Abstract
Under fasting conditions, activation of several hepatic genes sets the stage for gluconeogenesis in the liver. cAMP response element–binding protein (CREB), CREB-regulated transcription coactivator 2 (CRTC2), and peroxisome proliferator–activated receptor γ coactivator 1-alpha (PGC-1α) are essential for this transcriptional induction of gluconeogenic genes. PGC-1α induction is mediated by activation of a CREB/CRTC2 signaling complex, and recent findings have revealed that small heterodimer partner–interacting leucine zipper protein (SMILE), a member of the CREB/ATF family of basic region–leucine zipper (bZIP) transcription factors, is an insulin-inducible corepressor that decreases PGC-1α expression and abrogates its stimulatory effect on hepatic gluconeogenesis. However, the molecular mechanism whereby SMILE suppresses PGC-1α expression is unknown. Here, we investigated SMILE's effects on the CREB/CRTC2 signaling pathway and glucose metabolism. We found that SMILE significantly inhibits CREB/CRTC2-induced PGC-1α expression by interacting with and disrupting the CREB/CRTC2 complex. Consequently, SMILE decreased PGC-1α–induced hepatic gluconeogenic gene expression. Furthermore, SMILE inhibited CREB/CRTC2-induced phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) gene expression by directly repressing the expression of these genes and by indirectly inhibiting the expression of PGC-1α via CREB/CRTC2 repression. Indeed, enhanced gluconeogenesis and circulating blood glucose levels in mice injected with an adenovirus construct containing a constitutively active CRTC2 variant (CRTC2–S171A) were significantly reduced by WT SMILE, but not by leucine zipper–mutated SMILE. These results reveal that SMILE represses CREB/CRTC2-induced PGC-1α expression, an insight that may help inform potential therapeutic approaches targeting PGC-1α–mediated regulation of hepatic glucose metabolism.
Keywords: cAMP response element-binding protein (CREB), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1a) (PPARGC1A), gluconeogenesis, transcription corepressor, basic helix-loop-helix transcription factor (bHLH), glucose metabolism, diabetes, small heterodimer partner interacting leucine zipper protein (SMILE)
Introduction
Under fasting conditions, protein kinase A (PKA) stimulates phosphorylation of CREB4 (serine 133) and dephosphorylation of the CRTCs to increase hepatic gluconeogenic gene expression in the liver (1–3). Knockdown of CRTC2 in liver decreases gluconeogenic gene expression as well as the blood glucose concentration (4). CREB indirectly induces gluconeogenic gene expression via induction of the nuclear receptor coactivator PGC-1α, a direct target of the CREB/CRTC2 pathway. On the other hand, CREB also induces gluconeogenic gene expression via its direct binding to the cAMP response element (CRE) site on the promoter of phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) (5). During refeeding conditions, insulin increases the phosphorylation and ubiquitin-dependent degradation of CRTC2 by induction of SIK2 (6). In the liver, PGC-1α increases the expression of PEPCK and G6Pase via direct interaction with transcription factors including hepatocyte nuclear factor 4α (HNF4α), FOXO1, and glucocorticoid receptor (GR) (7, 8). Conversely, knockdown or knockout of PGC-1α results in lower blood glucose levels as a result of reduced gluconeogenesis.
Small heterodimer partner–interacting leucine zipper protein (SMILE), including two alternative translation-derived isoforms, SMILE-L (CREBZF; long form of SMILE) and SMILE-S (Zhangfei; short form of SMILE), has been classified as a member of the CREB/ATF family of basic region–leucine zipper (bZIP) transcription factors. However, SMILE cannot bind to DNA as a homodimer (9–11). Previously, we reported that SMILE is a corepressor of the estrogen receptor–related receptor gamma (ERRγ), GR, constitutive androstane receptor (CAR), HNF4α, and cAMP responsive element-binding protein H (CREBH) (12–14). A recent study demonstrated that SMILE is an insulin-inducible corepressor that decreases not only the stimulatory effect of PGC-1α on hepatic gluconeogenesis but also PGC-1α expression (15). In this study, we examined the mechanism by which SMILE inhibits PGC-1α gene expression and reduces hepatic gluconeogenesis.
Results
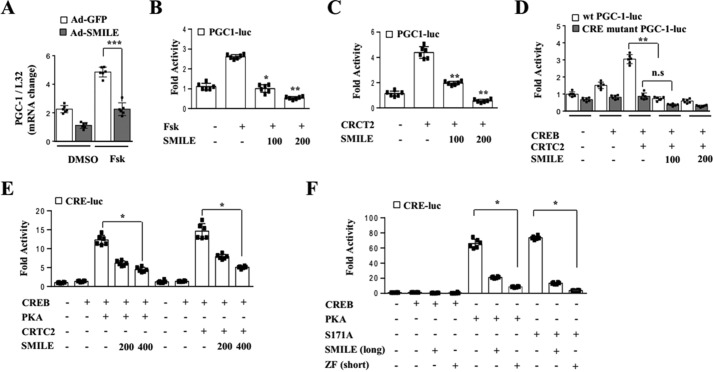
SMILE inhibits the induction of PGC-1α gene expression by CREB/CRTC2 signaling
A previous report has demonstrated that SMILE decreases blood glucose levels as well as PGC-1α gene expression in db/db and HFD-fed mice (15). Based on these findings, we hypothesized that SMILE would affect CREB/CRTC2-induced PGC-1α gene expression. To examine this hypothesis, we measured the effect of adenoviral overexpression of SMILE on PGC-1α gene expression in mouse primary hepatocytes. The long form of SMILE significantly inhibited forskolin-induced PGC-1α gene expression (Fig. 1A). Consistent with the decrease in PGC-1α gene expression, overexpression of SMILE significantly inhibited forskolin- or CRTC2-induced PGC-1α promoter activity (Fig. 1, B and C). Moreover, mutation of the CREB response element in the PGC-1α promoter completely blocked CREB/CRTC2-induced promoter activity and also diminished inhibitory effect of SMILE (Fig. 1D). To further confirm the inhibitory mechanism of SMILE in CREB/CRTC2-mediated PGC-1α gene expression, we measured CRE-Luc reporter activity. As expected, SMILE significantly inhibited PKA- and CRTC2-induced CRE-Luc reporter activity (Fig. 1E). Next, we examined whether the short form of SMILE also inhibits CREB/CRTC2 transcriptional activity. Indeed, the activation of the transcriptional activity of CREB by both PKA and constitutively active CRTC2 (S171A) was significantly prevented by the short form of SMILE (Fig. 1F). Taken together, these results indicate that SMILE inhibits the transcriptional activation of PGC-1α by CREB/CRTC2 signaling.
Figure 1.
SMILE decreases Pgc-1α gene expression via inhibition of CREB/CRTC2. A, Ad-GFP and Ad-SMILE were infected with primary hepatocytes and treated with forskolin. Expression of Pgc-1α was analyzed by quantitative PCR. B, AML12 cells were cotransfected with Pgc-1 promoter and SMILE expression vector. The cells were treated with vehicle or forskolin 24 h after the transfection. C, AML12 cells were cotransfected with Pgc-1–Luc, CRTC2, and SMILE expression vector. D, AML12 cells were cotransfected with WT Pgc-1–Luc, CRE site-mutated Pgc-1–Luc, CREB, CRTC2, and SMILE expression vector. E, AML12 cells were cotransfected with CRE-Luc, CREB, PKA, CRTC2, and SMILE. F, AML12 cells were cotransfected with CRE-Luc, PKA, S171A (constitutive active CRTC2), SMILE (long form), and ZF (short form of SMILE) expression vector. Error bars show mean ± S.D. n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.005 by two-tailed Student's t test. n.s. means not significant. Results are representative of three independently performed experiments.
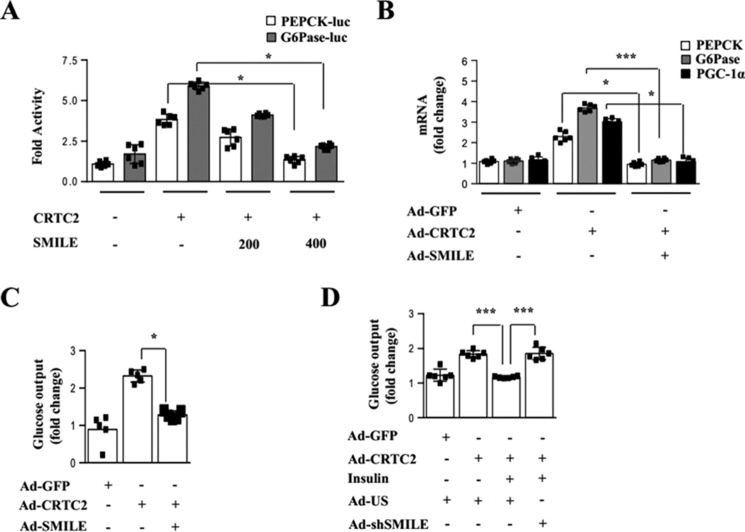
SMILE inhibits CREB/CRTC2-mediated hepatic gluconeogenesis in vitro
CREB stimulates the gluconeogenic program by binding directly to the promoters of the PEPCK and G6Pase genes (16, 17). Based on the ability of SMILE to decrease CREB/CRTC2-induced PGC-1α gene expression, we examined whether SMILE inhibits the stimulation of PEPCK and G6Pase genes expression by CREB/CRTC2. SMILE significantly inhibited stimulation of PEPCK and G6Pase promoter activity as well as gene expression of PEPCK, G6Pase, and PGC-1α in primary hepatocytes by CREB/CRTC2 signaling (Fig. 2, A and B). Consistent with decreases of gluconeogenic enzyme gene expression, SMILE markedly decreased CREB/CRTC2-induced hepatic glucose production in primary hepatocytes (Fig. 2C). Conversely, knockdown of SMILE prevented insulin-mediated repression of hepatic glucose production (Fig. 2D). Taken together, these results indicate that SMILE inhibits CREB/CRTC2-induced PEPCK and G6Pase gene expression directly via repressor effects upon the expression of these genes and indirectly via repressor effect upon the expression of PGC-1α.
Figure 2.
SMILE inhibits CRTC2-induced gluconeogenic enzyme gene expression in hepatocytes. A, AML12 cells were cotransfected with PEPCK-Luc, G6Pase-Luc, CRTC2, and SMILE expression vectors. B, primary hepatocytes were infected with Ad-GFP, Ad-CRTC2, and Ad-SMILE for 24 h, and gluconeogenic gene expression was analyzed by quantitative PCR. C, primary hepatocytes were infected with Ad-CRTC2 and Ad-SMILE for 24 h, and glucose output assay was performed using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mmol/liter) and sodium pyruvate (1 mmol/liter). D, primary hepatocytes were infected with Ad-CRTC2, Ad-US, or Ad-shSMILE for 24 h and treated with insulin for 9 h. Glucose output assay was performed using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mmol/liter) and sodium pyruvate (1 mmol/liter). Error bars show mean ± S.D. *, p < 0.05; ***, p < 0.005 by two-tailed Student's t test. n.s. means not significant. Results are representative of three independently performed experiments.
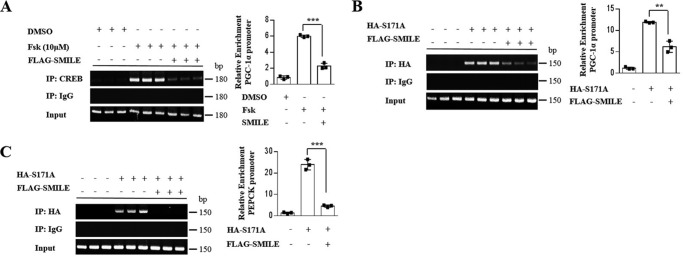
SMILE inhibits the binding of CREB/CRTC2 to the PGC-1α and PEPCK promoters
To determine whether SMILE attenuates CREB/CRTC2 occupancy on the PGC-1α promoter, we performed chromatin immunoprecipitation (ChIP) assays. DNA occupancy of both CREB and constitutively active CRTC2 (S171A) in the PGC-1α promoter was significantly decreased by SMILE (Fig. 3, A and B). SMILE also decreased CREB DNA occupancy on the PEPCK promoter (Fig. 3C). Taken together, these results indicate that SMILE inhibits the CREB/CRTC2 target gene transcription via attenuation of CREB/CRTC2 complex recruitment.
Figure 3.
SMILE decreases DNA occupancy of CREB/CRTC2. A, AML12 cells were cotransfected with FLAG-SMILE and treated with vehicle (DMSO) or forskolin. Protein extracts were co-immunoprecipitated using CREB or IgG antibody. B and C, AML12 cells were infected with Ad-SMILE (FLAG-tagged) and Ad-S171A (HA-tagged). Protein extracts were co-immunoprecipitated using HA or IgG antibody. PCR amplification of immunoprecipitated chromatin fragments was conducted using primer pairs specific for proximal, regulatory region of the Pgc-1α (B) and Pepck (C) gene promoter. Error bars show mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.005 by two-tailed Student's t test. Results are representative of three independently performed experiments.
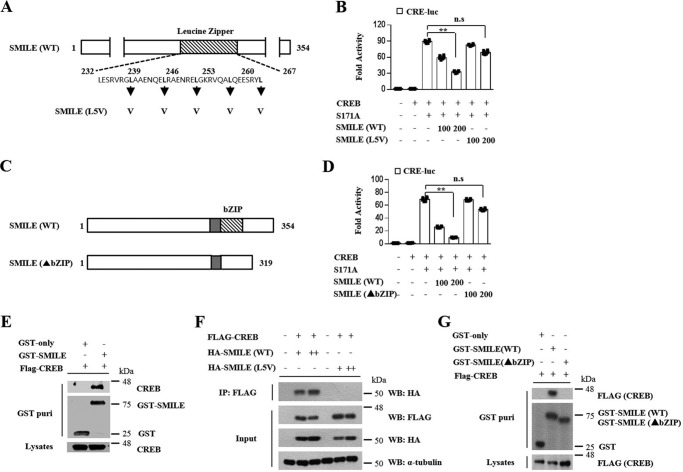
SMILE inhibits CREB activity via interaction with CREB
Phosphorylation of CREB by PKA activates CREB transcriptional activity because phosphorylated CREB binds in a dimeric form to CREs located in the target gene promoters. SMILE has been classified as a member of the CREB/ATF family of bZIP transcription factors. Even though SMILE can self-associate to form a dimer, similar to other members of the bZIP family, homodimerization of SMILE is not essential for repression of a nuclear receptor (13). However, it is not clear whether SMILE could be associated with another bZIP family protein as a heterodimer or whether the heterodimerization has a deterrent effect on bZIP family protein activity. To determine whether the leucine zipper region of SMILE is essential for inhibition of CREB/CRTC2 signaling, we used a mutant SMILE (L5V) in which the five consecutive leucine residues in the leucine zipper region were mutated to valine (Fig. 4A). The leucine zipper–mutated SMILE did not show a repressive effect on CREB/CRTC2-induced CRE-Luc reporter activity (Fig. 4B). To ensure that the leucine zipper region of SMILE is a critical factor for inhibition of CREB/CRTC2 transcriptional activity, we employed SMILE in which the leucine zipper region was deleted (Fig. 4C). Although wild type SMILE significantly inhibited CREB/CRTC2-induced CRE-Luc reporter activity, SMILE lacking the leucine zipper region (▴bZIP) did not show any effect on CREB/CRTC2 transcriptional activity (Fig. 4D). To further investigate the mechanism responsible for the repressive effect of SMILE on CREB transcriptional activity, we performed both in vivo GST interaction assay and endogenous co-immunoprecipitation assays between SMILE and CREB. We observed that SMILE directly interacts with CREB (Fig. 4E and Fig. S1). Moreover, to investigate whether the leucine zipper of SMILE is essential for interaction with CREB, co-immunoprecipitation assays were performed. The leucine zipper–mutated SMILE (L5V) did not co-precipitate with CREB, whereas WT SMILE interacted with CREB in a dose-dependent manner (Fig. 4F). To confirm the importance of leucine zipper in the interaction between SMILE and CREB, in vivo GST pulldown assays were performed. As expected, WT SMILE physically interacted with CREB but the leucine zipper region–deleted SMILE (▴bZIP) did not (Fig. 4G). Taken together, these results indicate that the leucine zipper of SMILE is essential for its repressive function on CREB/CRTC2 transcriptional activity.
Figure 4.
Leucine zipper of SMILE is essential for inhibition of CREB/CRTC2 signaling. A, the basic region and leucine zipper domains of WT SMILE. The leucine zipper mutant SMILE (L5V) indicates the leucine residues between positions 239 and 267 were mutated to valine, as indicated by the arrows. B, AML12 cells were cotransfected with CRE-Luc, CREB, S171A, SMILE (WT), and SMILE (L5V). C, schematic representation of the truncated structures of the leucine zipper domains of SMILE (▴bZIP). D, AML12 cells were cotransfected with CRE-Luc, CREB, S171A, SMILE (WT), and SMILE (▴bZIP). E, 293T cells were cotransfected with expression vectors for FLAG-CREB with pEBG-SMILE (GST-SMILE) or pEBG alone (GST-only). Complex formation (upper two panels, GST purification) and the amount of FLAG-CREB (lower panel, lysate) used for the in vivo binding assay determined the interaction with the anti-FLAG antibody. F, 293T cells were cotransfected with expression vectors for HA-SMILE and FLAG-CREB. Co-immunoprecipitation assays were performed using cell extract from 293T cells using indicated antibodies. G, 293T cells were cotransfected with expression vectors for FLAG-CREB with WT SMILE expressing (WT) or leucine zipper domain truncated SMILE (▴bZIP). Complex formation (upper two panels, GST purification) and the amount of FLAG-CREB (lower panel, lysate) used for the in vivo binding assay. Error bars show mean ± S.D. n.s., not significant; **, p < 0.01 by two-tailed Student's t test. Results are representative of three independently performed experiments.
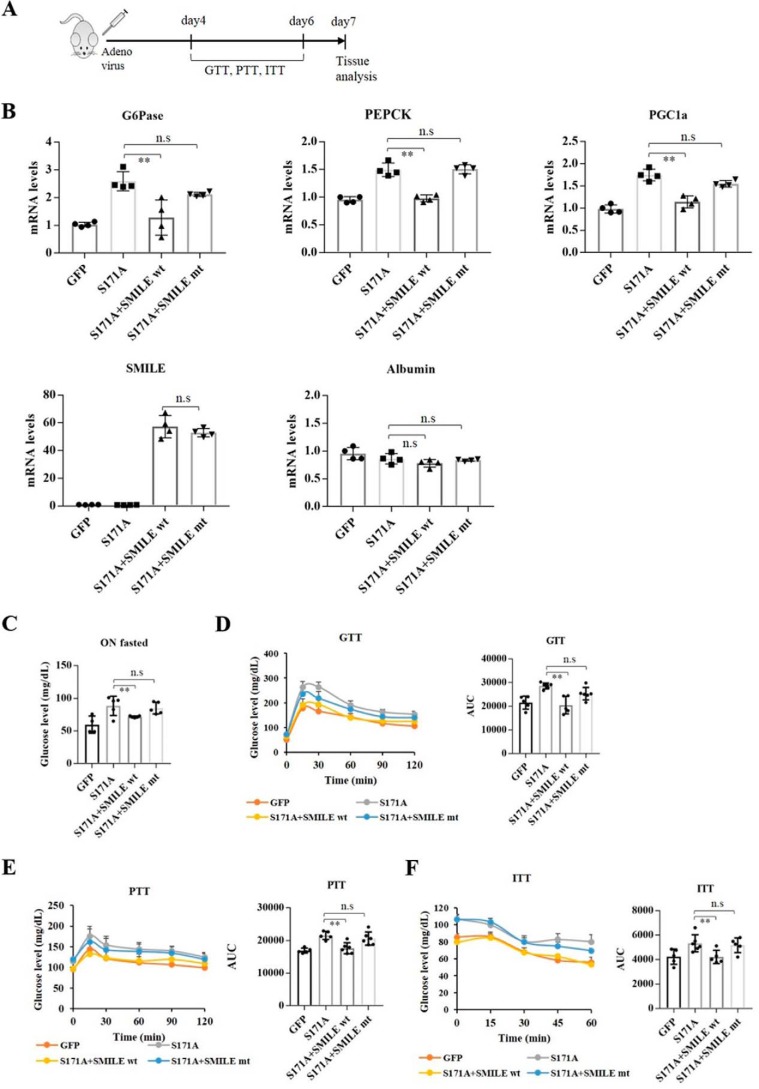
SMILE reduces high blood glucose levels and gluconeogenic gene expression induced by CREB/CRTC2 in mice
Our data showed that SMILE represses CREB/CRTC2-mediated hepatic gluconeogenic gene expression by directly interacting with CREB. We wondered whether CREB/CRTC2-induced increase in blood glucose level would be prevented by SMILE and whether the leucine zipper region of SMILE would be crucial for this effect. To this end, we treated mice with adenoviruses for SMILE, S171A, and the mutant form of SMILE (L5V) (Fig. 5A and Fig. S2). As predicted by the in vitro data, adenoviral overexpression of SMILE significantly repressed gluconeogenic gene expression but had no effect on albumin expression (Fig. 5B). Moreover, SMILE significantly blunted S171A-induced fasting blood glucose levels, whereas L5V did not (Fig. 5C). Consistent with the gluconeogenic gene expression, forced expression of S171A increased the blood glucose level in the pyruvate tolerance test as well as in the glucose tolerance test, and WT SMILE, but not L5V, opposed this effect (Fig. 5, D and E). In addition, adenoviral overexpression of SMILE in mice livers also showed increased insulin sensitivity compared with S171A-alone injected mice (Fig. 5F). Collectively, these results suggest that SMILE protects against hyperglycemia triggered by CREB/CRTC2 signaling and that the leucine zipper of SMILE is essential for inhibition of CREB/CRTC2 signaling pathway.
Figure 5.
CREB/CRTC2-induced increase in blood glucose level is reduced by WT SMILE, but not by leucine zipper–mutated SMILE. A–E, recombinant Ad-GFP, Ad-S171A, Ad-SMILE (WT SMILE), and Ad-L5V (leucine zipper mutant SMILE) were delivered by tail vein injection into C57BL/6 mice (n = 6). A, in vivo experiment scheme. B, expression of G6Pase, Pepck, Pgc-1α, albumin, and SMILE was analyzed by real-time quantitative RT-PCR. Expression was normalized to that of ribosomal L32. C, fasting glucose measured after 16 h of fasting, n = 6. D, glucose tolerance test in mice, glucose (2g/kg), right panel area under curve (AUC), n = 6. E, pyruvate tolerance test in mice, pyruvate (2g/kg), right panel, AUC, n = 6. F, insulin tolerance test (0.75 units/kg) in mice, right panel AUC, n = 6. Statistical test for B–E: One-way ANOVA with Tukey's multiple comparison test. n.s., not significant; **, p < 0.01. Data are represented as mean ± S.E.
Discussion
In this study, we have shown that SMILE inhibits CREB-mediated transactivation of gluconeogenic genes. Moreover, we revealed that the leucine zipper region of SMILE is critical to the protein–protein interaction with CREB. We further found that SMILE reduces hyperglycemia by inhibition of CREB/CRTC2 signaling. These metabolic improvements by SMILE can be attributed to the physical interaction between SMILE and CREB. Whether homodimerization of CREB is affected by SMILE remains uncertain.
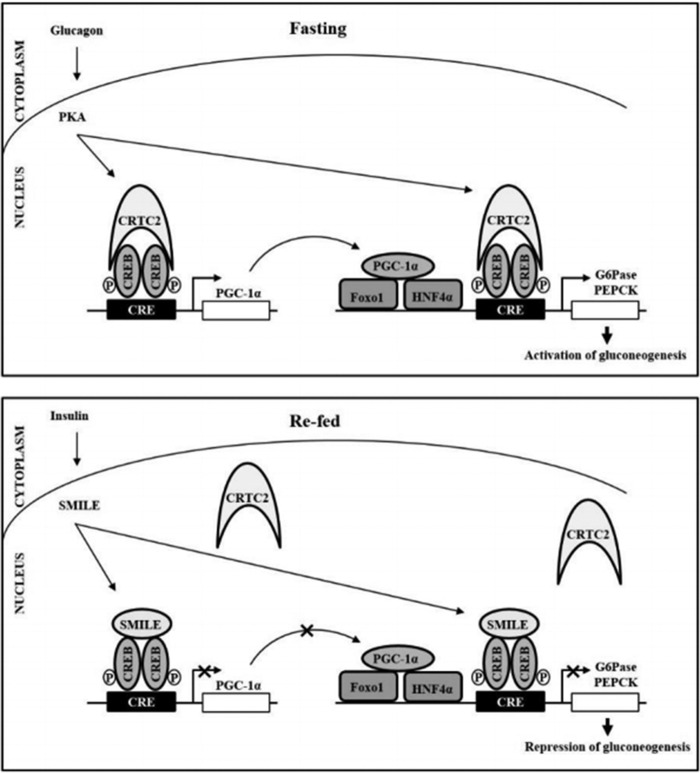
A previous report revealed that CREB is largely a monomer in solution but assembles cooperatively as a dimer on DNA, and CREB dimerization is unaffected by the phosphorylation of CREB by PKA (18). Both the basic region and the “leucine zipper” domain of CREB are essential for transcriptional activation (19). SMILE belongs to basic region leucine zipper (bZIP) family and has the ability to homodimerize like other bZIP proteins (9, 11). These reports suggest that SMILE might form a heterodimer with CREB. Although SMILE can self-associate as a dimer, homodimerization of SMILE is not essential for repression of nuclear receptors (13). Here, we found that the long form of SMILE also physically interacted with CREB in a leucine zipper–dependent manner. Moreover, both transcriptional activity and DNA recruitment of CREB coactivator CRTC2 on CRE were significantly diminished by SMILE, but not by leucine zipper–mutated SMILE. Consistent with our observation, the short form of SMILE (Zhangfei) has been shown to cause the proteasomal degradation of the basic leucine zipper motif–containing transcription factor Xbp1s in a leucine zipper–dependent interaction (20). We also found that the short form of SMILE inhibited CREB/CRTC2 transcriptional activity (Fig. 1F). Furthermore, results suggest that heterodimerization between CREB and SMILE may dissociate CRTC2 from the CREB complex. Additionally, SMILE repressed not only CRTC2 but also CREB DNA recruitments to CRE of a gluconeogenic gene promoter. Binding of CRTCs to the bZIP domain of CREB leads to increased occupancy of CREB on target gene promoters and CREB occupancy is reduced in CRTC2 knockout cells, and these effects are reversed when CRTC2 expression is restored (21). Taken together, our observations suggest that the ability of SMILE to associate with CREB decreases CREB/CRTC2-induced hepatic gluconeogenesis by preventing CREB occupancy on DNA and by competition with CTRC2 (Fig. 6).
Figure 6.
A proposed model for transcriptional regulation of CREB-mediated hepatic gluconeogenesis by SMILE. Under fasting conditions, glucagon triggers activation of PKA, which phosphorylates CREB at the serine 133 residue. These events lead to the increased expression of gluconeogenic genes such as PEPCK and G6Pase via direct and indirect mechanisms, promoting hepatic gluconeogenesis during fasting. By contrast, feeding leads to the increases plasma insulin, which leads to the activation of insulin signaling pathways in the liver. Activation of SMILE gene expression inhibited CREB/CRTC2-induced PEPCK and G6Pase gene expression directly via repressor effects upon the expression of these genes and indirectly via repressor effect upon the expression of PGC-1α.
CREB stimulates gluconeogenic gene expression through direct and feedforward mechanisms. During short-term fasting, CREB activity increases in response to glucagon and directly stimulates the expression of the phosphoenolpyruvate carboxykinase 1 (PEPCK1) and glucose-6-phosphatase (G6Pase) genes by binding to CREs within their promoters. Our studies reveal that SMILE prevents direct activation of PEPCK and G6Pase by CREB/CRTC2. SMILE decreased CREB/CRTC2-induced promoter activity of PEPCK and G6Pase, as well as DNA occupancy of CRTC2 on the PEPCK promoter (Figs. 2A and 3C). In long-term fasting conditions, CREB also increases the expression of the gluconeogenic program through a feedforward mechanism involving the induction of PGC-1α. The expression of PGC-1α is markedly induced in the liver upon fasting via a CREB-dependent transcriptional mechanism and is critical to maintaining prolonged gluconeogenesis under starvation (5, 7). The up-regulation of PGC-1α by CREB provides a mechanism to further amplify gluconeogenic gene expression in response to prolonged fasting. In fact, CREB/CRTC2 activity is increased in hyperglycemic models, and knockdown of CRTC2 efficiently reduces hyperglycemia (4). PGC1-α stimulates hepatic glucose production through its role as a co-activator for glucocorticoid receptor, hepatocyte nuclear factor 4, and the forkhead box (FOXO) transcription factor (7, 8). Our previous report demonstrated that SMILE also inhibits transcriptional activity promoted by GR and HNF4 (13). Taken together, these observations indicate that SMILE is involved in regulation of gluconeogenesis in both short-term and prolonged fasting. Moreover, we suggest that SMILE is a critical repressor for gluconeogenesis because cooperative stimulation by the major transcription factors, including CREB, GR, and HNF4, results in a strong synergistic effect that massively increases the enzymatic capacity for gluconeogenesis. Up-regulation of SMILE expression by insulin represses stimulation of the expression of gluconeogenic genes by a multitude of transcription factors.
In the current work, we found that SMILE improves hepatic glucose parameters by decreasing CREB/CRTC2-induced hepatic gluconeogenesis. Based on its role as a gluconeogenic regulator, SMILE may provide a therapeutic target for the treatment of hyperglycemia. A small molecule that induces SMILE expression in the liver would have a therapeutic potential for type 2 diabetes.
Experimental procedures
Materials and plasmids
Insulin was purchased from Sigma-Aldrich. Forskolin (FSK) was purchased from Calbiochem and dissolved in the recommended solvents. The 8× CRE-Luc, pGL3basic-PGC-1-Luc, pGL3basic-PGC-1-Luc (CRE mutant), pGL3basic-G6Pase-Luc, pGL3basic-PEPCK-Luc, pcDNA3-FLAG-CREB, pcDNA3-HA-CRTC2, pcDNA3-FLAG-S171A, pRC-RSV-PKA constructs were previously described (22, 23). The plasmids of pcDNA3-FLAG-SMILE, pcDNA3-FLAG-ZF (short form of SMILE), pcDNA3-HA-SMILE, pcDNA3-HA-SMILE (L5V), pEBG, pEBG-SMILE (WT), pEBG-SMILE (▴bZIP) constructs were previously described (13). All plasmids were confirmed via sequencing analysis.
Animal experiments
Eight- to 12-week-old male C57BL/6 mice were purchased from Charles River Laboratories. All mice were maintained in a pathogen-free environment and housed in clear cages in groups of five animals per cage with constant temperature (20–22 °C) and humidity and 12 h:12 h light-dark cycles. All animals had access to water at all times. Recombinant adenovirus (0.5 × 109 pfu) was delivered by tail vein injection to mice. To measure fasting blood glucose levels, animals were fasted for 16 h with free access to water. For the glucose or pyruvate tolerance tests, mice were fasted for 16 h or 6 h and then injected intraperitoneally with 2 g/kg glucose or 2 g/kg pyruvate. For the insulin tolerance tests, mice were fasted for 6 h and then injected with 0.75 units/kg of insulin. Plasma glucose levels were measured using blood drawn from the tail vein at designated time points using an automatic glucose monitor (One Touch, LifeScan Ltd., Milpitas, CA). Seven days later, total RNA and protein were extracted from liver, adipose, and muscle tissues for quantitative PCR, and Western blot analysis, respectively. Animal experiments were used with five to six mice per group. All animal experimental procedures were performed in a specific pathogen-free facility at the Korea University Institutional Animal Care, based on the protocols that were approved by the Korea University Institutional Animal Care and Use Committee.
Glucose output assay
Primary hepatocytes were infected with indicated virus for 1–2 days and the medium was then replaced with Krebs-Ringer buffer (115 mm NaCl, 5.9 mm KCl, 1.2 m MgCl2, 1.2 mm NaH2PO4, 2.5 mm CaCl2, and 25 mm NaHCO3, pH 7.4), supplemented with 10 mm lactate and 1 mm pyruvate. The cells were cultured for 24 h with or without 100 nm insulin. The glucose level in the media was measured using a QuantiChrom Glucose Assay Kit (Bioassay Systems, Hayward, CA).
Cell culture and transient transfection assay
AML12 and 293T cells were maintained in Dulbecco's modified Eagle's medium/F-12 (Invitrogen) or Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal bovine serum (Cambrex Bio Science Walkersville Inc., Walkersville, MD) and antibiotics (Invitrogen). Cells were split in 24-well plates (2–8 × 104 cells/well) at the day before transfection. Transient transfections were performed using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instruction. Total DNA used in each transfection was adjusted to 1 μg/well by adding appropriate amount of empty vector, and CMV-β-gal plasmids were cotransfected as an internal control. The cells were harvested 48 h after the transfection, and luciferase activity was measured and normalized to β-gal activity.
Culture of primary hepatocytes
Primary hepatocytes were prepared from C57BL/6 mice by the collagenase perfusion method as described previously (15). After attachment, cells were infected with adenoviruses for 18 h. Subsequently, cells were maintained in the serum-free Medium 199 media (Mediatech) overnight and treated with 10 μm forskolin and/or 100 nm insulin.
Recombinant adenovirus
Adenoviruses expressing unspecific shRNA, shSMILE, control GFP, SMILE, CRTC2, and S171A were described previously (24). Adenovirus-expressing leucine mutant of SMILE (SMILE L5V) was subcloned into the KpnI-XhoI site of the pAdtrack-CMV vector. pAdtrack-CMV-SMILE (L5V) vector was linearized with PmeI and then introduced into BJ5183-AD-1-competent cells (Agilent Technologies) by electroporation. Recombinate adenoviral vectors were transfected into AD-293 cells by TransIT-LT1 reagent (Mirus Bio, Madison, WI). All viruses were purified by using CsCl2 or the Adeno-X Maxi Purification Kit (Clontech).
Chromatin immunoprecipitation assay
The ChIP assay was performed as described previously (15). In brief, mouse livers (40 mg/sample) were fixed with 1% formaldehyde and sonicated. The soluble chromatin was then subjected to immunoprecipitation using anti-HA (2367, Cell Signaling Technology), anti-FLAG (2368, Cell Signaling Technology), anti-CREB (9197, Cell Signaling Technology), and anti-SMILE (C11755, Assay Biotechnology) followed by treatment with Protein A-agarose/salmon sperm DNA (Upstate Biotechnology, Upstate, NY). DNA samples were quantified by quantitative real time PCR using two pairs primers of the mouse Pepck promoter follows: forward (5′-GATGTAGACTCTGTCCTG-3′) and reverse (5′-GATTGCTCTGCTATGAGT-3′) or mouse PGC-1α promoter follows: forward (5′-GCCTATGAGCACGAAAGGCT-3′) and reverse (5′-GCGCTCTTCAATTGCTTTCT-3′).
Western blot analysis
Whole-cell extracts were prepared using RIPA buffer (Elpis-Biotech, Daejeon, Korea). Proteins from whole-cell lysates were separated by 10% SDS-PAGE and then transferred to nitrocellulose membranes. The membranes were probed with indicated antibodies. Immunoreactive proteins were visualized using an Amersham Biosciences ECL kit (GE Healthcare), according to the manufacturer's instructions.
Co-immunoprecipitation (co-IP)
Co-IP analysis was performed as described previously (13). Antibodies used for co-IPs were anti-CREB (9197, Cell Signaling Technology). Control co-IPs were carried out using rabbit IgG (Santa Cruz Technology, sc-2027). In Western blotting assays, the following antibodies were used at dilution of 1:1000: anti-CREB (9197, Cell Signaling Technology), anti-SMILE (C11755, Assay Biotechnology), and anti-tubulin (Cell Signaling Technology, 2146) antibodies. In Western blot analysis of immunoprecipitated proteins, conventional HRP conjugated anti-rabbit IgG was replaced with rabbit IgG TrueBlot (eBioscience, 18–8816) to eliminate signal interference by the immunoglobulin heavy and light chains.
Quantitative PCR
Total RNA from primary hepatocytes or liver was extracted using an RNeasy Mini Kit (Qiagen). cDNA was generated by Superscript II enzyme (Invitrogen) and analyzed by quantitative PCR using a SYBR Green PCR kit and an TP800 Thermal Cycler DICE Real Time system (Takara). All data were normalized to ribosomal L32 expression.
Statistical analyses
Data were expressed as mean ± S.D. In most of the in vivo experiments, an n of 6 mice was the minimum number used. One-way analysis of variance (ANOVA) with a Tukey's multiple comparison test was used when comparing four groups, as indicated in the figure legends. A two-tailed unpaired Student's t test was used for comparison of treatments in vitro experiments. In vivo metabolic experiments were repeated four times, and in vitro assays were performed on at least two independent occasions. Analysis was performed using Microsoft Excel and/or GraphPad Prism software version 7.0. *, p < 0.05, **, p < 0.01, ***, p < 0.
Author contributions
J.-M. L. and H.-S. H. formal analysis; J.-M. L., H.-S. H., and Y. S. J. investigation; J.-M. L. writing-original draft; H.-S. H., R. A. H., S.-H. K., and H.-S. C. writing-review and editing; R. A. H. data curation; S.-H. K. and H.-S. C. supervision; S.-H. K. and H.-S. C. funding acquisition; S.-H. K. and H.-S. C. project administration.
Supplementary Material
Acknowledgments
We thank Sun Myung Park for technical assistance and Seok-Yong Choi for critical reading of manuscript.
This work was supported through the National Research Foundation of Korea (NRF) by National Creative Research Initiatives Grant 20110018305 (to H.-S. C) and National Research Foundation of Korea Grants 2015R1A2A1A01006687, 2017M3A9D5A01052447, and 2015R1A5A1009024 to (S.-H. K). The work was also supported by a grant from Korea University, Seoul, Korea (to S.-H. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- CREB
- cAMP response element–binding protein
- CRE
- cAMP response element
- GR
- glucocorticoid receptor
- HNF4
- hepatocyte nuclear factor 4
- co-IP
- co-immunoprecipitation
- AUC
- area under curve
References
- 1. Hagiwara M., Brindle P. K., Harootunian A., Armstrong R., Rivier J., Vale W., Tsien R., and Montminy M. R. (1993) Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol. Cell Biol. 13, 4852–4859 10.1128/MCB.13.8.4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koo S.-H., Flechner L., Qi L., Zhang X., Screaton R. A., Jeffries S., Hedrick S., Xu W., Boussouar F., Brindle P. K., Takemori H., and Montminy M. R. (2005) The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437, 1109–1111 10.1038/nature03967 [DOI] [PubMed] [Google Scholar]
- 3. Dentin R., Hedrick S., Xie J., Yates J. 3rd, and Montminy M. (2008) Hepatic glucose sensing via the CREB coactivator CRTC2. Science 319, 1402–1405 10.1126/science.1151363 [DOI] [PubMed] [Google Scholar]
- 4. Saberi M., Bjelica D., Schenk S., Imamura T., Bandyopadhyay G. K., Li P., Jadhar V., Vargeese C., Wang W., Bowman K., Zhang Y., Polisky B., and Olefsky J. M. (2009) Novel liver-specific TORC2 siRNA corrects hyperglycemia in rodent models of type 2 diabetes. Am. J. Physiol. Endocrinol. Metabol. 297, E1137–E1146 10.1152/ajpendo.00158.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., and Montminy M. (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183 10.1038/35093131 [DOI] [PubMed] [Google Scholar]
- 6. Dentin R., Liu Y., Koo S. H., Hedrick S., Vargas T., Heredia J., Yates J. 3rd, and Montminy M. (2007) Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449, 366–369 10.1038/nature06128 [DOI] [PubMed] [Google Scholar]
- 7. Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., Newgard C. B., and Spiegelman B. M. (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 10.1038/35093050 [DOI] [PubMed] [Google Scholar]
- 8. Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., and Spiegelman B. M. (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423, 550–555 10.1038/nature01667 [DOI] [PubMed] [Google Scholar]
- 9. Lu R., and Misra V. (2000) Zhangfei: A second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 28, 2446–2454 10.1093/nar/28.12.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie Y. B., Lee O. H., Nedumaran B., Seong H. A., Lee K. M., Ha H., Lee I. K., Yun Y., and Choi H. S. (2008) SMILE, a new orphan nuclear receptor SHP-interacting protein, regulates SHP-repressed estrogen receptor transactivation. Biochem. J. 416, 463–473 10.1042/BJ20080782 [DOI] [PubMed] [Google Scholar]
- 11. Akhova O., Bainbridge M., and Misra V. (2005) The neuronal host cell factor-binding protein Zhangfei inhibits herpes simplex virus replication. J. Virol. 79, 14708–14718 10.1128/JVI.79.23.14708-14718.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xie Y. B., Park J. H., Kim D. K., Hwang J. H., Oh S., Park S. B., Shong M., Lee I. K., and Choi H. S. (2009) Transcriptional corepressor SMILE recruits SIRT1 to inhibit nuclear receptor estrogen receptor-related receptor γ transactivation. J. Biol. Chem. 284, 28762–28774 10.1074/jbc.M109.034165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xie Y. B., Nedumaran B., and Choi H. S. (2009) Molecular characterization of SMILE as a novel corepressor of nuclear receptors. Nucleic Acids Res. 37, 4100–4115 10.1093/nar/gkp333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Misra J., Chanda D., Kim D. K., Li T., Koo S. H., Back S. H., Chiang J. Y., and Choi H. S. (2011) Curcumin differentially regulates endoplasmic reticulum stress through transcriptional corepressor SMILE (small heterodimer partner-interacting leucine zipper protein)-mediated inhibition of CREBH (cAMP responsive element-binding protein H). J. Biol. Chem. 286, 41972–41984 10.1074/jbc.M111.274514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J. M., Seo W. Y., Han H. S., Oh K. J., Lee Y. S., Kim D. K., Choi S., Choi B. H., Harris R. A., Lee C. H., Koo S. H., and Choi H. S. (2016) Insulin-inducible SMILE inhibits hepatic gluconeogenesis. Diabetes 65, 62–73 10.2337/db15-0249 [DOI] [PubMed] [Google Scholar]
- 16. Quinn P. G., and Granner D. K. (1990) Cyclic AMP-dependent protein kinase regulates transcription of the phosphoenolpyruvate carboxykinase gene but not binding of nuclear factors to the cyclic AMP regulatory element. Mol. Cell Biol. 10, 3357–3364 10.1128/MCB.10.7.3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wynshaw-Boris A., Short J. M., Loose D. S., and Hanson R. W. (1986) Characterization of the phosphoenolpyruvate carboxykinase (GTP) promoter-regulatory region. I. Multiple hormone regulatory elements and the effects of enhancers. J. Biol. Chem. 261, 9714–9720 [PubMed] [Google Scholar]
- 18. Wu X., Spiro C., Owen W. G., and McMurray C. T. (1998) cAMP response element-binding protein monomers cooperatively assemble to form dimers on DNA. J. Biol. Chem. 273, 20820–20827 10.1074/jbc.273.33.20820 [DOI] [PubMed] [Google Scholar]
- 19. Dwarki V. J., Montminy M., and Verma I. M. (1990) Both the basic region and the “leucine zipper” domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 9, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang R., Rapin N., Ying Z., Shklanka E., Bodnarchuk T. W., Verge V. M., and Misra V. (2013) Zhangfei/CREB-ZF—a potential regulator of the unfolded protein response. PLoS One 8, e77256 10.1371/journal.pone.0077256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Inoue H., Ravnskjaer K., Viste K., Miller N., Liu Y., Hedrick S., Vera L., and Montminy M. (2010) Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 107, 3087–3092 10.1073/pnas.0914897107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J. M., Seo W. Y., Song K. H., Chanda D., Kim Y. D., Kim D. K., Lee M. W., Ryu D., Kim Y. H., Noh J. R., Lee C. H., Chiang J. Y., Koo S. H., and Choi H. S. (2010) AMPK-dependent repression of hepatic gluconeogenesis via disruption of CREB · CRTC2 complex by orphan nuclear receptor small heterodimer partner. J. Biol. Chem. 285, 32182–32191 10.1074/jbc.M110.134890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koo S.-H., Satoh H., Herzig S., Lee C.-H., Hedrick S., Kulkarni R., Evans R. M., Olefsky J., and Montminy M. (2004) PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nat. Med. 10, 530–534 10.1038/nm1044 [DOI] [PubMed] [Google Scholar]
- 24. Kim D. K., Ryu D., Koh M., Lee M. W., Lim D., Kim M. J., Kim Y. H., Cho W. J., Lee C. H., Park S. B., Koo S. H., and Choi H. S. (2012) Orphan nuclear receptor estrogen-related receptor γ (ERRγ) is key regulator of hepatic gluconeogenesis. J. Biol. Chem. 287, 21628–21639 10.1074/jbc.M111.315168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.