Abstract
Apolipoprotein E (ApoE) is a secreted apolipoprotein with three isoforms, E2, E3, and E4, that binds to lipids and facilitates their transport in the extracellular environment of the brain and the periphery. The E4 allele is a major genetic risk factor for the sporadic form of Alzheimer's disease (AD), and studies of human brain and mouse models have revealed that E4 significantly exacerbates the deposition of amyloid beta (Aβ). It has been suggested that this deposition could be attributed to the formation of soluble ApoE isoform–specific ApoE–Aβ complexes. However, previous studies have reported conflicting results regarding the directionality and strength of those interactions. In this study, using a series of flow cytometry assays that maintain the physiological integrity of ApoE–Aβ complexes, we systematically assessed the association of Aβ with ApoE2, E3, or E4. We used ApoE secreted from HEK cells or astrocytes overexpressing ApoE fused with a GFP tag. As a source of soluble Aβ peptide, we used synthetic Aβ40 or Aβ42 or physiological Aβ secreted from CHO cell lines overexpressing WT or V717F variant amyloid precursor protein (APP). We observed significant interactions between the different ApoE isoforms and Aβ, with E4 interacting with Aβ more strongly than the E2 and E3 isoforms. We also found subtle differences depending on the Aβ type and the ApoE-producing cell type. In conclusion, these results indicate that the strength of the ApoE–Aβ association depends on the source of Aβ or ApoE.
Keywords: Alzheimer disease, apolipoprotein E (ApoE), amyloid beta (AB), flow cytometry, astrocyte
Introduction
Apolipoprotein E (ApoE)4 is an apolipoprotein that is mainly secreted from astrocytes within the brain and liver cells in the periphery. In humans, ApoE exists in three isoforms: E2, E3, and E4. It has been found that E4 increases the risk for development of Alzheimer's disease (AD), whereas E2 has a protective role and E3 a neutral effect (1). Conversely, E2 has been associated with an increased risk for hyperlipidemia (2). Although the genetic data suggesting an association between ApoE genotype and risk for development of AD is strong, the mechanism underlying this association is unknown. It has been suggested that structural differences caused by the Cys–Arg interchanges at positions 112 and 158 affecting the conformation and lipidation of ApoE could be responsible for the differential effect of the isoforms on disease risk (3, 4).
There is a lot of evidence suggesting that ApoE genotype influences the aggregation and deposition of β-amyloid (Aβ) in the form of plaques. It has been shown that AD patients with an E4 allele have a higher burden of Aβ plaques in the brain compared with carriers of an E3 or E2 allele (5–7). A similar effect has also been observed in transgenic mouse models (8). In addition, in vivo studies have shown that gene therapy leading to overexpression of E2 within the brains of APP/PS1 transgenic mice leads to a reduction of Aβ plaque size (9). Plaque formation is also greatly reduced in APP transgenic mice with ApoE knockout (10). A similar effect has also been seen when treating mice with an anti-ApoE antibody in vivo (11, 12).
To decipher the mechanism underlying those observations, the formation of complexes between ApoE and soluble Aβ has been studied extensively. However, the results have been controversial (13, 14). Several studies have found that Aβ binds to cell-secreted cerebrospinal fluid- or plasma-derived ApoE (15–17). However, other studies have concluded that ApoE–Aβ interactions are minimal and thus do not have an important role in the pathogenesis of AD (18). These discrepancies have been attributed to the different methods used to isolate ApoE–Aβ complexes in each study (13, 14). In addition, some studies have suggested that the strength of Aβ–ApoE association is isoform-specific, although they do not agree on the directionality of the association (17, 19–23); other studies have not found a difference between isoforms (24–26). Similarly, the study of ApoE-induced oligomerization of Aβ has given inconsistent results, as some studies have indicated an increase in oligomerization (27) and others a decrease (28, 29) in the presence of ApoE. These controversies have been previously reviewed in detail (13, 14).
Given the uncertainty surrounding the ApoE–Aβ interactions, we decided to study this topic systematically using a novel flow cytometry–based assay that maintains the physiological integrity of ApoE–Aβ complexes. We aimed to determine whether ApoE associated with soluble Aβ and whether there were isoform-specific differences in ApoE–Aβ complex formation. As a secondary aim, we sought to assess whether the cell type of origin of ApoE and the type of Aβ influenced the ApoE–Aβ interactions. We used soluble Aβ from several sources and of several types: synthetic Aβ42 and Aβ40, physiological Aβ secreted from CHO cells expressing WT Aβ precursor protein (APP), physiological Aβ secreted from CHO cells expressing V717F mutant APP exhibiting an increased Aβ42/Aβ40 ratio, and Aβ secreted from primary neurons derived from Tg2576 mice carrying the Swedish mutation in APP. As a source of ApoE, we used recombinant ApoE and physiological HEK cell- and astrocyte-secreted ApoE. The results from those experiments showed that, in general, ApoE associated with Aβ and followed a directional pattern of E2 < E3 < E4. E4 was the isoform that associated the strongest with Aβ, E2 associated less strongly, and E3 associated at intermediate levels. Differences were seen between HEK- and astrocyte-secreted ApoE and between the various types of Aβ studied.
Results
Secreted ApoE interacts with synthetic Aβ42 and Aβ40
As a first step, we assessed whether synthetic Aβ can interact with ApoE and whether there are any isoform-specific differences in the interaction. Previous studies have shown that synthetic Aβ can form complexes with ApoE from a variety of sources, such as cell-secreted in tissue culture (15) and human ApoE isolated from cerebrospinal fluid (16) or plasma (17). We used synthetic Aβ42 and Aβ40 to see whether there was a difference in the interaction with ApoE between the two forms. As a source of ApoE, we used physiological HEK- and astrocyte-secreted ApoE fused to a GFP tag. Study of peripheral ApoE is important, as it has been suggested that it can cross the blood–brain barrier (30, 31).
Conditioned medium (CM) from HEK cells or astrocytes transfected with ApoE was attached overnight to Dynabeads. The following day, either synthetic Aβ42 or synthetic Aβ40 fused to the 647Hilyte fluorophore was added to each sample and incubated for 5.5 h before flow cytometry (Fig. 1A). To determine whether Aβ associates preferentially with ApoE, the mean fluorescence intensity (MFI) of 647Hilyte-Aβ was compared between ApoE+ and ApoE− populations within each sample. Indeed, the MFIs for 647Hilyte-Aβ were higher in ApoE+ populations compared with ApoE− populations for all ApoE isoforms secreted from HEK cells and astrocytes (Fig. S1, A–D).
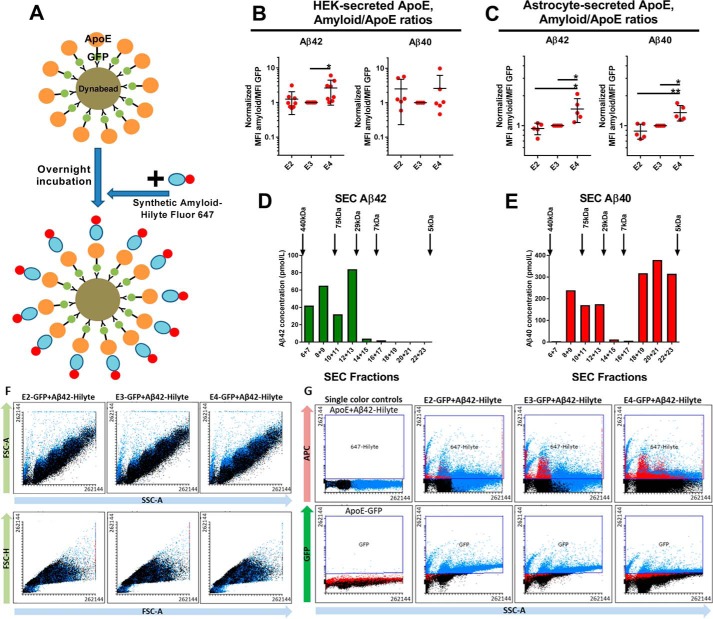
Figure 1.
Assessment of the interaction between ApoE and synthetic Aβ using flow cytometry. A, diagram illustrating the process of the experiment. First, ApoE is attached to Dynabeads. After overnight incubation, synthetic Aβ fused with 647Hilyte is added to the sample, and they are analyzed through flow cytometry 4 h later. B, plot indicating the Aβ/ApoE ratios for HEK-secreted ApoE. E4 interacts more strongly with Aβ42 compared with E2 and E3. However, this pattern is not present in the case of Aβ40. Number of independent experiments: Aβ42, 8; Aβ40, 6. C, plot indicating the Aβ/ApoE ratios for astrocyte-secreted ApoE. E4 interacts more strongly with Aβ42 and Aβ40 compared with E2 and E3. Number of independent experiments: Aβ42, 5; Aβ40, 5. D, SEC for synthetic Aβ42 shows that it forms medium- to high-molecular-mass complexes ranging from ∼7–440 kDa. E, SEC for synthetic Aβ40 shows primarily low-molecular-mass complexes from 5–75 kDa, with little formation of medium-sized complexes. F, forward scatter (FSC) and side scatter (SSC) plots for E2, E3, and E4, indicating that there are no differences between isoforms. G, fluorescence plots for E2, E3, E4, and single-color controls. In these plots, each dot represents the mean value from one independent experiment. The data were normalized to E3 before meta-analysis. Statistical analysis was done with one-way ANOVA with Tukey's post hoc correction. Mean ± S.D. is shown in the plots. **, p ≤ 0.01; *, p ≤ 0.05.
Next, we assessed whether there are isoform-specific differences in the interaction between ApoE and Aβ. To study this aspect of ApoE biology, we determined the ratio of 647Hilyte-Aβ MFI to ApoE-GFP MFI for each ApoE isoform to approximate the number of Aβ molecules attached to each ApoE particle. For HEK-secreted ApoE, we found that E4 interacted more strongly than E2 and E3 with Aβ42, but no statistically significant differences between isoforms were seen in terms of interaction with Aβ40 (Fig. 1B). For astrocyte-secreted ApoE, E4 interacted more strongly than E2 and E3 with both Aβ42 and Aβ40 (Fig. 1C).
Size exclusion chromatography (SEC)–based characterization of synthetic Aβ complexes formed at the concentration used in our experiments showed that Aβ42 forms complexes of higher molecular weight compared with Aβ40 (Fig. 1, D and E). Representative scatterplots and fluorescence plots derived from a flow cytometry experiment are shown in Fig. 1, F and G.
Physiological Aβ produced from cell lines overexpressing WT or mutant APP interacts with HEK-secreted ApoE in vitro
Even though it has been consistently shown that ApoE can interact with synthetic Aβ, the results have been controversial regarding the interactions of ApoE with physiological Aβ; some studies have shown that a significant interaction occurs (15–17), whereas others suggest that the interaction is minimal (18). To study the interactions between ApoE and physiological Aβ, we used CM from two CHO cell lines, 7w and 7PA2, overexpressing APP. The use of these cell lines allowed us to study Aβ with two different ratios of Aβ42/Aβ40. The 7w CHO cell line overexpressed WT APP (32), whereas the 7PA2 CHO cell line overexpressed the V717F mutant APP (33, 34). It has been shown that, independent of APP mutation status, Aβ is primarily of the Aβ40 species, although there is a relative increase of the Aβ42 species in the presence of APP mutations (35–37). We initially characterized the Aβ species secreted by the 7w and 7PA2 cell lines by size exclusion chromatography. We found that, in both cell lines, Aβ was present mainly in low-molecular-mass complexes, although the distribution ranged from 7–75 kDa (Fig. 2, A and B). This is consistent with what is seen in Aβ secreted from primary neurons from Tg2576 mice (38, 39). Aβ40 was the main species secreted by both cell lines; however, the Aβ42/Aβ40 ratio was higher in 7PA2 cells (10 versus 6.5), indicating that Aβ42 levels were relatively higher in 7PA2 cells compared with 7w cells (Fig. 2C), consistent with previous studies (35).
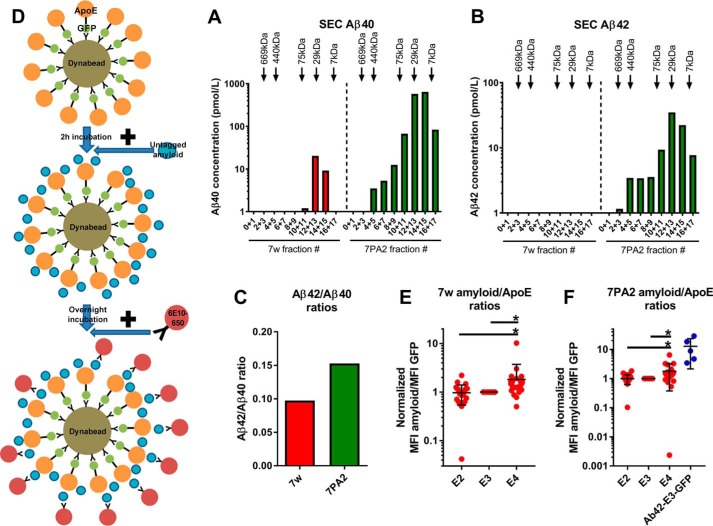
Figure 2.
Interactions between HEK-secreted ApoE and physiological Aβ as determined by flow cytometry. A–C, characterization of Aβ secreted from the 7w and 7PA2 cell lines. Results from one representative experiment are shown in the plots. A and B, Aβ40 (A) and Aβ42 (B) concentrations in 15-fold concentrated CM from 7w and 7PA2 cell lines after SEC. The Aβ secreted from both cell lines forms mainly low to intermediate-molecular-mass complexes, but the size of those complexes ranges between 7–75 kDa. C, ratios of Aβ42/Aβ40 concentrations in the CM of 7w and 7PA2 cell lines. The ratio is higher for the 7PA2 than for the 7w line, suggesting that 7PA2 cells secrete proportionately more Aβ42. D, diagram illustrating the process of the experiment. First, ApoE is attached to Dynabeads. 2 h later, CM from the 7w or 7PA2 cell line is added to the ApoE–Dynabead complex. After overnight incubation, the 6E10 anti-Aβ antibody that was first conjugated to 650Dylight is added to the samples and incubated for 5.5 h before flow cytometry. E, plot indicating the Aβ/ApoE ratios for HEK-secreted ApoE incubated with 7w CM. E4 interacts more strongly than E2 and E3 with 7w Aβ. Number of independent experiments: E2, 24; E3, 24; E4, 23. F, plot indicating the Aβ/ApoE ratios for HEK-secreted ApoE incubated with 7PA2 CM. E4 interacts more strongly than E2 and E3 with 7PA2 Aβ. Number of independent experiments: E2, 20; E3, 20; E4, 20; Aβ42-E3-GFP, 5. In these plots, each dot represents the mean value from one independent experiment. The data were normalized to E3 before meta-analysis. Statistical analysis was done with one-way ANOVA with Tukey's post hoc correction. Mean ± S.D. is shown in the plots *, p ≤ 0.05.
In this assay, secreted ApoE was attached to Dynabeads for 2 h, followed by overnight incubation with either 7w or 7PA2 CM. The following day, the anti-Aβ 6E10 antibody (recognizing the 1–16 N-terminal amino acids of Aβ) conjugated to the fluorescent dye 650Dylight was added to each sample and incubated for 5.5 h before flow cytometry (Fig. 2D). CM from cells transfected with a plasmid encoding a fusion protein of Aβ42-E3-GFP was used as a positive control. To determine whether physiological Aβ can interact with ApoE, we compared the 650Dylight MFI between GFP+ (i.e. ApoE+) and GFP− (i.e. ApoE−) populations within the same sample. The GFP− beads were used as a negative control because they indicate the background binding of Aβ and 6E10-650 in the absence of ApoE. For all isoforms, ApoE+ beads had a statistically significantly higher 650Dylight MFI compared with ApoE− beads for both 7w and 7PA2 Aβ (Fig. S2, E and F).
We then calculated the ratios of Aβ-6E10-650Dylight MFI to GFP MFI. E4 interacted significantly more than E2 and E3 with 7w and 7PA2 Aβ. There was no difference between E2 and E3 in terms of interaction with Aβ. The Aβ/GFP ratio was higher in the positive control Aβ42-E3-GFP compared with E4, E3, and E2 (Fig. 2, E and F).
The ApoE–Aβ flow cytometry interaction assay robustly and specifically detects ApoE–Aβ interactions
To confirm the results of our experiments on HEK-secreted ApoE, we undertook a series of control experiments. First, it was possible in our assay that the 6E10 antibody disrupted the formation of the ApoE–Aβ complexes because it targets the N terminus of Aβ (amino acids 1–16), which is thought to be the ApoE-binding region. To exclude this possibility, we used the 4G8 antibody to detect Aβ because it targets amino acids 17–24 (Fig. S2A). Second, it is possible that using an antibody against Aβ can disrupt its conformation and, thus, its interaction capabilities. To test this hypothesis, we transfected HEK cells with a plasmid encoding luciferase-Aβ42 and detected Aβ using an anti-luciferase antibody (i.e. the antibody was directed against the tag and not Aβ itself) (Fig. S2B). Third, there is also APP present in the 7w and 7PA2 CM, and this could be inadvertently detected by the N-terminal-specific 6E10 antibody. Thus, to rule out the possibility that the results of our assay were modified by the presence of APP in the samples, we transfected HEK cells with a plasmid encoding the complementary DNA of Aβ42, incubated this CM with ApoE-GFP attached to Dynabeads, and detected the Aβ using the 6E10-650 antibody (Fig. 2C). Fourth, we also tested the interaction between ApoE and Aβ secreted from primary neurons from transgenic mice carrying the Swedish mutation in APP (Fig. S2D). In all of those experiments, E4 interacted more strongly than E2 and E3 with Aβ, although the difference was not statistically significant.
It is possible that usage of an anti-GFP or an anti-ApoE antibody to capture ApoE on the Dynabeads selects for a specific species of ApoE. To ensure that all biologically relevant ApoE species were represented, we tried a modified version of the assay in which we first attached 7PA2 Aβ to the Dynabeads using the 4G8 antibody. 2 h later, we added ApoE-GFP, and the following day, we added 6E10-650 to detect the Aβ (Fig. S2E). The results of this experiment showed that Aβ interacted significantly with ApoE (data not shown), but there were no differences between the isoforms (Fig. S2E). It is possible that Aβ is inactivated by binding to the 4G8 antibody prior to the addition of ApoE to the sample, therefore resulting in no differences between the isoforms.
We configured the assay so that a minority of the beads would be decorated with ApoE, and the remainder could be used as nonspecific controls in the flow plots within each run. For example, typically fewer than 5% of Dynabeads were positive for ApoE, whereas over 95% of beads were negative for ApoE within a certain sample (Fig. S2F). Therefore, we decided to use the ApoE− beads as an internal negative control to determine the background signal originating from 6E10-650 binding to the beads in the absence of ApoE. To confirm that ApoE− beads were indeed equally functional as the ApoE+ beads, in other words, that they were coated with the anti-ApoE antibody and that they had the potential to bind to ApoE when the protein concentration is higher, we undertook two experiments. First, we performed a serial dilution of ApoE and found that the percentage of ApoE+ beads was proportional to the decrease in ApoE concentration (Fig. S2G). Second, we conjugated a goat anti-ApoE antibody to 594Dylight, attached it to Dynabeads, and found that ∼97% of the beads were positive for the antibody (Fig. S2H). Therefore, ApoE− beads are identical to ApoE+ beads in terms of functionality and are suitable to use as a control to determine the background signal.
An additional concern in our flow cytometry assay was whether the 650Dylight signal truly originated from the 6E10-650 antibody detecting Aβ that has attached to ApoE or whether it originates from 6E10-650 binding directly to ApoE, thus giving false positive results. To test this possibility, we performed two different experiments in the presence and absence of ApoE. First, we attached 7PA2 Aβ to Dynabeads using the 4G8 antibody; second, we attached ApoE-GFP to Dynabeads using an anti-GFP antibody, followed by adding 7PA2. We then added a concentration gradient of the 6E10-650 antibody and observed, in both situations, an increase in the 650 MFI signal that was commensurate with the increase in 6E10 concentration (Fig. S2, I and J). Second, we did the same experiments but used a serial dilution of 7PA2 (undiluted, 1:15, 1:35, and 1:50) and a stable concentration of 6E10-650. We observed that the 650 MFI signal decreased proportionately to the Aβ concentration in the experiment without ApoE; however, upon addition of ApoE these results were modified (Fig. S2, I and J), suggesting that the results of our assay are ApoE-specific.
We used the assay on recombinant E3, which is lipidated differently than physiological ApoE and is thus expected to interact with Aβ differently (40). Recombinant E3 was detected using the 3H1-488Dylight antibody. The results from this experiment suggested that recombinant ApoE interacts with Aβ (Fig. S2K) but at a much smaller degree than physiological ApoE (one significance asterisk versus three significance asterisks when comparing ApoE+ with ApoE− beads) (Fig. 1, E and F).
Finally, we wanted to determine whether the 6E10 antibody concentrations used in our experiments were optimal to allow the detection of isoform-specific differences for the interaction between ApoE and Aβ. To this end, we tried three different amounts of antibody, 1.25, 3, and 5 μg, and found that, although increased concentrations increased the Aβ/ApoE ratio, there were no differences in the interaction patterns for each isoform (Fig. S1L). We therefore decided to continue the experiments with the lowest amount of antibody (1 μg).
Astrocyte-secreted ApoE can interact with physiological Aβ in vitro
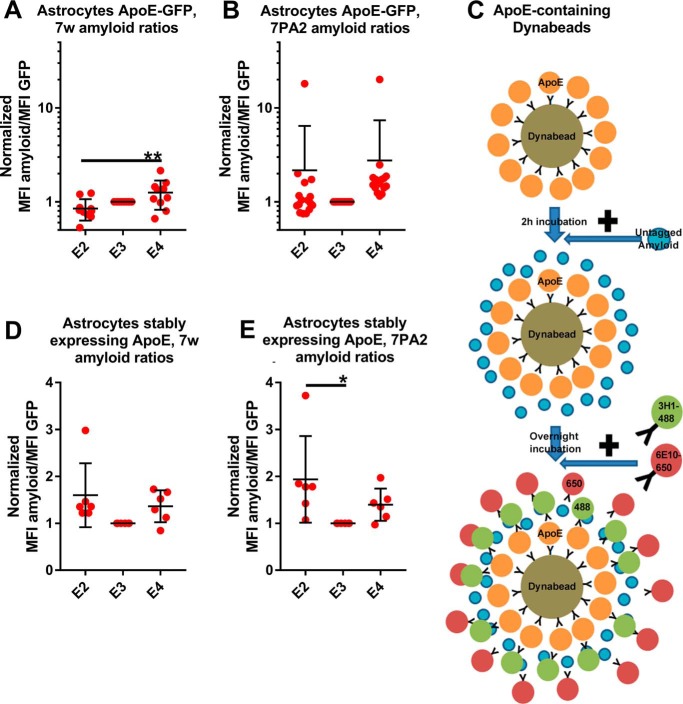
It has been found previously that astrocyte-secreted ApoE exhibits differences in its lipidation compared with HEK-secreted ApoE. It is also thought that it is a better approximation than HEK-secreted ApoE of brain-derived ApoE (3). We thus studied its interaction with physiological Aβ secreted from CHO cell lines overexpressing WT or V717F mutant APP. Astrocyte-secreted ApoE interacted significantly with both types of Aβ (Fig. S1, G and H). However, E4 exhibited significantly stronger interactions than E2 only with 7w Aβ but not with 7pa2 Aβ (Fig. 3, A and B).
Figure 3.
Interactions between astrocyte-secreted ApoE and physiological Aβ. A, plot indicating the Aβ/ApoE ratios for astrocyte-secreted ApoE incubated with 7w CM. E4 interacts more strongly than E2 and E3 with 7w Aβ. Number of independent experiments: E2, 10; E3, 10; E4, 10. B, plot indicating the Aβ/ApoE ratios for astrocyte-secreted ApoE incubated with 7PA2 CM. There are no differences between isoforms. Number of independent experiments: E2, 16; E3, 16; E4, 16. C, illustration of the process for the flow cytometry assay on untagged ApoE. ApoE is attached to Dynabeads using a goat anti-ApoE antibody. 2 h later, Aβ is added to the sample for overnight incubation. The following day, the 3H1-488 and the 6E10-650 antibodies are added to each sample. D, plot indicating the Aβ/ApoE ratios for astrocyte-secreted untagged ApoE incubated with 7w CM. There are no significant differences between the isoforms. Number of independent experiments: E2, 6; E3, 6; E4, 6. E, plot indicating the Aβ/ApoE ratios for astrocyte-secreted untagged ApoE incubated with 7PA2 CM. E2 interacts more strongly than E3 with Aβ. Number of independent experiments: E2, 6; E3, 6; E4, 6. In all samples, the concentrations of ApoE and Aβ were as described for the HEK experiments. In these plots, each dot represents the mean value from one independent experiment. The data were normalized to E3 before meta-analysis. Statistical analysis was done with one-way ANOVA with Tukey's post hoc correction. E2, E3, E4, and the negative control were included in the statistical analysis, which was done for each type of Aβ separately. Mean ± S.D. is shown in the plots. **, p ≤ 0.01; *, p ≤ 0.05.
It is possible that the presence of the large fluorophore tag on ApoE affects its ability to bind to Aβ. We thus tried the assay on ApoE secreted from primary immortalized astrocytes expressing untagged ApoE at endogenous levels (27). For this experiment, we first attached ApoE to Dynabeads using a goat polyclonal anti-ApoE antibody. 2 h later, we added 7w or 7pa2 Aβ for overnight incubation. The following morning, we added the 3H1-488 antibody to detect ApoE and the 6E10-650 antibody to detect Aβ (Fig. 3C). All isoforms interacted significantly with both types of Aβ (Fig. S1, I and J). E2 interacted more strongly than E3 with 7pa2 Aβ, but no other differences were observed between the isoforms (Fig. 3, D and E).
Discussion
Even though there are numerous studies suggesting that the ApoE genotype modifies the deposition of Aβ and plaque formation, with E4 being deleterious and E2 protective, the mechanism underlying this phenomenon is not clear. It has been hypothesized that direct interaction of Aβ with ApoE and the formation of ApoE–Aβ complexes are important components of this procedure. However, studies to date have given conflicting results; some have indicated that ApoE interacts with Aβ in an isoform-specific way, whereas others have not seen isoform-specific differences or have found only evidence of minimal interaction. To systematically examine the ApoE–Aβ interactions, we have developed a novel flow cytometry assay that does not include treatments and steps that could modify the physiological integrity of Aβ–ApoE complexes formed in vitro. As a source of Aβ, we used synthetic Aβ42 and Aβ40 as well as physiological Aβ secreted from cells overexpressing WT or mutant APP, giving us the opportunity to assess the effect of Aβ with different Aβ42/Aβ40 ratios. As a source of ApoE, we used physiological ApoE secreted from HEK cells or astrocytes. Using these assays, we demonstrated the following: physiological ApoE can interact significantly with physiological and synthetic Aβ; synthetic ApoE can interact only minimally with physiological Aβ; the strength of the interaction between ApoE and Aβ is isoform-dependent, in general with E4 exhibiting the strongest interactions but with E2 and E3 exhibiting weaker interactions; and there are differences in Aβ–ApoE interactions, depending on whether ApoE is secreted from HEK cells or astrocytes.
Our data indicate that there is great variability in ApoE–Aβ interactions that depend on the isoform of ApoE, the cell type of ApoE origin, the origin of Aβ, and the type of Aβ. This diversity could explain the discrepancies in previous studies that used various methods to isolate and examine the ApoE–Aβ complexes. Specifically, we found that astrocyte-secreted ApoE interacted significantly with synthetic Aβ40 and Aβ42 Aβ and that E4 interacted more strongly compared with E3 and E2. Three previous studies have assessed the same combination of ApoE and Aβ (18, 27, 41). Hashimoto et al. (27) found that ApoE increases the fibrilization of Aβ in an isoform specific way (E2 < E3 < E4), which is consistent with our current data. Verghese et al. (18) did not find a difference between E3 and E4, which associated only minimally with Aβ. Morikawa et al. (41) found that the results depended on the type of Western blotting (WB) used, with E3 associating more than E4 with Aβ on reducing WB but with no differences between isoforms when nonreducing WB was used.
In our study, HEK-secreted ApoE associated with synthetic Aβ42 and Aβ40; however, E4 interacted more strongly than E2 and E3 only with Aβ42 but not Aβ40. Five studies have previously assessed the Aβ–ApoE interactions using HEK-secreted ApoE and synthetic Aβ (15, 19, 42–44). Tai et al. (19) found no differences between the isoforms. LaDu et al. (15, 42), Bentley et al. (43), and Manelli et al. (44) found that E3 forms stronger interactions than E4 with Aβ. Given the variability in the results among the studies, it is possible that the association between HEK-secreted ApoE and synthetic Aβ is more stochastic in nature, given the nonphysiological nature of Aβ.
The only study to date that has studied the interaction between physiological Aβ and physiological ApoE is by Verghese et al. (18). In this study, the authors used Aβ secreted from H4 cells expressing APP with the Swedish mutation or CHO cells expressing the V717F APP mutation (7PA2 line) and ApoE secreted from primary immortalized astrocytes derived from mice with targeted replacement of murine ApoE with human ApoE (41). They measured, through ELISA, the amount of Aβ bound and unbound to ApoE in the density distribution of ApoE and Aβ following density gradient centrifugation. The results, which were also confirmed through SEC experiments, showed that the association between ApoE and Aβ is minimal and that there are no differences between the isoforms. However, we observed significant interactions between astrocyte-secreted ApoE and physiological Aβ using our flow cytometry assay. We also found that E4 associated more strongly than E2 with 7w Aβ. It is possible that the discrepancies are because we used a different method to isolate and examine the ApoE–Aβ complexes than Verghese et al., in that the centrifugation separation and isolation steps may have disrupted complexes that remained intact in the flow cytometry assay, which lacks these steps.
The consensus from all the studies to date is that ApoE interacts with Aβ, albeit to variable degrees and patterns. We have shown previously that HEK- and astrocyte-secreted E4 has a more closed conformation relative to E2 and E3 and it is more lipidated, and we hypothesized that the increased lipidation of E4 acts as a folding chaperone (3). It is possible that these two features can also increase the propensity of E4 to bind to Aβ through formation of a hydrophobic binding pocket. It is also possible that ApoE influences the deposition of Aβ without direct binding but through modulation of common receptors (18) or other mechanisms. It could also be that the modulation of Aβ aggregation occurs through a combination of those mechanisms. More studies are needed to understand the complex relationship between ApoE and Aβ. In addition, future studies should assess in more detail whether specific types of Aβ differentially interact with ApoE, which could also be a factor contributing to variable results between studies. We have already noted differences between Aβ40 and Aβ42 and between Aβ secreted from cells expressing WT versus mutant APP. Other Aβ species that could be assessed in the future include truncated Aβ, low- versus high-molecular-weight complexes, and aggregated versus nonaggregated Aβ.
In conclusion, it appears that Aβ can form complexes with secreted ApoE and that the structure of those complexes depends on the ApoE isoform, on the source of ApoE, and on the type of Aβ. It is possible that the formation of Aβ–ApoE complexes has a role in the pathogenesis of AD and constitutes a possible target of drugs attempting to modify the course of the disease. We speculate that ApoE conformation, lipidation, intermolecular interactions, and ability to interact with Aβ are interconnected features that are modulated during the pathogenesis of AD in an isoform-specific manner, with the contribution of independent modifying factors, such as receptor binding, to the process.
Experimental procedures
Tissue culture and transfections
HEK293 cells were maintained in OMEM (31985-088, Thermo Fisher), 5% fetal bovine serum (FBS) (16000044, Thermo Fisher), 100 units/ml penicillin, and 100 μg/ml streptomycin (15140122, Thermo Fisher). Primary immortalized astrocytes with ApoE knockout (27) were maintained in advanced DMEM (12491-015, Thermo Fisher), 10% FBS, 2 mm GlutaMAX (35050061, Thermo Fisher), and 100 μg/ml Geneticin (10131035, Thermo Fisher). WT CHO cells were maintained in DMEM, 10% FBS, 2 mm GlutaMAX, 100 μg/ml penicillin, and 100 μg/ml streptomycin. The CHO cell lines 7w and 7PA2 were used as a source of Aβ. 7w CHO cells (32) were maintained in OMEM, 5% FBS, and 200 μg/ml Geneticin, and 7PA2 cells (33, 34) were maintained in DMEM, 10% FBS, 150 μg/ml Geneticin, and 2 mm GlutaMAX.
HEK cells and astrocytes were plated in T75 flasks and transfected with ApoE2/3/4 tagged on either their C or N termini with GFP. The location of the tag did not affect the results (data not shown). Plasmids with the tag placed on the same terminus were used within each independent experiment. Cells were also transfected with a plasmid encoding untagged ApoE, and these CM samples were used as a basis for the single-color controls, as described below. The cloning of those plasmids has been described previously (3). Transfections were done using Lipofectamine 2000 as described previously (3). After transfection, HEK cells were fed with OMEM without phenol and 5% FBS and incubated for 3 days; the astrocytes were fed with DMEM without phenol, 10% FBS, and 2 mm GlutaMAX and incubated for 3 days.
Generation of CM from primary neurons from transgenic mice
Tg2576 male mice (Taconic Farms), which are heterozygous for the APP Swedish mutation and overexpress the protein under the PrP promoter, were bred with littermate females. Primary neuronal cultures were established at embryonic day 16 in 35-mm glass-bottom dishes, as described previously (39, 45). The conditioned medium was collected on the 14th day in vitro.
Measurement of ApoE and Aβ concentrations in the CM
The concentration of Aβ was measured using an Aβ ELISA kit (294-64701, Wako). The concentration of ApoE was measured using a commercial ELISA kit (KA-1031, Abnova).
ApoE–Aβ flow cytometry interaction assay
The CM from cells overexpressing GFP-tagged ApoE was collected after incubation for 3 days and centrifuged at 1500 rpm for 10 min to pellet floating cells. The supernatant was then concentrated down 15-fold using 10-kDa concentration columns (UFC901024, Millipore). Subsequently, the CM containing the GFP-tagged ApoE was attached to Dynabeads protein G (10007D, Thermo Fisher) using 1 μg of an anti-GFP antibody (AB6556, Abcam). The samples were then rotated at 4 °C. The procedure has been described previously in detail (3). Thereafter, three different variants of the assay were developed, depending on the origin of the soluble Aβ used.
For analysis of 7w and 7PA2 CHO-secreted Aβ and Aβ secreted from HEK cells transfected with a plasmid encoding pure Aβ42, the CM from those cell lines was collected after 3 days and concentrated down 15-fold. CM from Tg2576 cells was used unconcentrated and prepared as described above. 300 μl of each aforementioned sample containing ApoE plus Dynabeads was aliquoted after rotation for 2 h. 100 μl of Aβ-containing CM was added thereafter. Rotation then continued overnight at 4 °C. In the morning, 1.25 μg of 6E10 antibody (SIG-39320, Biolegend) conjugated with Dylight650 (62265, Invitrogen) was added to each sample, and they were rotated for another 5.5 h. In a separate set of experiments, the 4G8 antibody (SIG-39220, Biolegend) conjugated to 650Dylight was also used for the detection of bound Aβ. The conjugation of antibodies to Dylight has been described previously (3). As single color controls, ApoE that was singly tagged with GFP and untagged ApoE+Dynabeads that were incubated with 6E10-Dylight650 were used. The combination of GFP and far-red fluorophores was selected to minimize bleedthrough.
In the second version of the assay, synthetic Aβ (1–42) HiLyte Fluor 647- labeled (AS-64161, AnaSpec) or Aβ (1–40) HiLyte Fluor 647-labeled (AS-60493, AnaSpec) was reconstituted in 100 μl of DMSO to a stock concentration of 1 μg/μl in low-retention tubes, pipetted for 10 min at room temperature, aliquoted, and frozen overnight at −20 °C. The stock was used within a few days and was never thawed more than once. Synthetic Aβ was added at a final concentration of 0.03 μg/μl to the ApoE-GFP+Dynabead samples after overnight incubation. The samples were rotated at 4 °C for 4 h only to prevent aggregation of Aβ and quenching of its fluorescent tag (46).
In the third version of the assay, 600 μl of 7PA2 CM that was prepared as described previously described was attached to Dynabeads using 3 μg of the 4G8 antibody. 2 h later, 100 μl of that sample was mixed with 300 μl of ApoE-GFP CM that was prepared as described above. After overnight incubation, 1.25 μg of the 6E10-650 antibody were added to each sample. The samples were analyzed through flow cytometry ∼5 h later.
The concentrations of ApoE and Aβ used in this experiment are listed in Table S1. Schematic illustrations of the assays are provided in Figs. 1A and 2D and Figs. 2E and S3.
Flow cytometry measurements were performed using a custom Fortessa instrument (BD Biosciences). For detection of GFP fluorescence, a 488-nm laser and a 525/50-nm emission filter were used. For detection of 650Dylight and 647Hilyte fluorescence, a 640-nm laser and a 670/14-nm emission filter were used. Approximately 1,000,000 events were recorded for each sample. Voltages and compensations were adjusted using the single-color controls on the APC/FITC plot with the biexponential display. The gates for the green and far-red fluorescence were determined using the opposite single-color control on the fluorescence/side scatter-A plot.
For data analysis, Flowing Software 2 was used. The MFI of the far-red dye was determined in the GFP+ and GFP− populations. The MFI of GFP was determined in the ApoE+ population. The ratio of Aβ to ApoE was determined using the following formula:
| (Eq. 1) |
ApoE-Aβ flow cytometry interaction assay on untagged ApoE
CM from primary immortalized astrocytes expressing E2, E3, or E4 at physiological levels (27) was collected after 3 days of incubation in DMEM without phenol, 10% FBS, and 2 mm GlutaMAX and was prepared as described above. Recombinant human E3 protein (4144-AE-500, R&D Systems) was diluted to 1 μg/ml in PBS without calcium and magnesium, 300 μl of this suspension was attached to 25 μl of protein G Dynabeads using 4 μl of a goat anti-ApoE polyclonal antibody (AB947, Millipore). 2 h later, 100 μl of 7PA2 or 7w CM was added to each sample. The following morning, 3.3 μg of the 3H1 antibody against the C terminus of ApoE (47) that was conjugated to 488Dylight and 1.25 μg of the 6E10-650 antibody were added to the sample. The samples were analyzed through flow cytometry after incubation for 5 h. A schematic illustration of this assay is provided in Fig. 3C.
SEC
The 7w CHO and 7PA2 cell line CM was prepared as described in the previous section. Prior to the SEC experiment, the CM was centrifuged at 10,000 × g for 10 min, and the supernatant was collected. A Superdex75 column (GE Healthcare) was used for SEC. The samples were separated in 50 mm ammonium acetate (pH 8.5) at a flow rate of 0.5 ml/min. 400 μl of CM was used as starting material. The samples from two neighboring fractions were pooled, and the concentrations of Aβ40 and Aβ42 were determined using Aβ40-specific (292-62301, Wako) and Aβ42-specific (290-62601, Wako) ELISAs (48).
Statistical analysis
All experiments were performed independently at least three times. For each sample within the same experiment, the mean value was determined. Then, the values from each sample were normalized internally prior to meta-analysis of the data from independent experiments. Thus, one sample in each plot does not have error bars. Each dot on the plots represents the mean value for one sample/condition from one independent experiment. Statistical analysis was completed using GraphPad Prism version 5 software. A one-way ANOVA with Tukey's post hoc correction was used.
Author contributions
E. K., J. D. M., and B. T. H. conceptualization; E. K., J. D. M., and B. T. H. data curation; E. K., J. D. M., and B. T. H. formal analysis; E. K. and B. T. H. funding acquisition; E. K., J. D. M., and B. T. H. validation; E. K., J. D. M., A. D. R., C. C., Z. F., M. C.-R., and B. T. H. investigation; E. K., J. D. M., S. W., E. H., and B. T. H. visualization; E. K., J. D. M., A. D. R., C. C., Z. F., M. C.-R., S. W., E. H., and B. T. H. methodology; E. K., J. D. M., and B. T. H. writing-original draft; E. K. and B. T. H. project administration; A. D. R., C. C., Z. F., M. C.-R., S. W., E. H., and B. T. H. writing-review and editing; M. C.-R., S. W., E. H., and B. T. H. resources; B. T. H. supervision.
Supplementary Material
Acknowledgments
The cytometric findings reported here were obtained in the Massachusetts General Hospital Department of Pathology Flow and Image Cytometry Research Core, which obtained support from the National Institutes of Health Shared Instrumentation Program with Grants 1S10OD012027-01A1, 1S10OD016372-01, 1S10RR020936-01, and 1S10RR023440-01A1. We thank Prof. Dennis Selkoe (Harvard Medical School) for donating the 7w and 7PA2 cell lines.
This work was supported by EMBO Long-Term Fellowship ATLF-815-2014, which is co-funded by the Marie Curie Actions of the European Commission (LTFCOFUND2013, GA-2013-609409) (to E. K.) and the JPB Foundation (to B. T. H.). This work was also supported by a grant from F Prime. Financial support was partially obtained from a sponsored research agreement from Abbvie. Abbvie was not involved in assay design, data acquisition, data analysis, preparation of the manuscript or decision to publish the results. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Table S1.
- ApoE
- apolipoprotein E
- AD
- Alzheimer's disease
- APP
- amyloid precursor protein
- Aβ
- amyloid β
- CHO
- Chinese hamster ovary
- HEK
- human embryonic kidney
- CM
- conditioned medium
- MFI
- mean fluorescence intensity
- SEC
- size exclusion chromatography
- WB
- Western blotting
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- ANOVA
- analysis of variance
- OMEM
- Opti-modified Eagle's medium.
References
- 1. Corder E. H., Saunders A. M., Risch N. J., Strittmatter W. J., Schmechel D. E., Gaskell P. C. Jr., Rimmler J. B., Locke P. A., Conneally P. M., and Schmader K. E. (1994) Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 7, 180–184 10.1038/ng0694-180 [DOI] [PubMed] [Google Scholar]
- 2. Utermann G., Kindermann I., Kaffarnik H., and Steinmetz A. (1984) Apolipoprotein E phenotypes and hyperlipidemia. Hum. Genet. 65, 232–236 10.1007/BF00286508 [DOI] [PubMed] [Google Scholar]
- 3. Kara E., Marks J. D., Fan Z., Klickstein J. A., Roe A. D., Krogh K. A., Wegmann S., Maesako M., Luo C. C., Mylvaganam R., Berezovska O., Hudry E., and Hyman B. T. (2017) Isoform and cell type-specific structure of apolipoprotein E lipoparticles as revealed by a novel Forster resonance energy transfer assay. J. Biol. Chem. 292, 14720–14729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hatters D. M., Peters-Libeu C. A., and Weisgraber K. H. (2006) Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 31, 445–454 10.1016/j.tibs.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 5. Drzezga A., Grimmer T., Henriksen G., Mühlau M., Perneczky R., Miederer I., Praus C., Sorg C., Wohlschläger A., Riemenschneider M., Wester H. J., Foerstl H., Schwaiger M., and Kurz A. (2009) Effect of APOE genotype on Aβ plaque load and gray matter volume in Alzheimer disease. Neurology 72, 1487–1494 10.1212/WNL.0b013e3181a2e8d0 [DOI] [PubMed] [Google Scholar]
- 6. Grimmer T., Tholen S., Yousefi B. H., Alexopoulos P., Förschler A., Förstl H., Henriksen G., Klunk W. E., Mathis C. A., Perneczky R., Sorg C., Kurz A., and Drzezga A. (2010) Progression of cerebral Aβ load is associated with the apolipoprotein E ϵ4 genotype in Alzheimer's disease. Biol. Psychiatry 68, 879–884 10.1016/j.biopsych.2010.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rebeck G. W., Reiter J. S., Strickland D. K., and Hyman B. T. (1993) Apolipoprotein E in sporadic Alzheimer's disease: allelic variation and receptor interactions. Neuron 11, 575–580 10.1016/0896-6273(93)90070-8 [DOI] [PubMed] [Google Scholar]
- 8. Bales K. R., Liu F., Wu S., Lin S., Koger D., DeLong C., Hansen J. C., Sullivan P. M., and Paul S. M. (2009) Human APOE isoform-dependent effects on brain β-Aβ levels in PDAPP transgenic mice. J. Neurosci. 29, 6771–6779 10.1523/JNEUROSCI.0887-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hudry E., Dashkoff J., Roe A. D., Takeda S., Koffie R. M., Hashimoto T., Scheel M., Spires-Jones T., Arbel-Ornath M., Betensky R., Davidson B. L., and Hyman B. T. (2013) Gene transfer of human Apoe isoforms results in differential modulation of Aβ deposition and neurotoxicity in mouse brain. Sci. Transl. Med. 5, 212ra161 10.1126/scitranslmed.3007000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Irizarry M. C., Rebeck G. W., Cheung B., Bales K., Paul S. M., Holzman D., and Hyman B. T. (2000) Modulation of Aβ deposition in APP transgenic mice by an apolipoprotein E null background. Ann. N.Y. Acad. Sci. 920, 171–178 [DOI] [PubMed] [Google Scholar]
- 11. Kim J., Eltorai A. E., Jiang H., Liao F., Verghese P. B., Kim J., Stewart F. R., Basak J. M., and Holtzman D. M. (2012) Anti-apoE immunotherapy inhibits Aβ accumulation in a transgenic mouse model of Aβ Aβosis. J. Exp. Med. 209, 2149–2156 10.1084/jem.20121274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liao F., Hori Y., Hudry E., Bauer A. Q., Jiang H., Mahan T. E., Lefton K. B., Zhang T. J., Dearborn J. T., Kim J., Culver J. P., Betensky R., Wozniak D. F., Hyman B. T., and Holtzman D. M. (2014) Anti-ApoE antibody given after plaque onset decreases Aβ accumulation and improves brain function in a mouse model of Aβ Aβosis. J. Neurosci. 34, 7281–7292 10.1523/JNEUROSCI.0646-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanekiyo T., Xu H., and Bu G. (2014) ApoE and Aβ in Alzheimer's disease: accidental encounters or partners? Neuron 81, 740–754 10.1016/j.neuron.2014.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tai L. M., Mehra S., Shete V., Estus S., Rebeck G. W., Bu G., and LaDu M. J. (2014) Soluble apoE/Aβ complex: mechanism and therapeutic target for APOE4-induced AD risk. Mol. Neurodegener. 9, 2 10.1186/1750-1326-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. LaDu M. J., Falduto M. T., Manelli A. M., Reardon C. A., Getz G. S., and Frail D. E. (1994) Isoform-specific binding of apolipoprotein E to β-Aβ. The J. Biol. Chem. 269, 23403–23406 [PubMed] [Google Scholar]
- 16. Wisniewski T., Golabek A., Matsubara E., Ghiso J., and Frangione B. (1993) Apolipoprotein E: binding to soluble Alzheimer's β-Aβ. Biochem. Biophys. Res. Commun. 192, 359–365 10.1006/bbrc.1993.1423 [DOI] [PubMed] [Google Scholar]
- 17. Strittmatter W. J., Weisgraber K. H., Huang D. Y., Dong L. M., Salvesen G. S., Pericak-Vance M., Schmechel D., Saunders A. M., Goldgaber D., and Roses A. D. (1993) Binding of human apolipoprotein E to synthetic Aβ β peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 90, 8098–8102 10.1073/pnas.90.17.8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verghese P. B., Castellano J. M., Garai K., Wang Y., Jiang H., Shah A., Bu G., Frieden C., and Holtzman D. M. (2013) ApoE influences Aβ-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc. Natl. Acad. Sci. U.S.A. 110, E1807–E1816 10.1073/pnas.1220484110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tai L. M., Bilousova T., Jungbauer L., Roeske S. K., Youmans K. L., Yu C., Poon W. W., Cornwell L. B., Miller C. A., Vinters H. V., Van Eldik L. J., Fardo D. W., Estus S., Bu G., Gylys K. H., and Ladu M. J. (2013) Levels of soluble apolipoprotein E/Aβ-β (Aβ) complex are reduced and oligomeric Aβ increased with APOE4 and Alzheimer disease in a transgenic mouse model and human samples. J. Biol. Chem. 288, 5914–5926 10.1074/jbc.M112.442103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shuvaev V. V., and Siest G. (1996) Interaction between human amphipathic apolipoproteins and Aβ β-peptide: surface plasmon resonance studies. FEBS Lett. 383, 9–12 10.1016/0014-5793(96)00206-2 [DOI] [PubMed] [Google Scholar]
- 21. Wellnitz S., Friedlein A., Bonanni C., Anquez V., Goepfert F., Loetscher H., Adessi C., and Czech C. (2005) A 13 kDa carboxy-terminal fragment of ApoE stabilizes Aβ hexamers. J. Neurochem. 94, 1351–1360 10.1111/j.1471-4159.2005.03295.x [DOI] [PubMed] [Google Scholar]
- 22. Gylys K. H., Fein J. A., Tan A. M., and Cole G. M. (2003) Apolipoprotein E enhances uptake of soluble but not aggregated Aβ-β protein into synaptic terminals. J. Neurochem. 84, 1442–1451 10.1046/j.1471-4159.2003.01643.x [DOI] [PubMed] [Google Scholar]
- 23. Pillot T., Goethals M., Vanloo B., Lins L., Brasseur R., Vandekerckhove J., and Rosseneu M. (1997) Specific modulation of the fusogenic properties of the Alzheimer β-Aβ peptide by apolipoprotein E isoforms. Eur. J. Biochem. 243, 650–659 10.1111/j.1432-1033.1997.00650.x [DOI] [PubMed] [Google Scholar]
- 24. Golabek A. A., Soto C., Vogel T., and Wisniewski T. (1996) The interaction between apolipoprotein E and Alzheimer's Aβ β-peptide is dependent on β-peptide conformation. J. Biol. Chem. 271, 10602–10606 10.1074/jbc.271.18.10602 [DOI] [PubMed] [Google Scholar]
- 25. Näslund J., Thyberg J., Tjernberg L. O., Wernstedt C., Karlström A. R., Bogdanovic N., Gandy S. E., Lannfelt L., Terenius L., and Nordstedt C. (1995) Characterization of stable complexes involving apolipoprotein E and the Aβ β peptide in Alzheimer's disease brain. Neuron 15, 219–228 10.1016/0896-6273(95)90079-9 [DOI] [PubMed] [Google Scholar]
- 26. Chan W., Fornwald J., Brawner M., and Wetzel R. (1996) Native complex formation between apolipoprotein E isoforms and the Alzheimer's disease peptide A β. Biochemistry 35, 7123–7130 10.1021/bi952852v [DOI] [PubMed] [Google Scholar]
- 27. Hashimoto T., Serrano-Pozo A., Hori Y., Adams K. W., Takeda S., Banerji A. O., Mitani A., Joyner D., Thyssen D. H., Bacskai B. J., Frosch M. P., Spires-Jones T. L., Finn M. B., Holtzman D. M., and Hyman B. T. (2012) Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of Aβ β peptide. J. Neurosci. 32, 15181–15192 10.1523/JNEUROSCI.1542-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wood S. J., Chan W., and Wetzel R. (1996) Seeding of A β fibril formation is inhibited by all three isotypes of apolipoprotein E. Biochemistry 35, 12623–12628 10.1021/bi961074j [DOI] [PubMed] [Google Scholar]
- 29. Wood S. J., Chan W., and Wetzel R. (1996) An ApoE-Aβ inhibition complex in Aβ fibril extension. Chem. Biol. 3, 949–956 10.1016/S1074-5521(96)90183-0 [DOI] [PubMed] [Google Scholar]
- 30. Koffie R. M., Hashimoto T., Tai H. C., Kay K. R., Serrano-Pozo A., Joyner D., Hou S., Kopeikina K. J., Frosch M. P., Lee V. M., Holtzman D. M., Hyman B. T., and Spires-Jones T. L. (2012) Apolipoprotein E4 effects in Alzheimer's disease are mediated by synaptotoxic oligomeric Aβ-β. Brain 135, 2155–2168 10.1093/brain/aws127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarkar G., Curran G. L., Sarkaria J. N., Lowe V. J., and Jenkins R. B. (2014) Peptide carrier-mediated non-covalent delivery of unmodified cisplatin, methotrexate and other agents via intravenous route to the brain. PLoS ONE 9, e97655 10.1371/journal.pone.0097655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koo E. H., and Squazzo S. L. (1994) Evidence that production and release of Aβ β-protein involves the endocytic pathway. J. Biol. Chem. 269, 17386–17389 [PubMed] [Google Scholar]
- 33. Welzel A. T., Maggio J. E., Shankar G. M., Walker D. E., Ostaszewski B. L., Li S., Klyubin I., Rowan M. J., Seubert P., Walsh D. M., and Selkoe D. J. (2014) Secreted Aβ β-proteins in a cell culture model include N-terminally extended peptides that impair synaptic plasticity. Biochemistry 53, 3908–3921 10.1021/bi5003053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shankar G. M., Welzel A. T., McDonald J. M., Selkoe D. J., and Walsh D. M. (2011) Isolation of low-n Aβ β-protein oligomers from cultured cells, CSF, and brain. Methods Mol. Biol. 670, 33–44 [DOI] [PubMed] [Google Scholar]
- 35. Hahn S., Brüning T., Ness J., Czirr E., Baches S., Gijsen H., Korth C., Pietrzik C. U., Bulic B., and Weggen S. (2011) Presenilin-1 but not Aβ precursor protein mutations present in mouse models of Alzheimer's disease attenuate the response of cultured cells to γ-secretase modulators regardless of their potency and structure. J. Neurochem. 116, 385–395 10.1111/j.1471-4159.2010.07118.x [DOI] [PubMed] [Google Scholar]
- 36. Suzuki N., Cheung T. T., Cai X. D., Odaka A., Otvos L. Jr, Eckman C., Golde T. E., and Younkin S. G. (1994) An increased percentage of long Aβ β protein secreted by familial Aβ β protein precursor (β APP717) mutants. Science 264, 1336–1340 10.1126/science.8191290 [DOI] [PubMed] [Google Scholar]
- 37. Tamaoka A., Odaka A., Ishibashi Y., Usami M., Sahara N., Suzuki N., Nukina N., Mizusawa H., Shoji S., and Kanazawa I. (1994) APP717 missense mutation affects the ratio of Aβ β protein species (A β 1–42/43 and A β 1–40) in familial Alzheimer's disease brain. J. Biol. Chem. 269, 32721–32724 [PubMed] [Google Scholar]
- 38. Wu H. Y., Hudry E., Hashimoto T., Uemura K., Fan Z. Y., Berezovska O., Grosskreutz C. L., Bacskai B. J., and Hyman B. T. (2012) Distinct dendritic spine and nuclear phases of calcineurin activation after exposure to Aβ-β revealed by a novel fluorescence resonance energy transfer assay. J. Neurosci. 32, 5298–5309 10.1523/JNEUROSCI.0227-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arbel-Ornath M., Hudry E., Boivin J. R., Hashimoto T., Takeda S., Kuchibhotla K. V., Hou S., Lattarulo C. R., Belcher A. M., Shakerdge N., Trujillo P. B., Muzikansky A., Betensky R. A., Hyman B. T., and Bacskai B. J. (2017) Soluble oligomeric Aβ-β induces calcium dyshomeostasis that precedes synapse loss in the living mouse brain. Mol. Neurodegener. 12, 27 10.1186/s13024-017-0169-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tokuda T., Calero M., Matsubara E., Vidal R., Kumar A., Permanne B., Zlokovic B., Smith J. D., Ladu M. J., Rostagno A., Frangione B., and Ghiso J. (2000) Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer's Aβ β peptides. Biochem. J. 348, 359–365 10.1042/bj3480359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morikawa M., Fryer J. D., Sullivan P. M., Christopher E. A., Wahrle S. E., DeMattos R. B., O'Dell M. A., Fagan A. M., Lashuel H. A., Walz T., Asai K., and Holtzman D. M. (2005) Production and characterization of astrocyte-derived human apolipoprotein E isoforms from immortalized astrocytes and their interactions with Aβ-β. Neurobiol. Dis. 19, 66–76 10.1016/j.nbd.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 42. LaDu M. J., Lukens J. R., Reardon C. A., and Getz G. S. (1997) Association of human, rat, and rabbit apolipoprotein E with β-Aβ. J. Neurosci. Res. 49, 9–18 10.1002/(SICI)1097-4547(19970701)49:1%3C9::AID-JNR2%3E3.0.CO%3B2-H [DOI] [PubMed] [Google Scholar]
- 43. Bentley N. M., Ladu M. J., Rajan C., Getz G. S., and Reardon C. A. (2002) Apolipoprotein E structural requirements for the formation of SDS-stable complexes with β-Aβ-(1–40): the role of salt bridges. Biochem. J. 366, 273–279 10.1042/bj20020207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manelli A. M., Stine W. B., Van Eldik L. J., and LaDu M. J. (2004) ApoE and Aβ1–42 interactions: effects of isoform and conformation on structure and function. J. Mol. Neurosci. 23, 235–246 10.1385/JMN:23:3:235 [DOI] [PubMed] [Google Scholar]
- 45. Hudry E., Wu H. Y., Arbel-Ornath M., Hashimoto T., Matsouaka R., Fan Z., Spires-Jones T. L., Betensky R. A., Bacskai B. J., and Hyman B. T. (2012) Inhibition of the NFAT pathway alleviates Aβ β neurotoxicity in a mouse model of Alzheimer's disease. J. Neurosci. 32, 3176–3192 10.1523/JNEUROSCI.6439-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Esbjörner E. K., Chan F., Rees E., Erdelyi M., Luheshi L. M., Bertoncini C. W., Kaminski C. F., Dobson C. M., and Kaminski Schierle G. S. (2014) Direct observations of Aβ β self-assembly in live cells provide insights into differences in the kinetics of Aβ(1–40) and Aβ(1–42) aggregation. Chem. Biol. 21, 732–742 10.1016/j.chembiol.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones P. B., Adams K. W., Rozkalne A., Spires-Jones T. L., Hshieh T. T., Hashimoto T., von Armin C. A., Mielke M., Bacskai B. J., and Hyman B. T. (2011) Apolipoprotein E: isoform specific differences in tertiary structure and interaction with Aβ-β in human Alzheimer brain. PLoS ONE 6, e14586 10.1371/journal.pone.0014586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takeda S., Hashimoto T., Roe A. D., Hori Y., Spires-Jones T. L., and Hyman B. T. (2013) Brain interstitial oligomeric Aβ β increases with age and is resistant to clearance from brain in a mouse model of Alzheimer's disease. FASEB J. 27, 3239–3248 10.1096/fj.13-229666 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.