Abstract
The epithelial-to-mesenchymal transdifferentiation (EMT) is crucial for tissue differentiation in development and drives essential steps in cancer and fibrosis. EMT is accompanied by reprogramming of gene expression and has been associated with the epithelial stem-cell state in normal and carcinoma cells. The cytokine transforming growth factor β (TGF-β) drives this program in cooperation with other signaling pathways and through TGF-β–activated SMAD3 as the major effector. TGF-β–induced SMAD3 activation is inhibited by SMAD7 and to a lesser extent by SMAD6, and SMAD6 and SMAD7 both inhibit SMAD1 and SMAD5 activation in response to the TGF-β–related bone morphogenetic proteins (BMPs). We previously reported that, in response to BMP, protein arginine methyltransferase 1 (PRMT1) methylates SMAD6 at the BMP receptor complex, thereby promoting its dissociation from the receptors and enabling BMP-induced SMAD1 and SMAD5 activation. We now provide evidence that PRMT1 also facilitates TGF-β signaling by methylating SMAD7, which complements SMAD6 methylation. We found that PRMT1 is required for TGF-β–induced SMAD3 activation, through a mechanism similar to that of BMP-induced SMAD6 methylation, and thus promotes the TGF-β–induced EMT and epithelial stem-cell generation. This critical mechanism positions PRMT1 as an essential mediator of TGF-β signaling that controls the EMT and epithelial cell stemness through SMAD7 methylation.
Keywords: SMAD transcription factor, cell surface receptor, epithelial cell, cancer stem cells, post-translational modification (PTM), epithelial-mesenchymal transition, stemness, epithelial stem cell, arginine methyltransferase
Introduction
Epithelial-to-mesenchymal transition (EMT)7 is a cellular transdifferentiation process whereby epithelial cells lose their apicobasal polarity, epithelial junctions, and other epithelial characteristics and acquire, to varying extents, changes in gene expression and behavior that are commonly associated with mesenchymal cells. EMT enables cells to become motile and migrate directionally, and this behavior is often accompanied by the cells' ability to invade through extracellular matrices (1). EMT is marked by and depends on the expression and activities of one or several “master” EMT transcription factors that direct many of the changes in gene expression but also depends on a variety of nontranscriptional changes, including changes in cytoskeletal and membrane organization and dynamics. The EMT process results from a convergence of signaling pathways and is often seen to be driven by, or at a minimum to depend on, TGF-β signaling (2). In this context, activation of TGF-β signaling leads to destabilization of epithelial gene expression and phenotype and activates the expression of key EMT transcription factors such as Snail and Slug/Snail2 while also directing changes in the expression of many other genes (1, 2). The EMT process plays crucial roles at many stages of tissue differentiation and development but also drives essential steps in pathological conditions, predominantly in cancer progression of carcinomas, and in fibrosis (1).
The EMT process has also been intricately associated with the epithelial stem-cell state as it promotes or accompanies stem cell–like features in both normal epithelial and carcinoma cells (3, 4). Thus, activation of genes encoding EMT transcription factors promotes the generation of cancer stem cells. For example, activation of ZEB1 is required for the tumor-initiating capacity of pancreatic, colorectal, and breast cancer cells (5, 6), and induction of Snail expression in colorectal cancer cells increases the number of cancer stem cells (7). The Snail-related transcription factor Slug and SOX9 both play central roles in the maintenance of normal breast epithelial stem cells, and perturbation of the expression of either impairs the generation of stem cells (8, 9). TGF-β has been shown to promote the generation of cancer stem cells able to initiate tumor formation in breast cancer and skin squamous cell carcinomas (5, 10, 11).
The ability of TGF-β to activate and drive the EMT program, or any differentiation program, results primarily from the activities of TGF-β–activated SMAD3 as the major effector. Following ligand binding to the cell-surface TGF-β receptor complex, the type I receptor C-terminally phosphorylates and thus activates SMAD2 and SMAD3, which then form heteromeric complexes with SMAD4, translocate into the nucleus, and cooperate with DNA-binding transcription factors in the activation or repression of TGF-β/SMAD target genes (12). In EMT, TGF-β–activated SMAD3 activates the expression of Snail and Slug, as well as other EMT transcription factors, and then cooperates with these EMT transcription factors to induce or repress their target genes, thus initiating changes in gene expression that lead to transcriptome reprogramming and differentiation (2). The SMAD-initiated gene reprogramming is complemented by non-SMAD signaling pathways that are activated by TGF-β and/or other classes of ligands and receptors and contribute to the loss of epithelial phenotype and to the behavior that characterize EMT (2). In addition to the effector SMADs SMAD2 and SMAD3, that direct changes in expression, the cells express inhibitory SMADs. These interact with the type I receptor as well as the effector SMADs, thus preventing SMAD activation, but are also thought to directly repress SMAD-mediated activation of target genes. SMAD6 and SMAD7 inhibit the activation of SMAD2 and SMAD3 in response to TGF-β and of SMAD1 and SMAD5 in the responses to the TGF-β–related bone morphogenetic proteins (BMPs). SMAD6 preferentially inhibits BMP signaling, whereas SMAD7 inhibits TGF-β signaling more efficiently than SMAD6 (13).
Protein arginine methyltransferases (PRMTs) methylate arginine residues in histones and thus control epigenetically the expression of an array of genes; however, they also modify nonhistone proteins, including signaling mediators, and thus control their functions. Among the PRMTs, PRMT1 is the most abundant and is responsible for 75% of all arginine methylation in cells (14). Besides the common histone 4 methylation at Arg-3, PRMT1 methylates and functionally regulates an extensive variety of proteins, including components of several signaling pathways (15). Increased PRMT1 expression has been observed in a variety of carcinomas, including breast carcinomas, and has been correlated with tumor growth and cancer progression and metastasis (16).
We reported that PRMT1 is required for BMP signaling activation. BMP induces PRMT1, in association with the type II BMP receptor (BMPRII), to methylate SMAD6 associated with the type I BMP receptor (BMPRI), leading to dissociation of methylated SMAD6 from the BMP receptor complex and enabling activation of the effector SMADs SMAD1 and SMAD5 (17). We now provide evidence that PRMT1 is also a critical mediator of TGF-β signaling through methylation of SMAD7, which complements SMAD6 methylation. PRMT1 is required for TGF-β–induced SMAD3 activation, through a similar mechanism as shown for BMP-induced SMAD6 methylation, and thus promotes TGF-β–induced EMT as well as epithelial stem-cell generation. This study defines a novel signaling pathway, from TGF-β through PRMT1 onto SMAD7, that controls EMT and epithelial stem-cell maintenance through arginine methylation.
Results
PRMT1 is required for TGF-β signaling
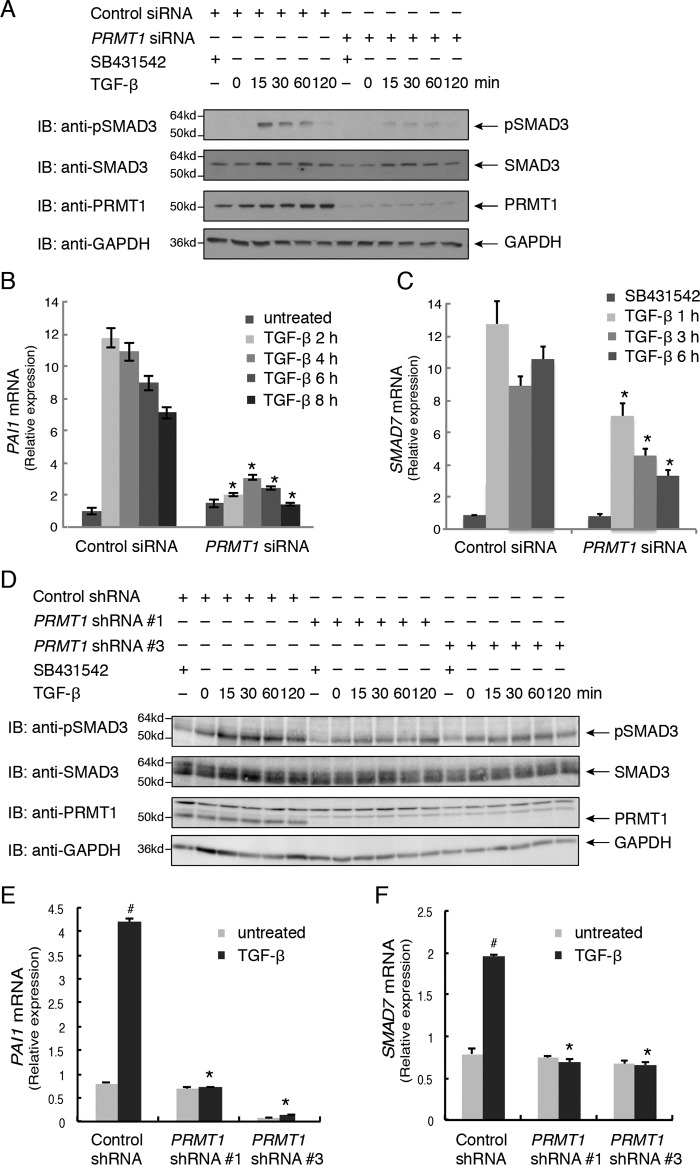
To evaluate whether PRMT1 controls TGF-β–induced SMAD activation, we silenced PRMT1 expression in human skin epithelial HaCaT cells using transfected siRNAs that target the expression of all PRMT1 isoforms. Silencing PRMT1 mRNA expression with >95% efficiency dramatically decreased the TGF-β–induced activation of SMAD3, detected by immunoblotting for C-terminally phosphorylated SMAD3 (Fig. 1A). Furthermore, silencing PRMT1 expression repressed the TGF-β–induced mRNA expression of known TGF-β/SMAD3 target genes such as the genes encoding plasminogen activator inhibitor type 1 (PAI1) (Fig. 1B) or SMAD7 (Fig. 1C). A similar inhibition of TGF-β responsiveness was apparent in human mammary epithelial (HMLE) cells. In these cells, silencing PRMT1 expression also impaired TGF-β–induced SMAD3 activation (Fig. 1D) and repressed TGF-β–induced PAI1 and SMAD7 mRNA expression (Fig. 1, E and F). These observations strongly suggest that PRMT1 is required for TGF-β–induced activation of SMAD3.
Figure 1.
PRMT1 is required for TGF-β–induced SMAD3 activation. A, down-regulating PRMT1 expression in HaCaT cells using transfected siRNA attenuates TGF-β–induced SMAD3 activation, assessed by immunoblotting (IB) for C-terminally phosphorylated SMAD3. HaCaT cells were transfected with PRMT1 siRNA or control siRNA and treated with the TGF-β signaling inhibitor SB431542 or with TGF-β1 for the indicated times or left untreated. B and C, down-regulating PRMT1 expression using siRNA reduced TGF-β–induced PAI1 (B) and SMAD7 (C) mRNA expression, assessed by qRT-PCR. HaCaT cells were transfected with siRNA and treated with TGF-β for the indicated times. *, p < 0.01 versus control siRNA at the same time point. D, down-regulating PRMT1 expression in HMLE cells using lentiviral shRNA attenuates TGF-β–induced SMAD3 activation, assessed by immunoblotting (IB) for C-terminally phosphorylated SMAD3. HMLE cells were infected with lentiviral constructs expressing one of two different PRMT1 shRNAs, shRNA 1 or shRNA 3, or a control shRNA and treated with the TGF-β signaling inhibitor SB431542 or with TGF-β1 for the indicated times or left untreated. E and F, down-regulating PRMT1 expression using lentiviral shRNA reduced TGF-β–induced PAI1 (E) and SMAD7 (F) mRNA expression, assessed by qRT-PCR. HMLE cells were infected with shRNAs and treated with TGF-β for 6 days. #, p < 0.01 versus untreated; *, p < 0.01 versus control shRNA. Error bars represent S.D.
TGF-β promotes SMAD7 methylation by PRMT1
We reported that PRMT1 facilitates BMP signaling through its ability to methylate SMAD6, resulting in its dissociation from the type I receptor, and thus enables BMP-induced SMAD1/5 activation (17). Because PRMT1 is also required for TGF-β–induced SMAD3 activation (Fig. 1) and SMAD7, in addition to SMAD6, inhibits TGF-β signaling (18), we evaluated whether PRMT1 methylates SMAD7, thus complementing the BMP-induced methylation of SMAD6 (17).
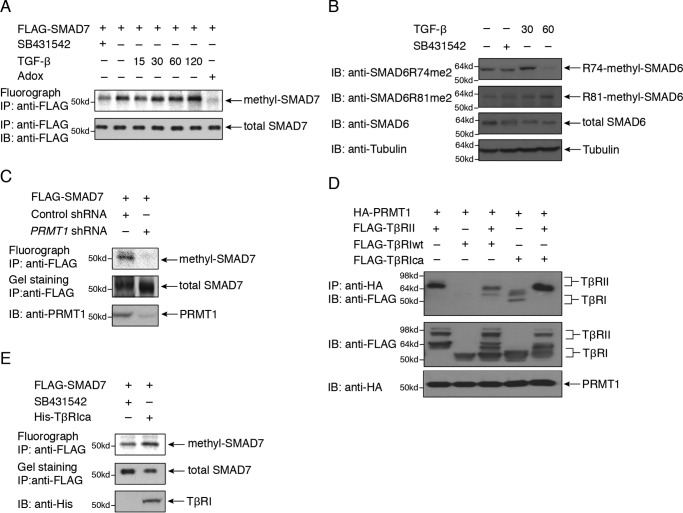
For this purpose, we evaluated by in vivo [3H]methylation whether TGF-β signaling induces SMAD7 methylation. This was indeed the case as shown in transfected HaCaT epithelial cells. In the absence of TGF-β signaling, i.e. in the presence of the type I TGF-β receptor kinase inhibitor SB431542, SMAD7 showed a low level of [3H]methylation, and TGF-β treatment enhanced the [3H]methylation of SMAD7, which was blocked by the general methyltransferase inhibitor adenosine dialdehyde (Fig. 2A). TGF-β also induced methylation of SMAD6, which could be probed using specific antibodies against either of the two dimethylated Arg residues (17), with strikingly different kinetics of Arg-74 and Arg-81 dimethylation (Fig. 2B). The increased methylation of SMAD6 and SMAD7 in response to TGF-β is consistent with the notion that both SMAD6 and SMAD7 act as inhibitory SMADs in TGF-β signaling (18). The [3H]methylation of endogenous SMAD7 could not be visualized due to the low level of SMAD7 expression, the limited quality of available SMAD7 antibodies, and the long exposures required to visualize [3H]methylated SMAD7. SMAD7 expressed in transfected 293T cells was also [3H]methylated, but its methylation was not enhanced in response to TGF-β, which is consistent with their low levels of TGF-β receptors and their poor responsiveness to TGF-β (data not shown). SMAD7 [3H]methylation was decreased when the expression of PRMT1 was silenced using shRNA (Fig. 2C).
Figure 2.
Role of SMAD7 in the control of TGF-β–induced SMAD activation by PRMT1. A, TGF-β treatment enhanced SMAD7 methylation in HaCaT cells, determined by in vivo labeling using [3H]methionine and fluorography, and this methylation was blocked by adenosine dialdehyde (Adox). B, TGF-β treatment enhanced SMAD6 methylation in HaCaT cells, determined by antibodies specific for SMAD6 dimethylation at Arg-74 and Arg-81. C, SMAD7 methylation is decreased following repression of PRMT1 expression. D, PRMT1 associates with TβRII but not with WT (wt) TβRI; however, PRMT1 associates with an activated form of TβRI (TβRIca). HA-tagged PRMT1 was coexpressed with WT or activated (ca) FLAG-tagged TβRI, TβRII, or both in transfected 293T cells. PRMT1 was immunoprecipitated, and the associated TβRII or TβRI was visualized by immunoblotting. E, expression of TβRIca enhances SMAD7 methylation in HaCaT cells expressing FLAG-SMAD7, determined by in vivo labeling using [3H]methionine and fluorography. FLAG-SMAD7 was immunoprecipitated and subjected to SDS-PAGE, staining, and autoradiography to visualize 3H incorporation. IP, immunoprecipitation; IB, immunoblotting.
Our previous findings on the role of PRMT1 in BMP signaling led to the model that PRMT1 associates with the BMP type II receptor, whereas SMAD6 binds to the BMP type I receptor, and that BMP-induced receptor heteromerization promotes PRMT1 interaction with SMAD6 and consequently SMAD6 methylation by PRMT1 (17). We hypothesized that a similar model holds for PRMT1-mediated SMAD7 methylation in TGF-β signaling and therefore examined the interaction of PRMT1 with the TGF-β type II receptor (TβRII) and TβRI. In transfected 293T cells, PRMT1 interacted with TβRII but not with the WT type I receptor (Fig. 2D, lanes 1 and 2). Thr-204 replacement by Asp is known to confer a partial activation of TβRI, leading to SMAD2/3 activation (19), which is thought to depend on autocrine TGF-β binding to endogenous TβRII. In contrast to the lack of PRMT1 association with WT TβRI, PRMT1 associated with the mutant constitutively activated TβRI (TβRIca) (Fig. 2D, lane 4), which may result from its association with endogenous TβRII. Coexpression of TβRII and TβRIca, thus promoting TβRII–TβRI heteromerization, illustrated the predominant association of PRMT1 with TβRII in the receptor complexes (Fig. 2D, lane 5). This scenario is similar to the recruitment of PRMT1 to the heteromeric BMP receptor complexes, although no PRMT1 association with the activated BMPRI was seen (17). Consistent with this scenario of PRMT1 interaction with TβRII and its recruitment in the TβRII–TβRI complex, introduction of the TβRIca mutant in HaCaT cells resulted in enhanced SMAD7 methylation (Fig. 2E).
PRMT1 methylates SMAD7 on Arg-57 and Arg-67
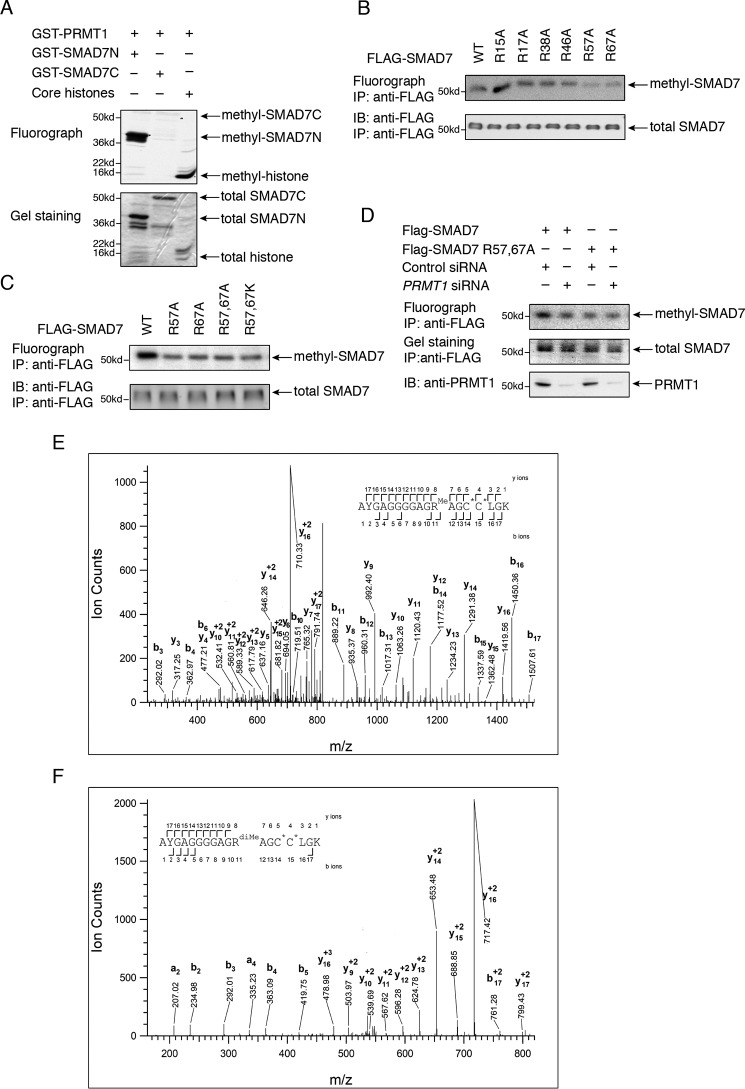
To define the sites in SMAD7 that are methylated by PRMT1, we first studied SMAD7 methylation in vitro. Deletion analyses revealed that PRMT1 methylates SMAD7 in its N-terminal region that spans amino acids 1–89 but not in the remaining large segment that comprises its MH2 domain and spans amino acids 90–427 (Fig. 3A). As a control for the activity of PRMT1 in vitro, PRMT1 methylated histone 4 (Fig. 3A, lane 3).
Figure 3.
PRMT1 methylates SMAD7. A, PRMT1 methylates SMAD7 in vitro in the N-terminal 89 amino acid sequence (SMAD7N) but not in the remaining segment spanning amino acids 90–427 (SMAD7C). GST-SMAD7N and GST-SMAD7C or core histones were incubated with PRMT1 in the presence of [3H]SAM, separated by SDS-PAGE, and visualized by GelCode Blue staining and 3H radiography. B, replacement of Arg-57 (R57A) or Arg-67 (R67A), but not other arginines, with alanine decreased SMAD7 methylation in vivo. 293T cells expressing WT or mutant FLAG-tagged SMAD7 were labeled in the presence of [3H]methionine, immunoprecipitated with anti-FLAG antibody, and visualized by SDS-PAGE, gel staining, and 3H autoradiography. C, replacement of Arg-57 and/or Arg-67 with alanine or lysine decreased SMAD7 methylation in vivo as determined in B using [3H]methionine labeling and autoradiography. D, down-regulating PRMT1 expression using transfected siRNA decreased the methylation of FLAG-tagged WT SMAD7 but not the double-mutant SMAD7. WT or mutant SMAD7 was transfected in 293T cells with PRMT1 siRNA or control siRNA, labeled using [3H]methionine, immunoprecipitated with anti-FLAG antibody, and visualized by SDS-PAGE, gel staining, and 3H autoradiography. E and F, CID tandem MS identified Arg-57 monomethylation (E) and dimethylation (F) in 293T cells expressing FLAG-SMAD7. Shown are CID tandem mass spectra obtained from precursor ions with m/z 827.3800+2 (E) or 556.5971+3 (F), corresponding to the mono- and dimethylated Arg-57 forms of the peptide spanning residues Ala-47 to Lys-64 of SMAD7. Theoretical masses and measured mass errors were: 827.3828+2, 3.4 ppm (E) and 556.5962+3, 1.6 ppm (F). The observed sequence ions are labeled in the figure and over the sequence. C*, carbamidomethyl cysteine residues; IP, immunoprecipitation; IB, immunoblotting.
To identify the residues of SMAD7 that are methylated by PRMT1 in vivo, we carried out mutagenesis analyses, focusing on Arg residues that are adjacent to Gly and thus represent the preferred methylation sites of PRMT1 (20). SMAD7 mutants, in which each of these arginines within the first 89 amino acids was individually replaced by alanine, were expressed in 293T cells, and their methylation was evaluated by in vivo [3H]methylation and immunoprecipitation (Fig. 3B). These analyses revealed that the R57A and R67A substitutions strongly decreased the methylation of SMAD7 (Fig. 3B), suggesting that these two arginine residues may be methylated. Substitutions of both Arg-57 and Arg-67 with alanine or lysine confirmed the decreased SMAD7 methylation when compared with WT SMAD7 but did not show a cumulative decrease of [3H]methylation (Fig. 3C). Furthermore, silencing PRMT1 expression did not further decrease the [3H]methylation of the double-mutant SMAD7 and kept it at a level comparable with the [3H]methylation of WT SMAD7 following PRMT1 silencing (Fig. 3D).
We also analyzed by MS the methylation of SMAD7 that was expressed in transfected 293T cells treated with TGF-β for 1 h and isolated by immunoaffinity purification. Mono- and dimethylation of Arg-57 of SMAD7 was detected in multiple independent samples (Fig. 3, E and F). However, we did not detect methylation of Arg-67 in these assays. This inability to detect Arg-67 methylation has limited value as a negative result but may suggest that in our methylation assays (Fig. 3, B–D) Arg-67 is required to enable Arg-57 methylation or that Arg-67 methylation occurs at a later stage of TGF-β signaling, i.e. after 1 h. MS also revealed lysine methylation of SMAD7, which is the subject of a separate study (data not shown), and explains the persistent [3H]methylation of SMAD7 after silencing PRMT1 expression (Fig. 3D).
Arg methylation regulates SMAD7 binding to TβRI
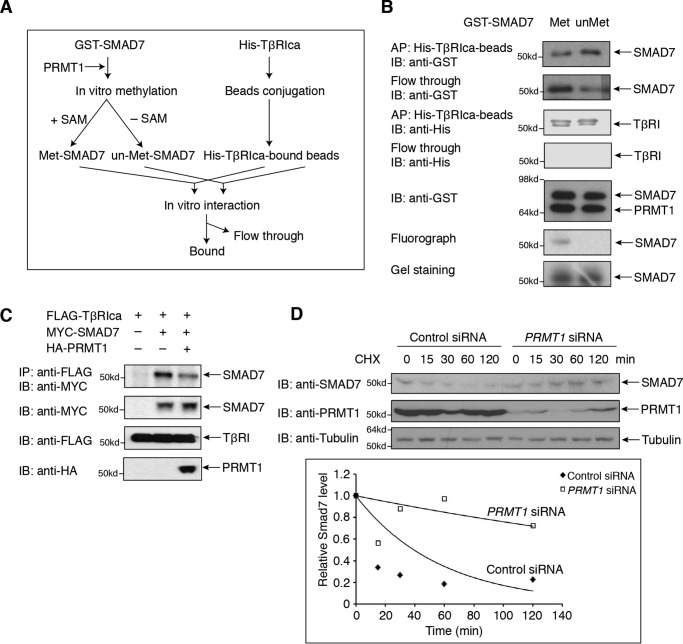
Further drawing on the parallel with the role of SMAD6 methylation in BMP signaling, we reported that BMP-induced methylation of SMAD6 decreases the SMAD6 association with the BMPRI, thus enabling SMAD6 dissociation from the activated BMPRI (17). By analogy, we examined whether arginine methylation by PRMT1 decreases the association efficiency of SMAD7 for TβRI using an in vitro binding assay (Fig. 4A). For this purpose, purified GST-SMAD7, methylated in vitro by PRMT1, was compared with unmethylated GST-SMAD7 for binding to immobilized, His-tagged TβRIca with its activating T204D mutation that was purified from transfected 293T cells (Fig. 4B). Unmethylated SMAD7 showed efficient binding to TβRIca (Fig. 4B, right lane), whereas methylated SMAD7 associated with a lower efficiency, apparent by the higher abundance of SMAD7 in the unbound fraction (Fig. 4B, left lane). These results provide evidence that arginine methylation decreases the binding of SMAD7 to the activated TβRI as was seen for the binding of SMAD6 to BMPRI (17). These results therefore suggest that SMAD7 methylation by PRMT1 enables SMAD7 dissociation from the activated TβRI receptor, similar to the effect of SMAD6 methylation leading to SMAD6 dissociation from the BMPRI receptor. Supporting this notion, increasing PRMT1 expression decreased the interaction of SMAD7 with TβRIca (Fig. 4C), again as observed for the binding of SMAD6 to BMPRI (17).These results support the model that the TGF-β–induced methylation of SMAD7 by TβRII-associated PRMT1 results in dissociation of SMAD7 from the TβRI receptor and thus controls the availability of TβRI receptor for subsequent SMAD2 and SMAD3 activation, similar to the BMP4-induced methylation of BMPRI-associated SMAD6 by BMPRII-associated PRMT1 to allow SMAD1 and SMAD5 activation in response to BMP (17). Protein methylation often alters protein stability, thus controlling the half-life of proteins. To determine whether Arg methylation controls the stability of SMAD7, we examined the half-life of SMAD7 in cycloheximide chase experiments in the presence and absence of PRMT1 (Fig. 4D). The estimated half-life of SMAD7 in control HaCaT cells was 38 min, whereas, following PRMT1 silencing, the estimated half-life was extended to 4 h, indicating that PRMT1-mediated methylation promotes degradation of SMAD7. Together, these data suggest that TGF-β–induced methylation of SMAD7 by TβRII-associated PRMT1 results in dissociation of SMAD7 from the TβRI receptor and subsequent degradation of SMAD7.
Figure 4.
SMAD7 methylation decreases the association with the TβRI receptor. A, flow chart of the experiments aimed at evaluating the effect of PRMT1-mediated methylation of SMAD7 on the association of SMAD7 with TβRIca. Bacterially expressed GST-fused SMAD7 was purified using GSH-Sepharose and incubated or not with purified PRMT1 in the presence or absence of [3H]SAM, thus generating Arg-methylated (Met) or unmethylated (un-Met) GST-SMAD7. In parallel, His-tagged TβRIca expressed in 293T cells was immunoprecipitated and coupled to Ni-Sepharose. The methylated and unmethylated GST-SMAD7 were then incubated with the Sepharose-bound TβRIca. Both the bound GST-SMAD7 and nonbound SMAD7 were analyzed by immunoblotting. B, results of the experiment shown in A. Methylation by PRMT1 decreases the binding efficiency of GST-SMAD7 for TβRIca. With equal amounts of GST-SMAD7 used, methylation by PRMT1 decreased the amount of SMAD7 bound to TβRIca, when compared with unmethylated SMAD7, and increased the fraction of nonbound SMAD7 (flow-through). The lower panels show methylation of SMAD7, assessed by 3H labeling. C, increased PRMT1 expression decreased the association of SMAD7 with TβRIca in 293T cells transfected to express FLAG-tagged TβRIca and MYC-tagged SMAD7. TβRIca association was revealed by immunoblotting. D, down-regulation of PRMT1 expression prolonged the half-life of SMAD7. HaCaT cells, transfected with control or PRMT1 siRNA for 48 h, were treated with 50 μg/ml cycloheximide (CHX) for the indicated times. SMAD7 was detected by immunoblotting, and its relative expression level was quantified by densitometry and plotted against time to determine the half-life of SMAD7. IB, immunoblotting; IP, immunoprecipitation.
PRMT1 controls TGF-β–induced epithelial cell dedifferentiation
TGF-β signaling is known to repress the epithelial-cell phenotype and, depending on the cell system and physiological conditions, to induce an EMT. In TGF-β–induced EMT, SMAD3 activates the expression of EMT master transcription factors such as Snail and cooperates with these transcription factors in the repression of epithelial genes and activation of mesenchymal genes (2). HaCaT cells are often used as model system to study the changes in gene expression during EMT. Considering the role of PRMT1 in the control of TGF-β–induced SMAD3 activation, we examined its role in TGF-β–induced EMT of HaCaT cells.
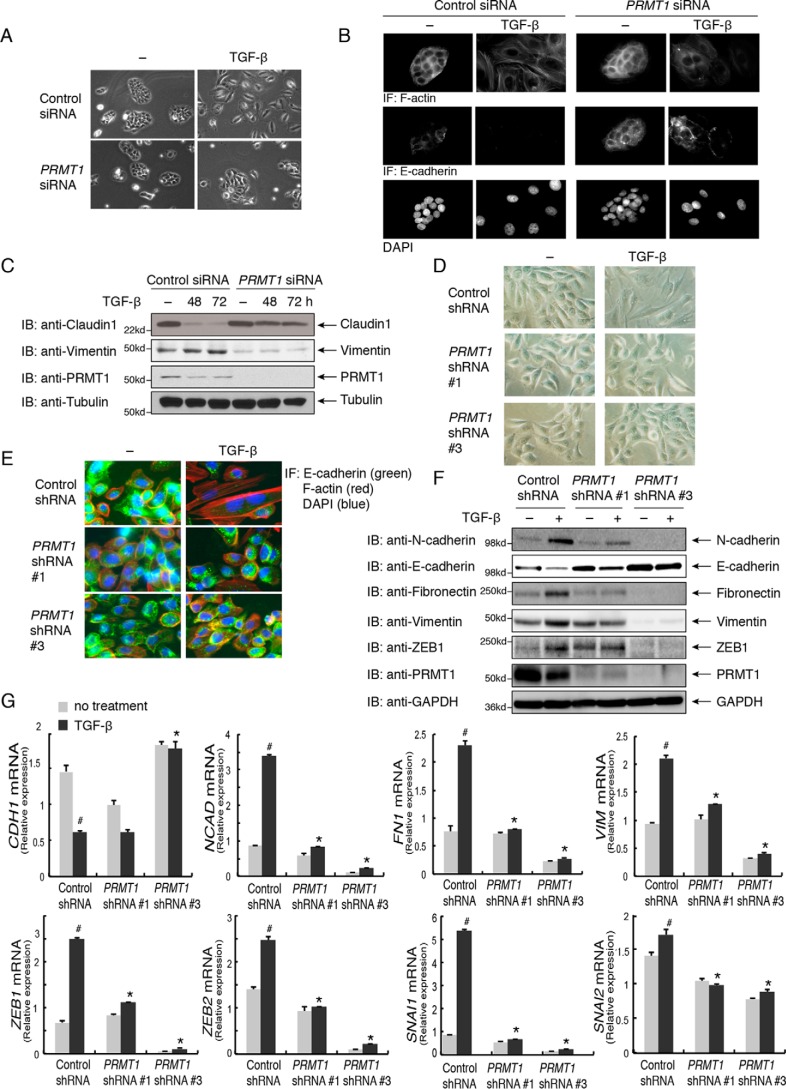
TGF-β induced a loss of the cobblestone-like cell shape that characterizes the epithelial identity and induced a more elongated EMT-like cell phenotype (Fig. 5A) with actin stress fibers rather than cortical actin organization, decreased E-cadherin immunostaining at cell junctions, decreased Claudin1 expression, and increased Vimentin expression (Fig. 5, B and C). Silencing PRMT1 expression using transfected siRNA inhibited this loss of epithelial phenotype (Fig. 5, A and B) and largely prevented these changes (Figs. 5, B and C, and S1).
Figure 5.
PRMT1 regulates TGF-β–induced EMT. A, repression of PRMT1 expression using transfected siRNA attenuated the TGF-β–induced EMT morphology of HaCaT cells. Cells transfected with PRMT1 siRNA or control siRNA were treated with TGF-β for 48 or 72 h to induce EMT or left untreated. B and C, depletion of PRMT1 expression using transfected siRNA prevented the TGF-β–induced change in actin organization and down-regulation of E-cadherin expression, shown by immunofluorescence (B), as well as the EMT-associated decrease in Claudin1 expression and increase in Vimentin expression, shown by immunoblotting (C). HaCaT cells were transfected with PRMT1 siRNA or control siRNA and treated with TGF-β as in A. D, down-regulating PRMT1 expression using lentiviral shRNA reduced the TGF-β–induced EMT morphology in HMLE cells. HMLE cells were infected with lentiviral vectors expressing one of two different PRMT1 shRNAs, shRNA 1 and shRNA 3, or a control shRNA and treated with TGF-β for 6 days to induce EMT. E and F, down-regulating PRMT1 expression using lentiviral shRNA reduced TGF-β–induced changes in EMT marker expression. The HMLE cells expressing PRMT1 shRNA or control shRNA were treated with TGF-β as in D. Down-regulation of PRMT1 expression reduced the actin reorganization into stress fibers and the junctional localization of epithelial E-cadherin, shown by immunofluorescence (IF) (E), and attenuated the decreased expression of epithelial E-cadherin and the increased expression of mesenchymal N-cadherin, Fibronectin, Vimentin, and ZEB1, shown by immunoblotting (IB) (F). G, down-regulating PRMT1 expression using lentiviral shRNA reduced the TGF-β–induced expression of EMT markers, assessed by qRT-PCR analyses of mRNA. HMLE cells expressing PRMT1 shRNA or control shRNA were treated with TGF-β as in D. Down-regulation of PRMT1 reduced the suppression of epithelial marker gene CDH1, which encodes E-cadherin, and induction of mRNAs encoding mesenchymal marker genes NCAD, FN1, VIM, ZEB1, ZEB2, SNAI1, and SNAI2. #, p < 0.01 versus no treatment; *, p < 0.01 versus control shRNA. Error bars represent S.D.
A similar role for PRMT1 was observed in the differentiation of HMLE cells. In these cells, TGF-β also induces a morphological transition from a cobblestone-like phenotype to a more elongated phenotype (Fig. 5D), although this transition in phenotype requires a longer time than in HaCaT cells. Silencing PRMT1 expression through lentiviral expression of either of two shRNAs inhibited this morphological transition (Fig. 5D) as well as the reorganization of actin from cortical to stress fibers and the down-regulation of E-cadherin at epithelial junctions (Fig. 5E). Consistent with these findings, TGF-β induced a significant decrease in E-cadherin expression and a significant increase in the levels of the mesenchymal markers ZEB1 and ZEB2, Snail and Slug, Vimentin, Fibronectin, and N-cadherin, and silencing PRMT1 expression dramatically inhibited these changes (Fig. 5, F and G).
PRMT1 controls TGF-β–induced stem-cell generation
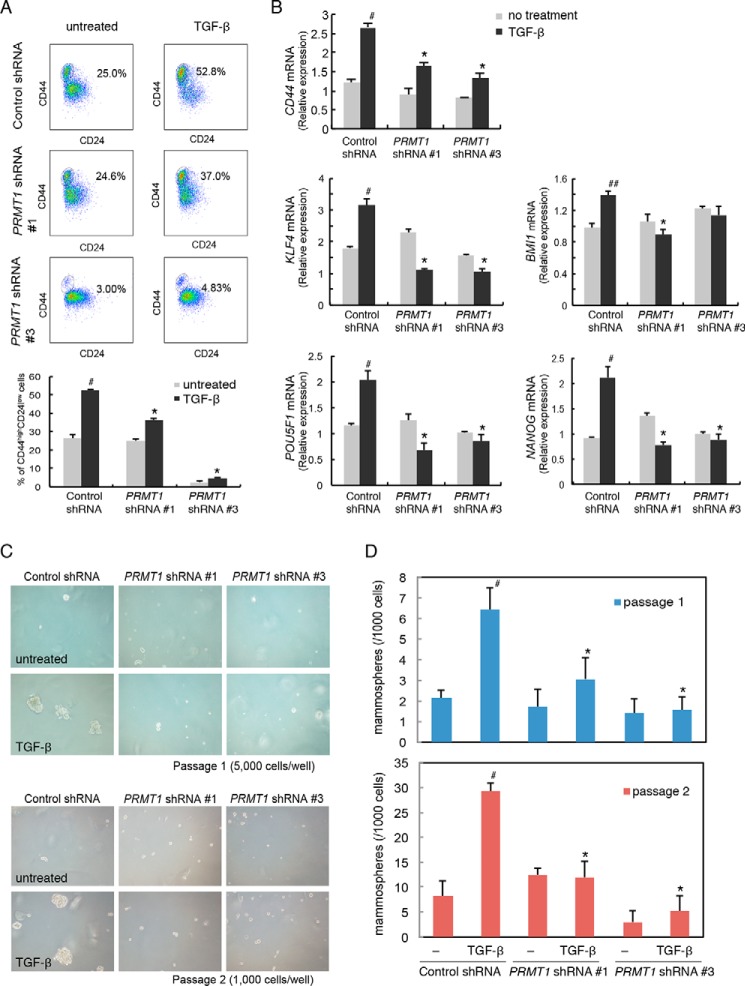
The acquisition of mesenchymal characteristics has been linked to the generation of epithelial and carcinoma stem cells, which, in the context of cancer progression, promotes the cancer reseeding capacity (3, 4). HMLE cells have been used as a model system to study the generation of epithelial stem cells and its correlation with EMT (10). Adding to our characterization of EMT marker expression (Fig. 5, E–G), we evaluated the CD44highCD24low cell population, which is the signature for epithelial stem cells. TGF-β treatment enhanced the percentage of CD44highCD24low cells from 25.0 to 52.8%, but silencing PRMT1 expression dramatically inhibited this increase (Fig. 6A). We further evaluated the expression of CD44, KLF4, BMI1, POU5F1, and NANOG, which correlate with stemness and pluripotency in normal and malignant mammary epithelial stem cells (21–23). TGF-β induced the mRNA expression of these markers, but silencing PRMT1 expression dramatically inhibited this induction (Fig. 6B). The repression of CD44 mRNA as a result of silencing PRMT1 expression correlated, both quantitatively and over time, with a concomitant repression of induced mesenchymal marker expression, specifically of N-cadherin, Fibronectin, and ZEB1 mRNAs (Figs. 6B and 5, F and G), which are encoded by direct TGF-β/SMAD3 target genes. Consistent with the repression of SMAD3 activation following silencing of PRMT1 expression (Fig. 1D), silencing PRMT1 decreased the expression of PAI1 and SMAD7 mRNAs, which are also transcribed from direct TGF-β/SMAD–responsive genes (Fig. 1, E and F).
Figure 6.
PRMT1 controls stem-cell generation. A, FACS analysis for the expression of the cell-surface markers CD44 and CD24 in HMLE cells infected with lentiviral vectors expressing PRMT1 shRNA 1 or 3 or control shRNA. HMLE cells were infected with lentiviral shRNA vectors and treated with TGF-β for 6 days. Down-regulating PRMT1 expression decreased the TGF-β–induced enhancement of the CD44high/CD24low population. The fractions of the CD44high/CD24low cell populations against the total viable cell population are shown in the panels and the graph below the panels. #, p < 0.01 versus untreated; *, p < 0.01 versus control shRNA. B, down-regulating PRMT1 expression using lentiviral shRNA reduced the TGF-β–induced expression of stemness markers, assessed by qRT-PCR analyses of mRNA. HMLE cells, infected with lentiviral constructs as shown, were treated with TGF-β as in A. Down-regulation of PRMT1 reduced the induction of stem-cell marker gene CD44 and the pluripotency genes POU5F1, NANOG, KLF4, SOX2, and BMI1. #, p < 0.01 versus no treatment; ##, p < 0.05 versus no treatment; *, p < 0.01 versus control shRNA. C and D, down-regulating PRMT1 expression using lentiviral shRNA reduced mammosphere formation. HMLE cells were infected with lentiviral shRNA constructs and treated with TGF-β as in A. Down-regulation of PRMT1 reduced mammosphere formation in primary and secondary passages. #, p < 0.01 versus untreated; *, p < 0.01 versus control shRNA. Error bars represent S.D.
To examine whether PRMT1 expression controls the functional properties of these stem-like cells, we tested their efficiency of mammosphere formation. The ability to form mammospheres in serial nonadherent passages correlates with the number of stem cells that have the ability to self-renew and thus reconstruct the gland structure (24). TGF-β treatment dramatically enhanced the efficiency of mammosphere formation of HMLE cells, and silencing PRMT1 expression almost completely blocked the enhancement in primary passages (Fig. 6, C and D). The requirement for PRMT1 was well maintained in secondary passages because silencing of PRMT1 expression prevented the TGF-β–induced mammosphere formation (Fig. 6, C and D). These results strongly suggest that PRMT1 controls the generation of mammary epithelial stem cells. Considering the role of PRMT1 in the dissociation of SMAD7 from TβRI and, consequently, in TGF-β–induced SMAD3 activation and considering the close correlation of the inhibition of stem-cell generation with the inhibition of TGF-β target gene expression and EMT, our data strongly suggest that PRMT1's control of stem-cell generation occurs through its effects on TGF-β–induced SMAD activation.
Discussion
SMAD6 and SMAD7 are seen as negative feedback regulators that constrain the activities of receptor-activated SMAD signaling. These inhibitory SMADs were shown to bind to effector SMADs and to the type I receptors, thus preventing the activation of the effector SMADs through C-terminal phosphorylation by the type I receptor kinases. SMAD6 is known to inhibit BMP-induced SMAD activation, whereas SMAD6 and SMAD7 both target TGF-β–, activin-, and BMP-induced SMAD signaling (13). We previously reported that SMAD6 associates with the cell-surface BMP type I receptors and that PRMT1 associated with the BMP type II receptor methylates SMAD6 on Arg-74 in response to ligand and consequently induces its dissociation. Thus, BMP-induced SMAD6 methylation by PRMT1 initiates BMP-induced SMAD1 and SMAD5 signaling by enabling the dissociation of the inhibitory SMAD6 and allowing the recruitment and activation of the effector SMADs SMAD1 and SMAD5 (17). Here, we provide evidence that a similar and parallel mechanism controls the function of SMAD7 at the TGF-β receptor and, thus, TGF-β–induced SMAD activation. Taken together, PRMT1 acts on inhibitory SMADs to control both the BMP and TGF-β/activin pathways and to enable effector SMAD activation by the type I receptors. We also show that, in addition to SMAD7, TGF-β induces SMAD6 methylation on Arg, supporting the notion that both inhibitory SMADs control TGF-β–induced SMAD activation.
We additionally provide evidence that PRMT1 controls the EMT program using two epithelial cell lines. With TGF-β/SMAD signaling driving the initiation and progression of this transdifferentiation program, we surmise that TGF-β signaling, through SMAD7 methylation by PRMT1, is a strong determinant of EMT. Indeed, silencing PRMT1 expression, and thus inhibiting TGF-β–induced SMAD3 activation, suppresses the EMT program. PRMT1 is a versatile enzyme that methylates histone and nonhistone substrates, raising the possibility that it acts at multiple levels. Accordingly, PRMT1 has been shown to modulate the roles of the EMT transcription factors Twist and ZEB1. PRMT1-mediated methylation of Twist contributes to the repression of epithelial E-cadherin expression in non-small-cell lung cancer cells (25), whereas PRMT1-mediated dimethylation of histone 4 at Arg-3 at the ZEB1 promoter activates ZEB1 expression in breast cancer cells, thus contributing to EMT (26). Therefore, PRMT1 may act at multiple levels in epithelial-to-mesenchymal transition.
The EMT program has been functionally linked to the generation of mammary epithelial stem cells and cancer stem cells. The EMT transcription factor Slug/Snail2 marks the basal mammary stem cells and cooperates with Sox9 to maintain the gland-reconstituting capacity of mammary epithelial cells (8). The expression of Snail, an EMT transcription factor related to Slug, marks the neoplastic population and is tightly associated with the tumor-invasive front (9). We illustrate the role of PRMT1 in both the EMT fate transition and the generation of mammary epithelial stem cells.
Increased PRMT1 expression has been documented in various types of cancer, including breast cancer. Its role in cancer progression has been linked to enhanced cell proliferation in breast, lung, liver, and colorectal cancer; squamous cell carcinomas; and leukemia (27–32). Although the underlying mechanisms remain to be further characterized, it has been proposed that PRMT1 methylates the EGF receptor to promote colorectal cancer growth (28). Also, PRMT1 was shown to methylate a splicing isoform of the AML1-ETO fusion protein that acts as a transcription factor, thus facilitating the expression of target genes while also epigenetically controlling their methylation on histone 4 (31). PRMT1 also regulates cellular functions such as senescence and genomic stability (26, 33).
Besides its roles in cancer, PRMT1 also controls development and tissue injury, which is consistent with its role as a histone methyltransferase and its ability to methylate a variety of signaling effectors (14, 15). Consequently, many activities of PRMT1 in development may not be related to TGF-β family signaling, and others might involve the methylation of inhibitory SMADs. For example, PRMT1 inactivation in periodontal epithelium aggravates inflammatory responses and periodontal tissue injury, and these effects appear to relate to the role of PRMT1-mediated SMAD6 methylation in repressing TLR–MyD88–NF-κB signaling (34). Whether SMAD7 plays a parallel role needs to be assessed. PRMT1 deletion in neural progenitors causes hypomyelination and defects in the central nervous system (35), which may in part relate to the roles of TGF-β family signaling in neural and neuronal-cell differentiation (36). Because the TGF-β family pathway proteins TGF-β and BMP, as well as SMAD7, are also involved in craniofacial development (36, 37), PRMT1-mediated SMAD7 methylation may contribute to developmental defects resulting from PRMT1 inactivation (38). Our evidence that SMAD7 methylation by PRMT1 enables TGF-β signaling to regulate EMT and stemness of mammary epithelial cells now provides an additional role for PRMT1 in promoting EMT and cancer stem-cell formation that may be relevant in cancer-cell dissemination and raises the expectation of normal roles in cell differentiation and development.
Experimental procedures
Plasmids
The expression plasmids pRK5-FLAG-PRMT1, pRK5-HA-PRMT1, pRK5-FLAG-TβRII, pRK5-FLAG-TβRIWT, pRK5-FLAG-TβRIca, pRK5-His-TβRIca, GST-PRMT1, GST-SMAD7N, GST-SMAD7C, pRK5-FLAG-SMAD7, and pRK-MYC-SMAD7 have been described (17, 39–41). Substitutions in the SMAD7 coding sequence were introduced using the QuikChange Lightening site-directed mutagenesis kit from Agilent, thus generating the expression plasmids for SMAD7 mutants, pRK5-FLAG-SMAD7 R57A, pRK5-FLAG-SMAD7 R67A, pRK5-FLAG-SMAD7 R57A,R67A, pRK5-FLAG-SMAD7 R57K,R67K,pRK5-FLAG-SMAD7 R15A, pRK5-FLAG-SMAD7 R17A, pRK5-FLAG-SMAD7 R38A, and pRK5-FLAG-SMAD7 R46A.
Reagents
Control and PRMT1 siRNAs were purchased from Qiagen. A mixture of two independent siRNAs, HRMT1L2_4 and HRMT1L2_7, was used to silence PRMT1 (17). Control shRNA and shRNAs targeting PRMT1 were purchased from Sigma and described previously (17). The 293T stable cells expressing control shRNA or shRNA targeting PRMT1 were described previously (17). Recombinant human TGF-β1 was purchased from Humanzyme. SB431542, cycloheximide, chloroquine, and MG132 were purchased from Sigma. Core histones were purchased from Millipore.
Cell culture
HaCaT and 293T cells were maintained in DMEM with 10% fetal bovine serum. Immortalized HMLE cells were maintained in mammary epithelial cell basal medium (Lonza) supplemented with 10 μg/ml insulin, 10 ng/ml EGF, and 0.5 μg/ml hydrocortisone.
Antibodies, immunoprecipitation, and immunoblotting
Antibodies to PRMT1, phospho-SMAD3, SMAD3, Claudin1, Snail, E-cadherin, and N-cadherin were purchased from Cell Signaling Technology. Antibodies to SMAD7 were purchased from Abcam and Santa Cruz Biotechnology. Antibodies to Vimentin, Fibronectin, and the FLAG, MYC, His, and HA epitope tags were purchased from Sigma. Antibodies to GST, tubulin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Santa Cruz Biotechnology. FLAG-conjugated M2 agarose and HA-conjugated Sepharose beads were purchased from Sigma-Aldrich. Rabbit polyclonal antibodies against asymmetric dimethyl-Arg-74 of SMAD6 (anti-S6R74me2) and asymmetric dimethyl-Arg-81 of SMAD6 (anti-S6R81me2) were characterized previously (17).
For immunoblotting, cells were lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.1% NP-40, 10% glycerol, and protease inhibitor mixture). Proteins were quantified using a Bio-Rad protein assay, and 20–80 μg of protein was separated by SDS-PAGE and transferred to 0.45-μm polyvinylidene difluoride (PVDF) membrane. Membranes were blocked in Tris-buffered saline with Tween 20 and 5% BSA (blocking solution) for 1 h followed by overnight incubation with primary antibody diluted at 1:500–1:5,000 in blocking solution and 1-h incubation with horseradish peroxidase–conjugated secondary antibody diluted at 1:5,000–1:20,000. Immunoreactive protein was detected using ECL (GE Healthcare) and film.
For immunoprecipitation of transfected proteins, 293T cells were harvested at 24 or 48 h after transfection and lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.1% NP-40, 10% glycerol, and protease inhibitor mixture). Lysates were subjected to immunoprecipitation with anti-MYC or anti-HA antibody and conjugation to protein G–Sepharose (GE Healthcare). Immune complexes were washed three times with immunoprecipitation wash buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.1% NP-40, and 10% glycerol) and subjected to immunoblotting.
Reversed-phase LC–electrospray tandem MS (LC-MS/MS) analysis
In-solution digestion
FLAG-SMAD7 (7–10 μg) eluted from the antibody column in 100 mm NH4HCO3 was concentrated by evaporation in a SpeedVac system, and the volume was adjusted to 25 μl. Samples were partially denatured by adding urea to a concentration of 2.7 m and reduced by incubating for 15 min at 60 °C in the presence of 2.5 mm DTT. Samples were then alkylated by incubation with 3 mm iodoacetamide for 1 h in the dark at room temperature. Remaining iodoacetamide was quenched by adding DTT to a final concentration of 3 mm and incubating at 37 °C for 15 min. Samples were then diluted to a final concentration of urea of 2 m and digested overnight at 37 °C using 100 ng of sequencing-grade modified trypsin (Promega, Madison, WI). Formic acid (4%) was added to the samples, and peptides were extracted using C18 OMIX tips (Agilent Technologies) according to the manufacturer's protocol. Eluates from the OMIX tips were vacuum-evaporated and resuspended in 20 μl of 0.1% formic acid in water.
Reverse-phase LC-MS/MS analysis
The digests were separated by nanoflow LC using a 75-μm × 150-mm reverse-phase 1.7-μm BEH130 C18 column (Waters) at a flow rate of 600 nl/min in a NanoAcquityTM ultraperformance UPLC system (Waters). Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. Following equilibration of the column in 5% solvent B, an aliquot of each digest (5 μl) was injected, and then the organic content of the mobile phase was increased linearly to 40% over 60 min and then to 50% in 1 min. The LC eluate was coupled to a hybrid linear ion trap-Orbitrap mass spectrometer (either an LTQ-Orbitrap XL or an LTQ-Orbitrap Velos, Thermo Scientific, San Jose, CA) equipped with a nanoelectrospray ion source. Spraying was from an uncoated 15-μm-inner-diameter spraying needle (New Objective, Woburn, MA). Peptides were analyzed in positive ion mode and in information-dependent acquisition mode to automatically switch between MS and MS/MS acquisition. MS spectra were acquired in profile mode using the Orbitrap analyzer in the m/z range between 300 and 1,800. For each MS spectrum, the six most intense multiple-charged ions over a threshold of 1,000 counts were selected to perform CID experiments. Product ions were analyzed on the linear ion trap in centroid mode. The CID collision energy was automatically set to 30%. A dynamic exclusion window of 0.5 Da was applied that prevented the same m/z from being selected for 60 s after its acquisition.
Peak lists were generated using PAVA in-house software (42). The peak lists were searched against the murine and human subsets of the Swiss-Prot database as of July 6, 2011 using in-house ProteinProspector version 5.8.0 (a public version is available online). Peptide tolerance in searches was 20 ppm for precursor and 0.8 Da for product ions. Peptides containing two miscleavages were allowed. Carbamidomethylation of cysteine was allowed as a constant modification; acetylation of the N terminus of the protein, pyroglutamate formation from N-terminal glutamine, and oxidation of methionine were allowed as variable modifications in initial searches. Some searches allowed for mono- and dimethylation of arginine and acetylation and mono-, di-, and trimethylation of lysine as variable modifications. In these cases, systematic error of the peptides masses observed on the initial searches was used to correct the search parameters. The number of modifications was limited to two per peptide. Protein hits were considered significant when two or more peptide sequences matched a protein entry and the Prospector score was above the significance level. A minimal ProteinProspector protein score of 20, a peptide score of 15, a maximum expectation value of 0.05, and a minimal discriminant score threshold of 0.0 were used for initial identification criteria. For identifications of posttranslational modification sites, the MS/MS spectrum was reinterpreted manually by matching all the observed fragment ions to a theoretical fragmentation obtained using MS Product (ProteinProspector) (43).
In vitro GST binding assays
Nonmethylated recombinant GST-tagged SMAD7 was generated in Escherichia coli, which is deficient in Arg methylation. Immunopurified GST-tagged SMAD7 was methylated in vitro using purified recombinant PRMT1 in the presence or absence of methylation cofactor S-adenosylmethionine (SAM). His-tagged TβRIca was purified from transfected 293T cells and conjugated to anti-His antibody–conjugated Sepharose beads. Then methylated or nonmethylated SMAD7 was incubated with His-tagged TβRIca for 2–4 h with agitation. After incubation, the bead-bound and flow-through fractions were separated by centrifugation and subjected to SDS-PAGE followed by immunoblotting.
In vitro methylation assay
GST-tagged SMAD7, SMAD7 mutants, and SMAD7 N-terminal and C-terminal segments were generated in E. coli transformed with pGEX expression vectors and purified using GSH-Sepharose 4B (GE Healthcare). FLAG-tagged PRMT1 was expressed in 293T cells, immunopurified, and incubated with GST-SMAD7 or core histone (Millipore) in reaction buffer (50 mm Tris-HCl, pH 8, 20 mm KCl, 5 mm MgCl2, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 10% glycerol), in the presence of 2 μCi of 3H-labeled SAM (PerkinElmer Life Sciences) at 30 °C for 90 min. The reaction mixture was quenched with 5× SDS buffer and separated by SDS-PAGE. The gel was stained with GelCode Blue (Pierce), scanned (Canoscan 9000F), fluorographed, dried, and exposed to Eastman Kodak Co. film at −80 °C.
In vivo methylation assay
Transfected 293T cells were pretreated with the proteasomal degradation inhibitor MG132 (20–50 μm) and the lysosomal degradation inhibitor chloroquine (100 μm) for 2–6 h. The medium was replaced with DMEM without methionine containing the same inhibitors and supplemented with the protein synthesis inhibitor cycloheximide (100 μg/ml). After 30 min, 10 μCi/ml of l-[methyl-3H]methionine (PerkinElmer Life Sciences) was added and incubated with the cells for an additional 1–6 h. The cells were lysed in lysis buffer. The SMAD7 complexes were immunopurified with anti-FLAG M2 (Sigma-Aldrich) and separated by SDS-PAGE. The protein gel was then stained with GelCode Blue, scanned, fluorographed, dried, and exposed to Kodak film at −80 °C.
Immunocytochemistry
Cells plated on chamber slides were fixed with ice-cold methanol for 20 min, permeabilized in PBS containing 0.3% Triton X-100 (PBT) for 15 min, and incubated in PBT and 5% serum blocking solution for 1 h. The slides were incubated with anti-E-cadherin antibody at 4 °C overnight and then stained for 2 h with Alexa Fluor secondary antibodies (1:500; Invitrogen) or incubated with Alexa Fluor 488 or 568 phalloidin for 1–3 h. The slides were then mounted using Prolong Gold antifade reagent and stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei (Invitrogen). The cells were viewed with an inverted light microscope (DMI5000, Leica Microsystems) or a laser-scanning confocal microscope (SP5, Leica Microsystems).
qRT-PCR analysis
To quantify mRNA expression, cells were treated as indicated, and RNA was isolated with TRIzol (Invitrogen) and used as a template for reverse transcriptase. The mRNAs of interest were quantified by real-time PCR with IQ SYBR Green Supermix (Bio-Rad) and normalized against RPL19 mRNA. The primer sequences are listed in Table S1.
Flow cytometry
Antibodies for the human CD24 (Clone ML5) and human CD44 (Clone G44-26) were purchased from BD Biosciences. Cells were dissociated into single cells and stained with antibodies for CD24 and CD44 in PBS. Flow cytometry analysis was done using a Gallios flow cytometer (Beckman Coulter).
Mammosphere assay
Mammosphere formation assays were performed as described (10). Cells were seeded in ultralow-attachment plates at a density of 5,000 cells/well in mammary epithelial cell growth medium (Lonza) supplemented with B27, 10 ng/ml basic fibroblast growth factor, 20 ng/ml EGF, and 1% methyl cellulose. After incubating the cells for 6 days, the mammospheres were observed by phase-contrast microscopy and quantified. For secondary sphere formation, primary spheres were collected by centrifugation, dissociated into single cells by trypsinization, and replated in ultralow-attachment plates at 2,000 cells/well.
Author contributions
Y. K., J. O.-P., and J. X. formal analysis; Y. K., J. Q., J. O.-P., H. W., O. J.-W., T. Z., S. L., J. W., and J. X. investigation; Y. K., S. L., A. L. B., and J. X. methodology; Y. K., J. O.-P., and J. X. writing-original draft; A. L. B., J. X., and R. D. supervision; J. X. and R. D. conceptualization; J. X. and R. D. resources; R. D. funding acquisition; R. D. writing-review and editing.
Supplementary Material
Acknowledgments
The MS analysis was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at University of California San Francisco (A. L. Burlingame, Director) supported by the Biomedical Research Technology Program of the National Institutes of Health National Center for Research Resources under Grant 5P41RR001614-29; NIGMS, National Institutes of Health Grant 8 P41 GM103481-29; and the Howard Hughes Medical Institute.
This work was supported in part by NCI, National Institutes of Health Grants RO1-CA63101 and RO1-CA136690 (to R. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1 and Table S1.
- EMT
- epithelial-to-mesenchymal transdifferentiation
- TGF-β
- transforming growth factor β
- BMP
- bone morphogenetic protein
- PRMT
- protein arginine methyltransferase
- BMPR
- BMP receptor
- PAI1
- plasminogen activator inhibitor type 1
- HMLE
- human mammary epithelial
- TβR
- TGF-β receptor
- TβRIca
- constitutively activated TβRI
- TLR
- Toll-like receptor
- GST
- glutathione S-transferase
- DMEM
- Dulbecco's modified Eagle's medium
- EGF
- epidermal growth factor
- HA
- hemagglutinin
- NP-40
- Nonidet P-40
- DTT
- dithiothreitol
- CID
- collision-induced dissociation
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu J., Lamouille S., and Derynck R. (2009) TGF-β-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172 10.1038/cr.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polyak K., and Weinberg R. A. (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9, 265–273 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- 4. Shibue T., and Weinberg R. A. (2017) EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14, 611–629 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaffer C. L., Marjanovic N. D., Lee T., Bell G., Kleer C. G., Reinhardt F., D'Alessio A. C., Young R. A., and Weinberg R. A. (2013) Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61–74 10.1016/j.cell.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., Brunton V. G., Morton J., Sansom O., Schüler J., Stemmler M. P., et al. (2009) The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487–1495 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- 7. Hwang W. L., Jiang J. K., Yang S. H., Huang T. S., Lan H. Y., Teng H. W., Yang C. Y., Tsai Y. P., Lin C. H., Wang H. W., and Yang M. H. (2014) MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 16, 268–280 10.1038/ncb2910 [DOI] [PubMed] [Google Scholar]
- 8. Guo W., Keckesova Z., Donaher J. L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zürrer-Härdi U., Bell G., Tam W. L., Mani S. A., van Oudenaarden A., and Weinberg R. A. (2012) Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ye X., Tam W. L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E., and Weinberg R. A. (2015) Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., and Weinberg R. A. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oshimori N., Oristian D., and Fuchs E. (2015) TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 10.1016/j.cell.2015.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu P., Lin X., and Feng X. H. (2016) Posttranslational regulation of Smads. Cold Spring Harb. Perspect. Biol. 8, a022087 10.1101/cshperspect.a022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miyazawa K., and Miyazono K. (2017) Regulation of TGF-β family signaling by inhibitory Smads. Cold Spring Harb. Perspect. Biol. 9, a022095 10.1101/cshperspect.a022095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blanc R. S., and Richard S. (2017) Arginine methylation: the coming of age. Mol. Cell 65, 8–24 10.1016/j.molcel.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 16. Yang Y., and Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 10.1038/nrc3409 [DOI] [PubMed] [Google Scholar]
- 17. Xu J., Wang A. H., Oses-Prieto J., Makhijani K., Katsuno Y., Pei M., Yan L., Zheng Y. G., Burlingame A., Brückner K., and Derynck R. (2013) Arginine methylation initiates BMP-induced Smad signaling. Mol. Cell 51, 5–19 10.1016/j.molcel.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng X. H., and Derynck R. (2005) Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 10.1146/annurev.cellbio.21.022404.142018 [DOI] [PubMed] [Google Scholar]
- 19. Wieser R., Wrana J. L., and Massagué J. (1995) GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J. 14, 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thandapani P., O'Connor T. R., Bailey T. L., and Richard S. (2013) Defining the RGG/RG motif. Mol. Cell 50, 613–623 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- 21. Visvader J. E., and Stingl J. (2014) Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 28, 1143–1158 10.1101/gad.242511.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu F., Li J., Chen H., Fu J., Ray S., Huang S., Zheng H., and Ai W. (2011) Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 30, 2161–2172 10.1038/onc.2010.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Itoh F., Watabe T., and Miyazono K. (2014) Roles of TGF-β family signals in the fate determination of pluripotent stem cells. Semin. Cell Dev. Biol. 32, 98–106 10.1016/j.semcdb.2014.05.017 [DOI] [PubMed] [Google Scholar]
- 24. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., and Wicha M. S. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 10.1101/gad.1061803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Avasarala S., Van Scoyk M., Karuppusamy Rathinam M. K., Zerayesus S., Zhao X., Zhang W., Pergande M. R., Borgia J. A., DeGregori J., Port J. D., Winn R. A., and Bikkavilli R. K. (2015) PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J. Biol. Chem. 290, 13479–13489 10.1074/jbc.M114.636050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao Y., Zhao Y., Zhang J., Lu Y., Liu X., Geng P., Huang B., Zhang Y., and Lu J. (2016) The dual function of PRMT1 in modulating epithelial-mesenchymal transition and cellular senescence in breast cancer cells through regulation of ZEB1. Sci. Rep. 6, 19874 10.1038/srep19874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chuang C. Y., Chang C. P., Lee Y. J., Lin W. L., Chang W. W., Wu J. S., Cheng Y. W., Lee H., and Li C. (2017) PRMT1 expression is elevated in head and neck cancer and inhibition of protein arginine methylation by adenosine dialdehyde or PRMT1 knockdown downregulates proliferation and migration of oral cancer cells. Oncol. Rep. 38, 1115–1123 10.3892/or.2017.5737 [DOI] [PubMed] [Google Scholar]
- 28. Nakai K., Xia W., Liao H. W., Saito M., Hung M. C., and Yamaguchi H. (2018) The role of PRMT1 in EGFR methylation and signaling in MDA-MB-468 triple-negative breast cancer cells. Breast Cancer 25, 74–80 10.1007/s12282-017-0790-z [DOI] [PubMed] [Google Scholar]
- 29. Zhou W., Yue H., Li C., Chen H., and Yuan Y. (2016) Protein arginine methyltransferase 1 promoted the growth and migration of cancer cells in esophageal squamous cell carcinoma. Tumour Biol. 37, 2613–2619 10.1007/s13277-015-4098-3 [DOI] [PubMed] [Google Scholar]
- 30. Liao H. W., Hsu J. M., Xia W., Wang H. L., Wang Y. N., Chang W. C., Arold S. T., Chou C. K., Tsou P. H., Yamaguchi H., Fang Y. F., Lee H. J., Lee H. H., Tai S. K., Yang M. H., et al. (2015) PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J. Clin. Investig. 125, 4529–4543 10.1172/JCI82826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shia W. J., Okumura A. J., Yan M., Sarkeshik A., Lo M. C., Matsuura S., Komeno Y., Zhao X., Nimer S. D., Yates J. R. 3rd, and Zhang D. E. (2012) PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 119, 4953–4962 10.1182/blood-2011-04-347476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng S., Moehlenbrink J., Lu Y. C., Zalmas L. P., Sagum C. A., Carr S., McGouran J. F., Alexander L., Fedorov O., Munro S., Kessler B., Bedford M. T., Yu Q., and La Thangue N. B. (2013) Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol. Cell 52, 37–51 10.1016/j.molcel.2013.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Z., Chen T., Hébert J., Li E., and Richard S. (2009) A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol. Cell. Biol. 29, 2982–2996 10.1128/MCB.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang T., Wu J., Ungvijanpunya N., Jackson-Weaver O., Gou Y., Feng J., Ho T. V., Shen Y., Liu J., Richard S., Jin J., Hajishengallis G., Chai Y., and Xu J. (2018) Smad6 methylation represses NFκB activation and periodontal inflammation. J. Dent. Res. 97, 810–819 10.1177/0022034518755688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hashimoto M., Murata K., Ishida J., Kanou A., Kasuya Y., and Fukamizu A. (2016) Severe hypomyelination and developmental defects are caused in mice lacking protein arginine methyltransferase 1 (PRMT1) in the central nervous system. J. Biol. Chem. 291, 2237–2245 10.1074/jbc.M115.684514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krampert M., Chirasani S. R., Wachs F. P., Aigner R., Bogdahn U., Yingling J. M., Heldin C. H., Aigner L., and Heuchel R. (2010) Smad7 regulates the adult neural stem/progenitor cell pool in a transforming growth factor β- and bone morphogenetic protein-independent manner. Mol. Cell. Biol. 30, 3685–3694 10.1128/MCB.00434-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang S., Snider P., Firulli A. B., and Conway S. J. (2010) Trigenic neural crest-restricted Smad7 over-expression results in congenital craniofacial and cardiovascular defects. Dev. Biol. 344, 233–247 10.1016/j.ydbio.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gou Y., Li J., Wu J., Gupta R., Cho I., Ho T. V., Chai Y., Merrill A., Wang J., and Xu J. (2018) Prmt1 regulates craniofacial bone formation upstream of Msx1. Mech. Dev. 152, 13–20 10.1016/j.mod.2018.05.001 [DOI] [PubMed] [Google Scholar]
- 39. Choy L., Skillington J., and Derynck R. (2000) Roles of autocrine TGF-β receptor and Smad signaling in adipocyte differentiation. J. Cell Biol. 149, 667–682 10.1083/jcb.149.3.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muthusamy B. P., Budi E. H., Katsuno Y., Lee M. K., Smith S. M., Mirza A. M., Akhurst R. J., and Derynck R. (2015) ShcA protects against epithelial-mesenchymal transition through compartmentalized inhibition of TGF-β-induced Smad activation. PLoS Biol. 13, e1002325 10.1371/journal.pbio.1002325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu P., Bailey-Bucktrout S., Xi Y., Xu D., Du D., Zhang Q., Xiang W., Liu J., Melton A., Sheppard D., Chapman H. A., Bluestone J. A., and Derynck R. (2014) Innate antiviral host defense attenuates TGF-β function through IRF3-mediated suppression of Smad signaling. Mol. Cell 56, 723–737 10.1016/j.molcel.2014.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guan S., Price J. C., Prusiner S. B., Ghaemmaghami S., and Burlingame A. L. (2011) A data processing pipeline for mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol. Cell. Proteomics 10, M111.010728 10.1074/mcp.M111.010728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clauser K. R., Baker P., and Burlingame A. L. (1999) Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871–2882 10.1021/ac9810516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.