Abstract
Four-stranded guanine tetraplexes in RNA have been identified to be involved in crucial biological functions, such as dimerization of retroviral RNA, translational repression, and mRNA turnover. However, the structural basis for these biological processes is still largely unknown. Here we report the RNA tetraplex structure (UGGGGU)4 at ultra-high resolution (0.61 Å). The space group is P4212, and cell constants are a = b = 36.16 Å and c = 74.09 Å. The structure was solved by the multiple-wavelength anomalous dispersion method using a set of three-wavelength data of the isomorphous bromo derivative brUGGGGU and refined to 0.61-Å resolution. Each of the four strands in the asymmetric unit forms a parallel tetraplex with symmetry-related molecules. The tetraplex molecules stack on one another in opposite polarity (head-to-head or tail-to-tail) to form a pseudocontinuous column. All of the 5′-end uridines rotate around the backbone of G2, swing out, and form unique octaplexes with the neighboring G tetraplexes, whereas the 3′-end uridines are stacked-in and form uridine tetrads. All of the bases are anti, and the riboses are in the mixed C2′- and C3′-puckering mode. Strontium ions are observed in every other guanine tetrad plane, sitting on the fourfold axis and associated to the eight O6 atoms of neighboring guanine bases in a bipyramidal-antiprism geometry. The hydrogens are clearly observed in the structure.
Keywords: ultra-high resolution|uridine tetrad|octad
For several decades, it has been known that guanine-rich repeats in nucleic acids can form four-stranded tetraplexes in the presence of monovalent ions (1–7) with various biological functions. For example, DNA guanine tetraplexes function in chromosome telomeres (8) and the site-specific recombination of immunoglobulin genes (9). The essential biological functions of RNA tetraplexes have been studied during the past several years. Intramolecular RNA G tetraplex was postulated for a guanine-rich segment adjacent to an endonucleolytic cleavage site in insulin-like growth factor II mRNA (10). Strong evidence has shown that interstrand RNA tetraplexes could be involved in dimerization of HIV 1 genomic RNA, which serves as a critical process in organization of genomic retroviral RNA into infectious virion particles (11). In addition, RNA G tetraplexes have recently been reported to be preferential substrates of bacteriophage fd gene 5 protein (12), which is responsible for the switch from double-stranded to single-stranded viral DNA replication after infection of the bacterial cell. These data indicate that RNA guanine tetraplexes could be crucially involved in translational repression. Furthermore, RNA G tetraplexes have been reported to be specifically targeted by mammalian 5′–3′ exoribonuclease (13), suggesting their potential role in mRNA turnover, which is important in determining the levels and regulation of gene expression. Despite the biological importance of RNA tetraplexes, their structural basis is not clear yet, and it could be much different from that of DNA tetraplexes. The RNA tetraplex structure UGGGGU was reported by NMR in solution in the presence of monovalent ions (14). To further explore the atomic structure of RNA tetraplexes, we have synthesized, purified, and crystallized the RNA hexamer UGGGGU tetraplex in the presence of divalent Sr2+ ions, and here we report its structure at ultra-high resolution (0.61 Å), which is the highest resolution so far for an RNA.
Methods
Synthesis and Crystallization.
The RNA hexamers UGGGGU and brUGGGGU were synthesized by the phosphoramidite method and cleaved from the solid support by using 3:1 (vol/vol) ammonia/ethanol and incubated overnight in the same solution at 55°C. The 2′-hydroxyl groups were deprotected by triethylamine tris(hydrofluoride) and purified by ion-exchange chromatography. Crystallization was carried out by using the hanging-drop vapor diffusion method at room temperature (293 K). The best crystals were obtained with a solution containing 40 mM sodium cacodylate buffer (pH 7.0), 20 mM magnesium chloride, 12.0 mM spermine chloride, 80 mM lithium chloride, 40 mM strontium chloride, 20 mM calcium chloride, 10% (vol/vol) methyl-2,4-pentanediol (MPD), and 35% MPD in the reservoir. Crystals of dimensions 0.2 mm × 0.2 mm × 0.2 mm were obtained in 2 weeks.
Date Collection and Structure Analysis.
Information about the crystal data, the multiple-wavelength anomalous dispersion (MAD) phasing, and the structure refinement is summarized in Table 1. A set of data was collected from a UGGGGU crystal at beamline 14BM-D at the Advanced Photon Source with a wavelength of 0.8 Å. A total of 111,103 independent reflections, corresponding to 94.7% of the theoretically possible data, were collected with an Rmerge of 6.8%. The completeness of the data for the last resolution bin, 0.62–0.61 Å, was 72.7%. The data were processed by using denzo and scalepack (15). The crystals belonged to the tetragonal space group P4212 with the cell constants of a = b = 36.13 Å and c = 74.15 Å. The asymmetric unit contains four strands of RNA (Fig. 1) with a tight volume per base pair of 1,008 Å3. The structure was solved by MAD phasing using the isomorphous crystal of the bromo derivative brUGGGGU. Three-wavelength MAD data were collected to 1.7 Å (Table 1); only one brUGGGGU crystal was used. The intensity data were integrated and scaled with denzo and scalepack. Four bromine atom sites in the asymmetric unit were solved by using the snb program (16). The anomalous data were collected at the bromine peak and the remote sites gave the same result. The phase refinement was carried out by using mlphare/ccp4 (17). Density modifications including noncrystallographic symmetry (ncs) averaging were performed with dm/ccp4 (17). The electron density map obtained could be clearly traced, with the exception of one disordered uridine residue (U18). The native data of UGGGGU were used in the refinement. After simulated annealing, iterative positional and B-refinement were performed by using cns (18) to 1.0-Å resolution. Model building was carried out with graphic package o (http://origo.imsb.au.dk/∼mok/o/). The R value and free R were 12.2% and 13.4%, respectively. shlex-97 [G. M. Sheldrick (1997) Institut für Anorganische Chemie, Gottingen, Germany] was then used for the subsequent refinement with all of the data. Aniostropic B refinement was performed for all of the atoms by using the data from 20- to 0.61-Å resolution. Hydrogen atoms were added, and their positions were refined in the final round of refinement. The final Rwork/Rfree was 10.32%/11.17%. The model in the asymmetric unit contains 500 non-hydrogen atoms and 253 hydrogen atoms, two Sr2+ ions, and one-half Na+ ion, three Ca2+ ions, one spermine molecule, and 110 water molecules. The atomic coordinates and the structure factors have been deposited in the Protein Data Bank, PDB code (19).
Table 1.
Crystal Data and MAD phasing statistics
| UGGGGU | brUGGGGU | |||
|---|---|---|---|---|
| Crystal data | ||||
| Wavelength, Å | 0.800 | 0.9195 | 0.9192 | 0.8984 |
| Space group | P4212 | P4212 | ||
| Unit cell dimensions, Å | a = b = 36.154, c = 74.133 | a = b = 36.13, c = 74.12 | ||
| Resolution, Å | 0.61 | 1.7 | 1.7 | 1.5 |
| Unique reflections | 207,845* | 10,427* | 10,424* | 14,969* |
| Completeness, % | 92.9 (61.5†)* | 100 | 100 | 100 |
| I/σ | 26 (2.0†)* | |||
| Rsym, % | 6.6 (33.9)* | 11.3 | 11.4 | 11.1 |
| Rwork/Rfree, % | 10.32/11.17 | |||
| Phasing statistics at 1.7-Å resolution | ||||
| Phasing power | Isomorphous centric | — | 0.81 | 1.68 |
| Isomorphous acentric | — | 1.29 | 2.16 | |
| Figure of merit | Before density modifications | 0.77 | ||
| After density modifications | 0.82 | |||
Friedel pairs unmerged.
Last reflection bin, 0.62–0.61 Å.
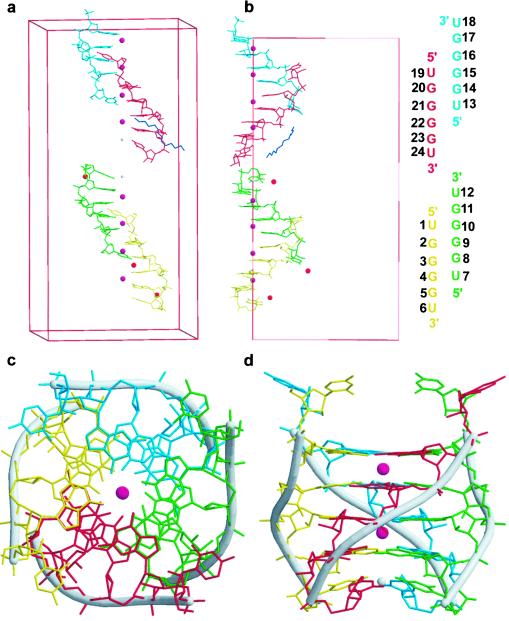
Figure 1.
The RNA tetraplex molecule. (a) The four independent RNA strands in the asymmetric unit represented in four different colors: strand 1, yellow; strand 2 green; strand 3 cyan; strand 4 red; Sr2+ ions are in purple and located on the fourfold symmetry axis; Na+ ions are in silver and are also on the fourfold axis; three Ca2+ ions are in general positions and shown in orange; and there is one spermine molecule in general position, which is shown in blue. Sr2+ ions are associated with every other guanine base plane. (b) Another view along crystal b axis. The numbering scheme is shown to the right. (c) Top view of one tetraplex molecule. The Sr2+ ions are sitting on the fourfold symmetry axis, which passes through the helix. The single strand forms a tetraplex with symmetry-related strands. (d) Side view. Notice the 5′-side uridines swing out of the column and the 3′-side uridines are stacked-in and form a U tetrad with a highly tilted angle (≈30°).
Results and Discussion
Crystallography and Conformation.
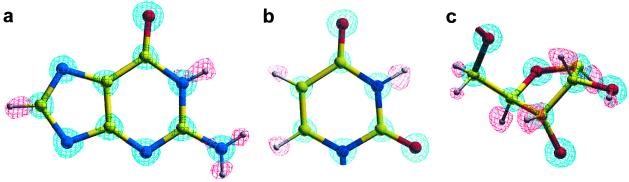
The RNA hexamer UGGGGU was solved by the MAD method using three-wavelength data collected from the isomorphous bromo derivative brUGGGGU. The model obtained from this procedure was used in refinement with high-resolution native data (0.61 Å). The average twist angle of the RNA tetraplex is 28.7°, corresponding to 12.5 residues per turn. There are four independent RNA strands in the asymmetric unit, and each strand forms a right-handed parallel RNA tetraplex with its symmetry-related molecules (Fig. 1). The tetraplex molecules stack in opposite polarity (tetraplex 1 and 2, as well as 3 and 4 are head-to-head; whereas 2 and 4 are tail-to-tail, Fig. 2). Most of the hydrogen atoms on the bases and sugars can be clearly discerned from the Fourier difference electron density map at ≈1.0 Å from the C, N, and O atoms (Fig. 3).
Figure 2.
Stereoview of the four tetraplexes: Tetraplex 1 (yellow), tetraplex 2 (green), tetraplex 3 (cyan), and tetraplex 4 (red). Each strand sits around the fourfold symmetry axis to form tetraplexes with symmetry-related molecules. The tetraplex molecules stack coaxially in opposite polarity. There are eight Sr2+ ions and two Na+ ions on the fourfold axis. Notice the 5′-end Us are unstacked with the Gs and protrude into neighboring G tetraplexes, forming octads as indicated by the arrows.
Figure 3.
Hydrogen atoms. 2Fo − Fc maps (2σ level, blue) and Fo − Fc maps (2.5σ level, red) are shown for guanine (a), uracil (b), and base and sugar (c) moieties.
The conformations of the RNA strands 1, 2, and 3 are similar, with an average rms deviation of 0.20 Å. The first five residues of strand 4 are close to those in strands 1, 2, and 3, but the 3′-end uridine adopts significantly different conformation with an average rms deviation of 1.9 Å. The overall RNA helix demonstrates a mixed sugar puckering: the second guanosines (G3, G9, G15, and G21) and the 3′-end uridines (U6, U12, and U24, with the exception of U18, which is disordered) are in C2′-endo, whereas the rest are in C3′-endo conformations. All of the nucleosides are in the anti glycosidic conformations.
RNA Tetraplex with Sr2+ Ions and Other Metal Ions.
The tetraplex molecules bind eight Sr2+ ions, two Na+ ions, three Ca2+ ions, and one spermine molecule (multiply charged organic cation) (Fig. 1 a and b). The Sr2+ ions (two Sr2+ ions per tetraplex, Fig. 1) and the Na+ ions sit on the fourfold symmetry axis, which passes through the center of the tetraplex helices. It is unique that the RNA tetraplexes are associated with divalent strontium ions between every other guanine tetrad plane. Each of them are coordinated to the eight neighboring guanine O6 atoms at a distance of 2.61 Å, forming a bipyramidal-distorted-antiprism geometry (Fig. 4), which mimics those of monovalent ions in DNA tetraplexes. Two disordered Na+ ions are found, and each is located in the center of the 3′-end uridine tetrads and coordinated to four O4 atoms. At the outside of the tetraplex column, three Ca2+ ions are clearly identified at both the 5′ and 3′ ends of the tetraplex 1 and 2 in the crystal lattice (Figs. 1 and 4), and they are coordinated to U1, U6, and U12 residues, respectively. In addition to the metal ions, one spermine molecule is observed in the structure (Fig. 1), located in between symmetry-related tetraplexes (tetraplexes 2 and 3, Figs. 2 and 4). The spermine N5 atom forms bifurcated hydrogen bonds with O2′ (G2, in a symmetry-related molecule) and O1P (G22), and the N10 atom forms a hydrogen bond with O1P (G9, in a symmetry-related molecule). The spermine carbon atoms C4, C6, and C11 are linked to O1P (G22), O2′, and O1P atoms of a symmetry-related guanine residue G8, respectively, by means of C-H—O interactions (average C—O distance 3.1 Å).
Figure 4.
The coordination geometries of the metal ions and the spermine molecule. (a) The Sr2+ ion (purple) is coordinated to eight guanine O6 atoms. (b) Three Ca2+ ions are coordinated to U6 (Ca1), U7 (Ca2), and U12 (Ca3) residues, respectively. (c) Coordination of the spermine molecule (blue) in stereoview. Hydrogen bonds are shown in white dashed lines. Individual RNA strands are depicted in different colors.
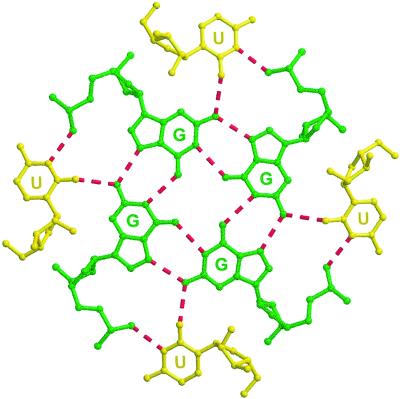
Unusual Octads (with Four Us and Four Gs) and Uridine Tetrads.
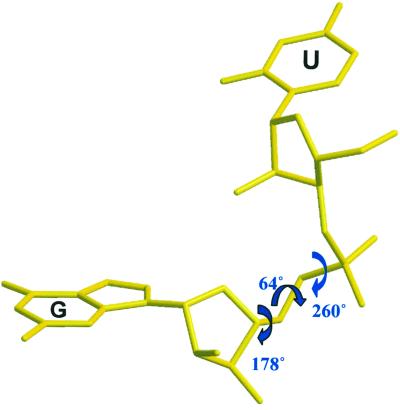
The first guanines (G2, G8, G14, and G19) of the four strands in the asymmetric unit display uncommon backbone conformations with the torsion angles P–O5′ (α), O5′–C5′ (β), and C5′–C4′ (γ) in gauche− (260°), gauche+ (64°), and trans (178°), respectively (Fig. 5). The 5′-end uridines are not stacked; instead, they rotate around the backbone of the 5′-terminal Gs and protrude into the grooves of the neighboring G tetraplexes (Fig. 2), and form hydrogen bonds to the second G bases and phosphate groups: O2(U1)—N2(G9) = 2.8 Å; N3H(U1)—O1P(G9) = 2.77 Å (Fig. 6). By this “embracing pattern,” two octads sandwiching two guanine tetrads are formed. The 5′-uridine bases are slightly tilted relative to the planes of the G tetrads. The 3′-end uridines (U6, U12, and U24) are stacked and form uridine tetrads. The phosphate groups of U6 and U12 have different conformations [O3′–P and P–O5′ torsions are gauche+ (52°) and trans (159°), respectively] from the normal geometry adopted by U24. Bases U6 and U12 are flipped over, compared to U24, and stack with the opposite face on the guanine tetrad. Thus U24 tetrads have a clockwise hydrogen bonding pattern (N3-H—O4, Fig. 7f), compared to the counterclockwise hydrogen bonding for U6 and U12 tetrads (O4—H-N3, Fig. 7e).
Figure 5.
The geometry of the 5′-end uridine. The torsion angles P–O5′ (α), O5′–C5′ (β), and C5′–C4′ (γ) are in gauche− (260°), gauche+ (64°), and trans (178°), respectively. As a result, the 5′-end uridines flip out of the tetraplex columns.
Figure 6.
The geometry of the octad. The G tetrad is from tetraplex 2, and the uridine bases (yellow) are from tetraplex 1. The O2 atom of uridine is hydrogen bonded to the amino group (N2) of guanine, and the N3H atom of uridine is hydrogen bonded to the oxygen atom of the guanosine 5′-phosphate group.
Figure 7.
Hydrogen bonding in the G and U tetrads. The 2Fo − Fc maps are calculated at 2σ level and shown in blue. Top views are shown in the upper row and side views in the lower row. (a and b) G tetrads. The guanines are hydrogen bonded to each other by clockwise Hoogsteen-like pairing (N1H → O4) with near ideal planarity. (c and d) The U6/U12 tetrad. It is formed with a single hydrogen bond between N3 and O4 atoms in a counterclockwise pattern. (e and f) The U24 tetrad is flipped over in a clockwise N3-H—O4 hydrogen-bonding pattern.
Base Pairing and Base Stacking Geometry.
The fourfold symmetry-related guanosine tetrads are associated with Hoogsteen-like pairing. The N1H is hydrogen bonded to the O6 atom and the N2H is hydrogen bonded to the N7 atom with an average distance of 2.85 Å and 2.80 Å, respectively (Fig. 7 a and b). The 3′-end uridines form U tetrads by means of a single hydrogen bond from the N3H to the O4 atom of a symmetry-related base with an average distance of 2.78 Å (Fig. 7 c and d). The average intermolecular distance between the C5 and O2 atoms of uridines is 3.54 Å; thus, there are no C5-H—O2 hydrogen bonds formed in the U tetrads. All four G tetraplexes in the asymmetric unit show the same base pair twist. The first two (G2 and G3), as well as the last two guanine bases (G4 and G5), have apparently low base pair twist angles (26° and 23°, respectively), whereas the middle two (G3 and G4) have relatively high twist angles (37°), which might be caused by the occurrence of the C2′-endo conformation at the second guanines. The base pair stacking within the tetraplex is similar to that in the parallel DNA G tetraplex, TGGGGT (20). The five-membered rings of the guanines overlap with the six-membered rings of the 5′-flanking guanine bases in both RNA and DNA tetraplexes. However, the guanine bases in our structure display a different stacking pattern at the 5′-end junction. The six-membered rings of the guanines stack extensively over the six-membered rings of the flanking guanines and the amino groups stack with the flanking five-membered rings. This stacking pattern is rather different from that in the DNA tetraplex, where the five-membered rings overlap with each other. The two different conformations of the uridines on the 3′-end (normal and flipped-over) result in two different types of base stacking patterns. The twist angle between U24 and G23 is very low (17°), compared to an average high value of 48° between G5 and U6, and between G11 and U12. Thus, U24 stacks well with G23, whereas U6 and U12 are weakly stacked.
Comparisons with the RNA NMR Model and the X-Ray Structure of the DNA Tetraplex.
Structures of guanine-rich tetraplexes have been determined by x-ray crystallography for DNA (20, 21) and by NMR for RNA in solution (14). Compared to these models, the G tetraplex region of the present structure has rms deviations of 0.97 Å to the DNA and 1.02 Å to the RNA (NMR). The detailed geometry differs significantly from either model. The present structure differs from the RNA NMR model in both the bases and sugar backbone conformations. Although the NMR model also predicted a mixed C3′-endo/C2′-endo sugar puckering, the C2′-endo sugars occur at different places (the second guanines in our x-ray structure vs. the first guanines in the NMR model). In addition, the guanine tetrad stacking distances in the present structure deviate from those in the NMR model by as much as 1 Å. In the case of the terminal uridines, 3′-end uridine tetrads were observed in the present structure as proposed in the NMR model (14). However, the geometries and orientations of the U tetrads differ significantly. Compared to the NMR model, the U24 tetrad has an rms deviation of 2.1 Å, whereas the value increases to 4.4 Å for the U6 and U12 tetrads with the “flipped-over” conformations. Compared to the DNA model, the base region of the current structure has very similar geometry, with major differences occurring at the backbone region. The present structure has sugar moieties in mixed C2′-endo/C3′-endo puckering, in contrast to the uniform C2′-endo puckering for the DNA tetraplex. The divalent Sr2+ ions are placed in between every other guanine plane in this structure, compared to the Na+/K+ ions in between each guanine plane in the DNA tetraplex. In our structure, each guanine O6 atom is coordinated to a single Sr2+ ion. In contrast, each guanine O6 atom in the DNA structure is shared by two Na+ ions above and below the G tetrad plane, except the terminal O6 atoms.
Acknowledgments
We thank Dr. H. Hauptman and Dr. C. Weeks for their initial assistance in attempting to perform the ab initio structure determination at 0.74 Å resolution by the direct method. We acknowledge the support of this work by National Institutes of Health Grant GM-17378 and the Board of Regents of Ohio for an Ohio Eminent Scholar Chair and Endowment to M.S. We also acknowledge the Hays Consortium Investment Fund by the Regents of Ohio for partial support for purchasing the R-axis IIc imaging plate. We thank Advanced Photon Source and Argonne National Laboratory for supporting our work on synchrotron data collection.
Abbreviation
- MAD
multiple-wavelength anomalous dispersion
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and the structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1J8G).
References
- 1.Gellert M, Lipsett M, Davies D. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman S B, Cohen G H, Davies D R. J Mol Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman S B. J Mol Biol. 1976;106:663–672. doi: 10.1016/0022-2836(76)90257-6. [DOI] [PubMed] [Google Scholar]
- 4.Williamson J R, Raghuraman M K, Cech T R. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 5.Sundquist W I, Klug A. Nature (London) 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Cheong C, Moore P B. Nature (London) 1991;351:331–332. doi: 10.1038/351331a0. [DOI] [PubMed] [Google Scholar]
- 7.Patel D, Bouaziz S, Kettani A, Wong Y. In: Nucleic Acid Structure. Neidle S, editor. New York: Oxford Science; 1999. pp. 389–453. [Google Scholar]
- 8.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 9.Sen D, Gilbert W. Nature (London) 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen J, Kofod M, Nielsen F C. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundquist W I, Heaphy S. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver A W, Kneale G G. Biochem J. 1999;339:525–531. [PMC free article] [PubMed] [Google Scholar]
- 13.Bashkirov V I, Scherthan H, Solinger J A, Buerstedde W D. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong C, Moore P B. Biochemistry. 1992;31:8406–8414. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Weeks C M, Miller R. J Appl Crystallogr. 1999;32:120–124. [Google Scholar]
- 17.Otwinowski Z. In: Isomorphous Replacement and Anomalous Scattering. Wolf W, Evans P R, Leslie A G W, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1991. pp. 80–86. [Google Scholar]
- 18.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Berman H M, Olson W K, Beveridge D L, Westbrook J, Gelbin A, Demeny T, Hsiesh S H, Srinivasan A R, Schneider B. Biophys J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laughlan G, Murchie A I H, Norman D G, Moore M H, Moody P C, Lilley D M J, Luisi B. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 21.Phillips K, Dauter Z, Murchie A I H, Lilley D M J, Luisi B. J Mol Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]