plant proteomes. BMC Genomics 14:772. 10.1186/1471-2164-14-772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yruela I., Oldfield C. J., Niklas K. J., Dunker A. K. (2017). Evidence for a strong correlation between transcription factor protein disorder and organismic complexity. Genome Biol. Evol. 9 1248–1265. 10.1093/gbe/evx073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Wang Y., Lin W., Li S., Li H., Zhou J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. 10.1371/journal.pbio.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yu W., Hu X., Shi Y., Liu Y., Zhong Y., et al. (2018). Physiological and transcriptomic analysis revealed the involvement of crucial factors in heat stress response of Rhododendron hainanense. Gene S037 30320–30322. 10.1016/j.gene.2018.03.082 [DOI] [PubMed] [Google Scholar]
- Zheng C., Chen E., Albert V. A., Lyons E., Sankoff D. (2013). Ancient eudicot hexaploidy meets ancestral eurosid gene order. BMC Genomics 14(Suppl. 7):S3. 10.1186/1471-2164-14-S7-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhao S., Dunker A. K. (2018). Intrinsically disordered proteins link alternative splicing and post-translational modifications to complex cell signaling and regulation. J. Mol. Biol. 430 2342–2359. 10.1016/j.jmb.2018.03.028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average fraction (A,B) and box-plot distribution (C,D) of total aligned IDRs (A,B) and aligned residues in ductile regions (L > 30 aa). Identical IDRs (blue), similar IDRs (red), and variable IDRs (green). The data represent the average of paralogs in the proteomes of three monocots (Z. mays, O. sativa, B. distachyon) and two eudicots (P. trichocarpa, A. thaliana). Disordered predictions are based on DisoPred v3.1.
Scatter plots of (A) number of chromosomes versus number of paralogous pairs and (B) proteome size versus number of paralogous pairs in monocots (Z. mays, O. sativa, B. distachyon) and eudicots (P. trichocarpa, A. thaliana). Disordered predictions are based on DisoPred v3.1.
Scatter plots of number of chromosomes versus the fraction of aligned identical IDRs (A), similar IDRs (B), and variable IDRs (C) in monocots Z. mays (n = 10), O. sativa (n = 12), B. distachyon (n = 5), and eudicots P. trichocarpa (n = 19) and A. thaliana (n = 5). Disordered predictions are based on DisoPred v3.1.
Characteristics of the plant species examined with DisoPred v3.1.