Abstract
Microtubule-associated protein 2c (MAP2c) is a 49-kDa intrinsically disordered protein regulating the dynamics of microtubules in developing neurons. MAP2c differs from its sequence homologue Tau in the pattern and kinetics of phosphorylation by cAMP-dependent protein kinase (PKA). Moreover, the mechanisms through which MAP2c interacts with its binding partners and the conformational changes and dynamics associated with these interactions remain unclear. Here, we used NMR relaxation and paramagnetic relaxation enhancement techniques to determine the dynamics and long-range interactions within MAP2c. The relaxation rates revealed large differences in flexibility of individual regions of MAP2c, with the lowest flexibility observed in the known and proposed binding sites. Quantitative conformational analyses of chemical shifts, small-angle X-ray scattering (SAXS), and paramagnetic relaxation enhancement measurements disclosed that MAP2c regions interacting with important protein partners, including Fyn tyrosine kinase, plectin, and PKA, adopt specific conformations. High populations of polyproline II and α-helices were found in Fyn- and plectin-binding sites of MAP2c, respectively. The region binding the regulatory subunit of PKA consists of two helical motifs bridged by a more extended conformation. Of note, although MAP2c and Tau did not differ substantially in their conformations in regions of high sequence identity, we found that they differ significantly in long-range interactions, dynamics, and local conformation motifs in their N-terminal domains. These results highlight that the N-terminal regions of MAP2c provide important specificity to its regulatory roles and indicate a close relationship between MAP2c's biological functions and conformational behavior.
Keywords: microtubule-associated protein (MAP), nuclear magnetic resonance (NMR), small-angle X-ray scattering (SAXS), Tau protein (Tau), protein conformation, NMR relaxation, paramagnetic relaxation enhancement (PRE)
Introduction
Cytoskeletal microtubule-associated proteins (MAPs)6 bind, stabilize, and regulate dynamics of microtubules, in a phosphorylation-dependent manner. MAP2 and Tau are neuronal MAPs, MAP2 being expressed mainly in dendrites, whereas Tau is found in axons (1). Both MAP2 and Tau are expressed as different spliced variants. Tau isoforms expressed in human brain differ in the number of near-N-terminal inserts as well as in the number of repeats in the microtubule-binding domain (MTBD), whereas MAP2 isoforms differ in the length of the N-terminal projection domain (2, 3). Expression of both MAP2 and Tau isoforms is regulated during development. The high-molecular weight isoforms, MAP2a and MAP2b, contain 1830 amino acids, and the two low-molecular weight variants, MAP2c and MAP2d, consist of 467 and 498 amino acids, respectively. MAP2c is the smallest functional isoform, expressed mainly during embryonal brain development. After birth, its expression is restricted to regions exhibiting post-natal plasticity, such as the olfactory bulb (4), suggesting a role in neuronal development.
Tau and MAP2c share a high sequence homology in the C-terminal part, containing MTBD, but differ in the N-terminal part, comprising the acidic and the proline-rich subdomains (Fig. 1) (5). The sequence diversity is further magnified by different phosphorylation patterns (6–13).
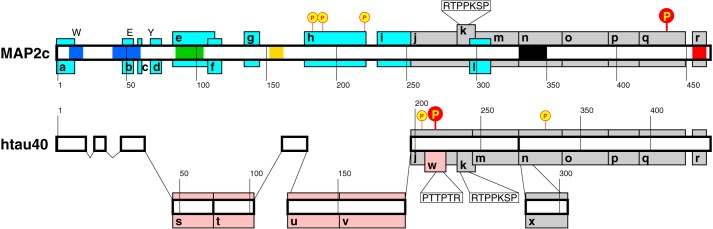
Figure 1.
Schematic drawing of the MAP2c and htau40 molecules. Regions associated with functions of MAP2c, htau40, and both proteins are shown as cyan, pink, and gray boxes, respectively, and labeled as follows. a–d, f, and l, proposed neurosteroid-binding site (57); e, RII site; g, proline-rich region coded by exon 7; h, proline-rich region coded by exon 14 and phosphorylated by PKA; i, short proline-rich region P1 of MAP2c; j, proline-rich region P2; k, Class I Fyn-binding site; m, MTBR1; n, MTBR3; o, MTBR4; p, region R′; q, region homologous to the muscarinic receptor binding site of Tau; r, C-terminal α-helix; s, near-N-terminal insert I1; t, near-N-terminal insert I2; u, region including α-helix 114LEDEAAGHVT123; v, long proline-rich region P1 of Tau; w, Class II Fyn-binding site; x, MTBR2. Regions of Tau with high homology with MAP2c are drawn closer to the scheme of MAP2c. Red and yellow circles, residues phosphorylated by PKA with high and medium rate, respectively (11, 13). SH3 recognition motifs are displayed in white boxes. Colored segments of the middle bar refer to clusters of spectral density values plotted in Fig. 7 (b and d). Positions of Trp14, Glu52, and Tyr67 of MAP2c are marked by W, E, and Y.
Tau, but not MAP2c, forms paired helical filaments observed in brains of patients suffering from Alzheimer's disease. The major difference in the aggregation properties seems to be encoded in a few amino acids of the aggregation seed motifs in the otherwise highly homologous C-terminal domains of Tau and MAP2c (14). In addition, interactions between the MTBD and the N-terminal domain of Tau are present in paired helical filaments (15).
Identification of structural motifs responsible for specific functions of MAP2c and Tau is complicated by the fact that MAP2c and Tau belong to the class of intrinsically disordered proteins (IDPs) lacking a unique structure and exist in multiple, quickly interconverting conformations (16–20). NMR has proved to be a reliable tool to study their dynamics and structural features (11, 13, 21–25). Despite their highly flexible nature, Tau (23) and MAP2c (25) adopt transient secondary structures, indicating that the structures of Tau and MAP2c are far from a random coil. Also, long-range intramolecular contacts have been observed in Tau (15), supporting the “paper-clip” model of the Tau molecule.
Direct studies of structure–function relations rely on the availability of atomistic structural models. Quantitative computational analysis of experimental data reflecting the local conformations and long-range contacts provided an atomic-resolution ensemble model describing molecular properties of Tau (26). In this paper, we built a similar model for MAP2c and investigated molecular motions of MAP2c using NMR relaxation. The results allowed us to correlate structural features and dynamics of MAP2c with its known and proposed binding sites. In particular, we addressed questions related to interactions of MAP2c with three important binding partners. The first of them was the Src-homology domain 3 (SH3) domain of the Fyn tyrosine kinase, recognizing different motifs of MAP2c and Tau (27, 28). It was not clear whether the conformation optimal for the SH3 binding is highly populated already in the free form of intrinsically disordered MAP2c or if it is formed only in the bound state. The second examined interaction partner was plectin. Its noncanonical SH3 domain (29) was reported to bind MAP2c, but the recognized motif was unknown (30). The third inspected interaction region was the site recognized by the RII regulatory subunit of cAMP-dependent protein kinase (PKA). This site was expected to adopt α-helical conformation, but populations of α-helix in MAP2c fragments covering the corresponding sequence were low (31).
Results
Overall shape from small-angle X-ray scattering
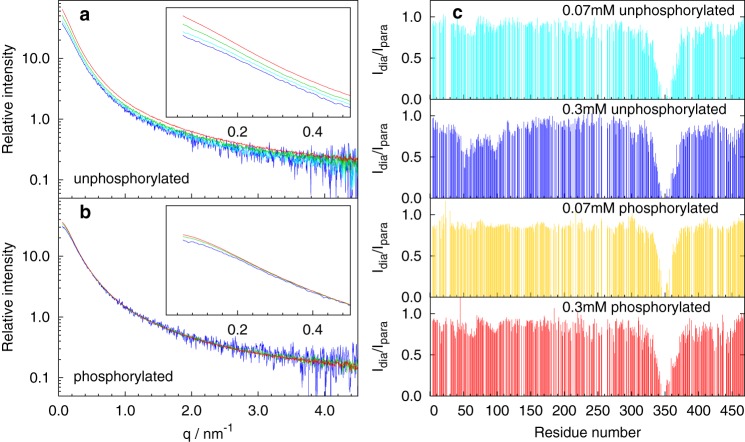
Sampling of the volume space by MAP2c was probed by small-angle X-ray scattering (SAXS). The scattering curves were acquired for various concentrations of unphosphorylated MAP2c and of MAP2c phosphorylated by PKA. A concentration dependence of the scattering intensity at low angles (corresponding to the momentum transfer q < 0.5 nm−1) was observed at concentrations of unphosphorylated MAP2c exceeding 0.1 mm (Fig. 2a). The concentration effects were greatly reduced when MAP2c was phosphorylated by PKA (Fig. 2b). This finding indicates that intermolecular interactions existing at submillimolar concentrations of MAP2c are suppressed by the PKA phosphorylation. The scattering data were further analyzed quantitatively using the ASTEROIDS approach (see below).
Figure 2.
Concentration dependence of SAXS curves of unphosphorylated MAP2c (a), phosphorylated MAP2c (b), and PRE data (c). In the SAXS dilution series, data for concentrated (∼0.3 mm) samples are shown in red, for 2-fold dilution in green, for 4-fold dilution in cyan, and for 8-fold dilution in blue. The initial regions of the scattering curves are shown in the insets. The relative intensity represents radially averaged scattering after buffer subtraction divided by the concentration. No additional scaling was applied. PRE values obtained for low (70 μm) and high (0.3 mm) concentration of unphosphorylated MAP2c are shown in cyan and blue, respectively. PRE values obtained for low (70 μm) and high (0.3 mm) concentration of PKA phosphorylated MAP2c are shown in gold and red, respectively.
Paramagnetic relaxation enhancement
Long-range interactions of MAP2c were monitored using paramagnetic relaxation enhancement (PRE) of NMR signals (32). The native MAP2c contains a single cysteine at position 348, located in MTBD, providing a convenient site for attachment of the paramagnetic nitroxide radical label (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl)-methanethiosulfonate (MTSL). Six mutants, with Cys348 replaced by serine and with a single cysteine located in the N-terminal acidic domain (E52C and Q110C), the proline-rich domain (T153C, E203C, and K282C), and in the C-terminal domain (L402C), were designed and labeled with MTSL.
To achieve high resolution, the peak intensities were monitored in 3D nonuniformly sampled HNCO spectra. Considering the lower sensitivity of the HNCO experiment compared with the 2D 1H-15N heteronuclear single quantum coherence spectra, we tested whether the lower tendency of MAP2c to aggregate allows us to work at higher concentrations than used in the PRE studies of Tau (23, 24). PRE observed for 70 μm unphosphorylated, 70 μm phosphorylated, and 0.3 mm phosphorylated WT MAP2c were comparable. In contrast, a strong concentration dependence of PRE was observed for 0.3 mm unphosphorylated WT MAP2c, in agreement with the SAXS data indicating possible intermolecular interactions at high concentrations of unphosphorylated MAP2c (Fig. 2c). Therefore, only data obtained at the 70 μm concentration were used for the quantitative conformational analysis (see below) in the case of unphosphorylated MAP2c. At this concentration, the sensitivity of the NMR experiment is still acceptable, and the concentration dependence of the SAXS curves is already weak.
The obtained PRE profiles (Fig. 3d) revealed the following interactions. Within the N-terminal domain, intramolecular interactions were observed between the vicinity of Glu52 (in the negatively charged stretch of glutamates 52EEEE55) and residues in the vicinity of Trp14 and Val95. In the C-terminal domain, weak intramolecular interactions were noticed between microtubule-binding repeats and proline-rich regions. Finally, long-range contacts were observed between the C-terminal domain and the N-terminal domain (namely in the vicinity of Trp14, Glu52, and Val95), with a strong contribution of intermolecular (concentration-dependent) interactions. The long-range interactions are in agreement with previous observations of bent conformations and antiparallel dimers of MAP2c molecules (33).
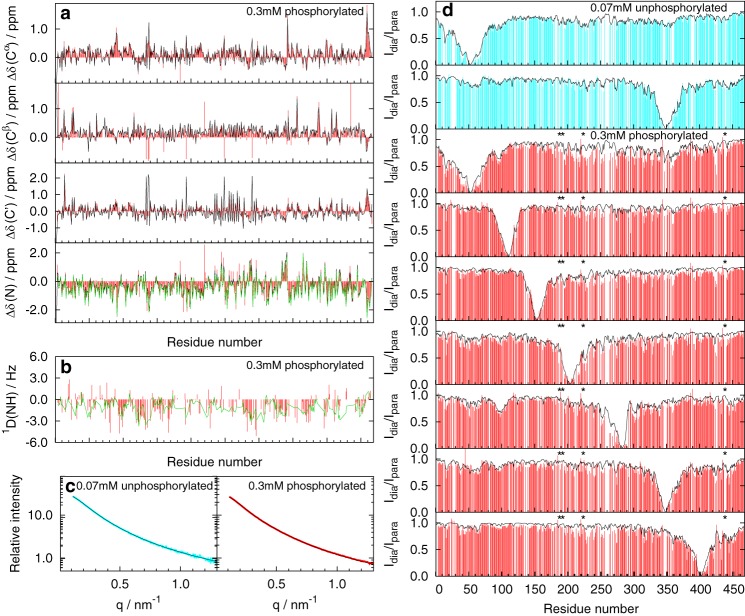
Figure 3.
Comparison of experimental data with values calculated from ensembles selected by the ASTEROIDS analysis. a, secondary chemical shifts; b, 1D(NH) RDCs; c, SAXS curves; d, PRE data (ratios of signal intensities in the absence and in the presence of the paramagnetic label). Experimental data are shown in cyan for 70 μm unphosphorylated MAP2c and in red for 0.3 mm phosphorylated MAP2c. The corresponding values calculated from the ensembles of 600 structures selected by the ASTEROIDS analysis are shown in black. The values calculated from the ensembles selected by the ASTEROIDS analysis of experimental data excluding 15N chemical shifts and 1D(NH) RDCs to validate the selection procedure are shown in green. Secondary chemical shifts are plotted only for 0.3 mm phosphorylated MAP2c (red) because the differences between the data for 70 μm unphosphorylated MAP2c and 0.3 mm phosphorylated MAP2c were negligible. Phosphorylated residues are marked by asterisks.
Conformational analysis
To convert the obtained experimental data into a structural model, conformational analysis was performed using the ASTEROIDS algorithm (34). Assigned chemical shifts (13, 25), SAXS, and PRE data served as an experimental input, reflecting local conformation, long-range contacts, and overall shape of the molecule, respectively. The potential presence of dimers (or higher oligomers) has a negligible effect on the description of local conformational behavior but may influence analysis of long-range interactions. To minimize the effect of intermolecular interactions, PRE and SAXS data measured at low concentrations were used in the case of unphosphorylated MAP2c. From a large set of 84,000 starting structures generated by the flexible-meccano program (see “Experimental procedures”), a set of 600 structures was selected to reproduce the experimental data. Despite the large conformational space of the 467-residue polypeptide chain of MAP2c, good agreement between the experimental data and values calculated from the selected ensemble was achieved, as documented in Fig. 3.
To validate the ensemble of the selected MAP2c conformers, (i) values of 15N secondary chemical shifts, Δδ(N), were removed from the selection procedure and back-calculated from the selected ensembles; and (ii) values of 1D(NH) residual dipolar couplings (RDCs), measured as described under “Experimental procedures” but not used in the ASTEROID analysis, were calculated from the ensembles selected based on chemical shift, PRE, and SAXS data of MAP2c phosphorylated by PKA. In both cases, the independent data were well reproduced by the selected ensembles (Table 1 and Fig. 3 (a and b)). Similarly to the data published for htau40, χ2 for the comparison of 259 experimental RDCs with the selected ensembles was a factor of 2 lower than χ2 for the comparison with the pool of statistical coil structures. The difference between χ2 values for the selected and statistical coil ensembles further increased when the compared values were limited to a subset of residues with a higher difference in local sampling between the selected ensemble and the statistical coil pool (measured by the parameter ΔRama, defined under “Experimental procedures”). The differences in χ2 were even greater for Δδ(N), measured with a high precision in this study. The validation confirmed that the conformational description provided by ASTEROIDS is meaningful and predictive, as reported for other proteins (26).
Table 1.
Validation of ASTEROIDS ensembles by prediction of independent observables
| Passivea | ΔRamab | χ2CV,ASTc | χ2CV,COILd |
|---|---|---|---|
| 1D(NH) | ≥0 | 1.81 (259)e | 3.71 (259) |
| 1D(NH) | >0.05 | 1.66 (134) | 4.05 (134) |
| 1D(NH) | >0.1 | 1.69 (57) | 4.09 (57) |
| Δδ(N) | ≥0 | 1.39 (463) | 17.59 (463) |
| Δδ(N) | >0.05 | 1.72 (207) | 19.79 (207) |
| Δδ(N) | >0.1 | 1.46 (90) | 25.14 (90) |
a Experimental data against which the validity of the ensemble is tested (not included in the selection).
b Amino acids are only included if ΔRama is greater than the reported threshold.
c Reduced χ2 of passive data set compared with the ASTEROIDS selection, calculated with the uncertainty of 1 Hz for 1D(NH) and 0.3 ppm for Δδ(N).
d Reduced χ2 of passive data set compared with the statistical coil description of the protein, calculated with the uncertainty of 1 Hz for 1D(NH) and 0.3 ppm for Δδ(N).
e Number in parentheses refers to number of residues included in the selection.
Local conformational propensities of MAP2c derived from the distribution of the φ and ψ torsion angles, expressed as populations of three typical regions of the Ramachandran diagram in Fig. 4, were in general agreement with previously reported (13, 25) secondary structure propensities calculated using the SSP program (35). Similarly to SSP, ASTEROIDS derives the local conformations mostly from chemical shifts. However, the ASTEROIDS analysis of chemical shifts, based on the SPARTA program (36), is more detailed. Of particular interest is the population of the polyproline II conformation, that can be estimated on the basis of the currently available experimental data (37). The propensities to form particular secondary structures were often identified for continuous sequences of residues, although the ensemble selection treats each amino acid independently. To better visualize the continuous segments of secondary structures, we also plotted populations of short stretches of amino acids in the same conformation (orange and red bars in Fig. 4). Note that the important information is not the absolute value of population of the continuous stretch (which reflects composition of the starting pool of structures with the statistical distribution of torsion angles), but the relative increase of the population in the selected ensemble.
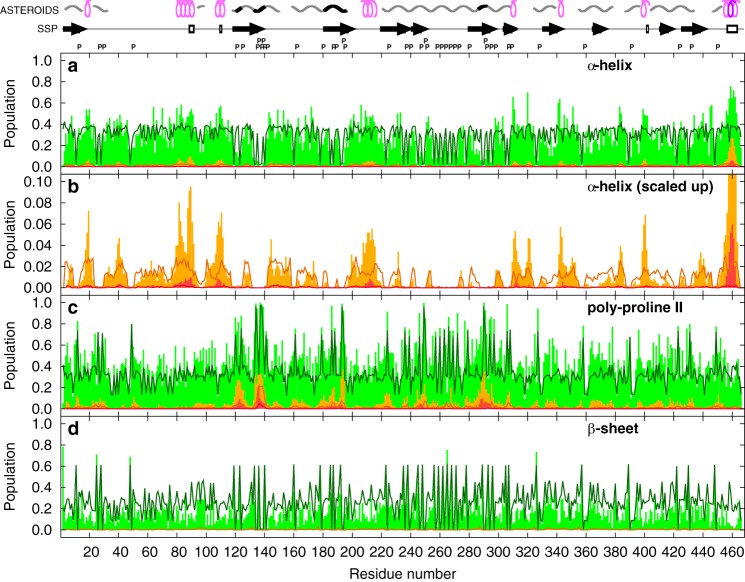
Figure 4.
Populations of torsion angles in the regions of the Ramachandran plot corresponding to the helical (a and b), polyproline-II (c), and β-sheet (d) conformations. Populations of individual residues and of continuous stretches of four and seven amino acids in the given conformation in the ensembles of 600 structures selected by the ASTEROIDS analysis of unphosphorylated MAP2c are shown as green, orange, and red bars, respectively. The orange and red bars are placed in the middle of the stretches. Populations in the statistical coil pool of structures are plotted as solid dark green, dark orange, and dark red lines, respectively. Letters P above the plots indicate positions of prolines. Regions of extended and helical conformations calculated using the SSP program (13, 35) are displayed as arrows and empty rectangles above the plots, respectively. Regions where the population of stretches of four amino acids in the α-helical conformation exceeds 5 and 25%, respectively, are displayed as pink and purple symbols in the top row above the plots, and regions where the population of stretches of four amino acids in the polyproline II conformation exceeds 5 and 25%, respectively, are displayed as gray and black symbols in the top row above the plots.
Highly populated continuous stretches of four and seven α-helical residues (corresponding to one and two turns of an α-helix, respectively) were identified in the C-terminal region (box r in Fig. 1). In addition, two regions were identified where a complete α-helical turn was observed in more than 5% structures of the selected ensembles, and at least five structures (0.8%) with two turns were selected. These regions correspond to the binding site of the regulatory RII subunit of PKA (RII site, Asp83–Glu113, box e in Fig. 1) (5, 31, 38) and the region 206SFSLNSSISSARR218 (box h in Fig. 1). Most of the extended regions (i.e. exhibiting a negative SSP score (35)) were found to highly populate the polyproline II region of the Ramachandran plot. This applies not only to the proline-rich sequences, but also to a major portion of MTBD and residues between Leu402 and Gln421. The highest population of continuous stretches of residues in the polyproline II conformation was observed for a short proline-rich segment encoded by exon 7 (box g in Fig. 1), both in the selected ensembles and in the statistical coil pool. In addition, two regions were identified where populations of four- and seven-residue stretches of polyproline II conformation exceed 30 and 5%, respectively, in the selected ensemble only, whereas the corresponding populations do not exceed 10 and 1% in the statistical coil pool, respectively. These regions are the sequence 120PAALPLAA127 and a binding site for the SH3 domain of the Fyn kinase (box k in Fig. 1) (27). The differences between secondary structures of unphosphorylated and phosphorylated MAP2c were very small.
We compared results of our conformational analysis with secondary structure predictions provided by several publicly available software tools. The predictors correctly identified the position of the C-terminal helix (box r in Fig. 1) and predicted the extended conformation in MTBD, but provided inconsistent results for the first 300 residues of MAP2c (data not shown). This finding is not surprising, considering that the predictors were developed for well-structured proteins, and it documents that experimental data are currently essential for conformational analysis of IDPs.
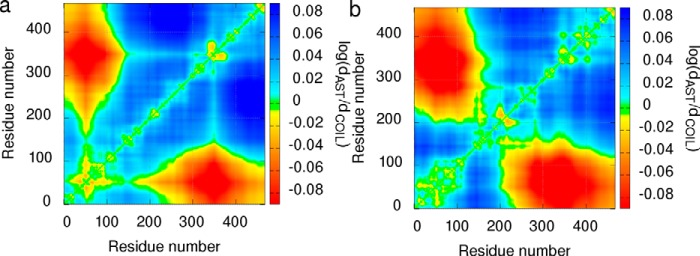
Long-range intramolecular interactions in the ensemble of structures selected by ASTEROIDS are presented in Fig. 5 as a map of relative distances between individual amino acids. The map shows that the average distances in the N-terminal region (including the vicinity of Trp14, the vicinity of Glu52, and the region Asp83–Glu113) and between the N-terminal region and MTBD are shorter in the selected ensembles than in the statistical coil structures, reflecting only distribution of torsion angles typical for individual amino acids. As mentioned above, the analyzed SAXS and PRE data were obtained at conditions where formation of oligomeric species is greatly reduced but cannot be completely excluded. Therefore, we do not interpret the ASTEROIDS distance maps quantitatively under “Discussion,” but we take into account also possible dimerization.
Figure 5.
Distance maps of 70 μm unphosphorylated (a) and 0.3 mm phosphorylated (b) MAP2c. The distances between individual residues in 600 structures selected by the ASTEROIDS analysis are expressed as a logarithm of the ratio of distances observed in the selected ensemble (dAST) to the distances in the statistical coil pool (dCOIL) and plotted using the displayed color scale (red, shorter average distance in the selected ensemble; blue, longer average distance in the selected ensemble).
NMR relaxation analysis
Backbone dynamics of MAP2c were investigated by 15N spin relaxation. To achieve sufficient resolution, relaxation rates were measured using 13C,15N-labeled MAP2c and nonuniformly sampled 3D relaxation experiments based on the HNCO correlation (39). The R1, R2 relaxation rates and steady-state heteronuclear Overhauser enhancement were measured with 600- and 950-MHz spectrometers. Because the R2 rate is potentially influenced by chemical and/or conformational exchange and by the presence of 13C (39), the R2 experiment was complemented by a less sensitive, but more accurate, measurement of the transverse cross-correlated relaxation rate Γx (a result of interference between 15N chemical shift anisotropy and 1H–15N dipole–dipole interaction (40)) at 950 MHz.
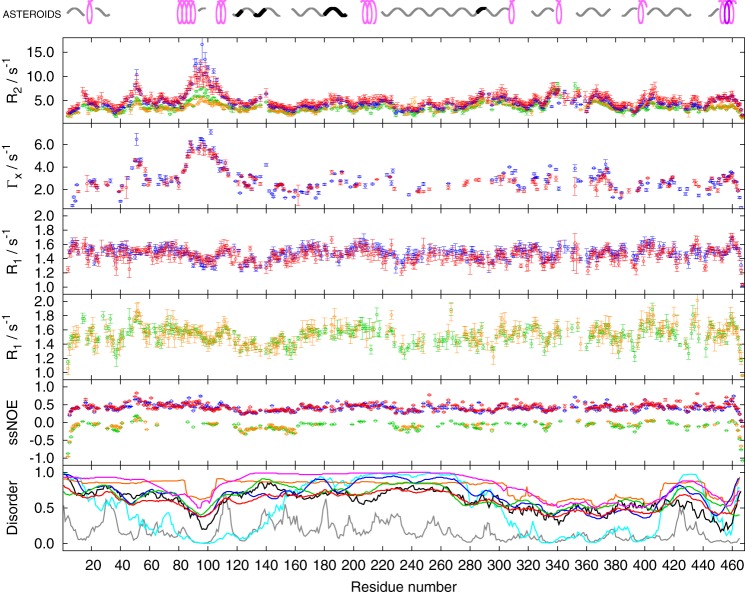
The relaxation rates obtained for unphosphorylated and PKA-phosphorylated MAP2c are presented in Fig. 6. Because MAP2c is an IDP, the measured relaxation rates represent average values for ensembles of all conformational and oligomeric states present at the given temperature. With the exception of the region Asp83–Glu113, phosphorylation by PKA has little effect on the measured rates. This indicates that the obtained values reflect mostly internal dynamics and not intermolecular interactions. The more ordered (more rigid) regions can be distinguished by higher values of the R2 and Γx relaxation rates. Such regions include the vicinity of Trp14, the vicinity of Glu52, residues Asp83–Glu113, serines in the proline-rich sequence 134PPSPPPSP141, residues Arg187–Gly197 and Glu204–Arg226, N-terminal regions of microtubule binding repeats and a homologous sequence following the microtubule-binding repeat 4 (MTBR4), the vicinity of the major phosphorylation site Ser435, and the C-terminal helix 456EDVTAALAK464. The obtained relaxation data are in a good agreement with outputs of some disorder predictors (bottom panel in Fig. 6).
Figure 6.
Values of the 15N relaxation rates of unphosphorylated (green and blue) and phosphorylated (orange and red) MAP2c measured at 600 MHz (green and orange) and 950 MHz (blue and red). Outputs of the following disorder predictors are plotted in the bottom panel: DISOPRED2 (78) (gray), DISpro (79) (cyan), IUPred short (80) (black), MetaDisorderMD2 (81) (orange), PrDOS (82) (blue), RONN (83) (green), and Spritz (84) (red). Secondary structure motifs determined by the ASTEROIDS analysis are shown above the plot (cf. Fig. 4).
The measured relaxation rates were further analyzed to obtain values of the spectral density function J(ω), which is a Fourier transform of the time-correlation function (TCF) directly describing stochastic motions of the molecule (41–45). TCF can be rather complex in the case of disordered proteins, but it can be approximated as a series of weighted mono-exponential functions characterized by correlation times. Various relaxation rates are different linear combinations of values of the spectral density function J(ω), where the values of ω are given by the Larmor frequencies of the observed nuclei (1H and 15N). In this work, we calculated J(0) from R1, Γx, and NOE measured at 950 MHz (46); J(ω) for 61 and 515 MHz from R1 and NOE measured at 600 MHz; and J(ω) for 96 and 815 MHz from R1 and NOE measured at 950 MHz (47, 48). Note that the obtained set of spectral density values is not affected by the aforementioned inaccuracy of the measured R2 rates.
A simple graphical analysis (Figs. 7 and 8) of the J(ω) values calculated from the relaxation rates provides insight into dynamics of individual residues (46, 49–51). Values clustered close to the limits of a single mono-exponential TCF (gray curves in Figs. 7 and 8) suggest dynamics dominated by a single mode of motion, whereas deviations from the curves indicate that more motional modes with different correlation times contribute to the dynamics.
Figure 7.
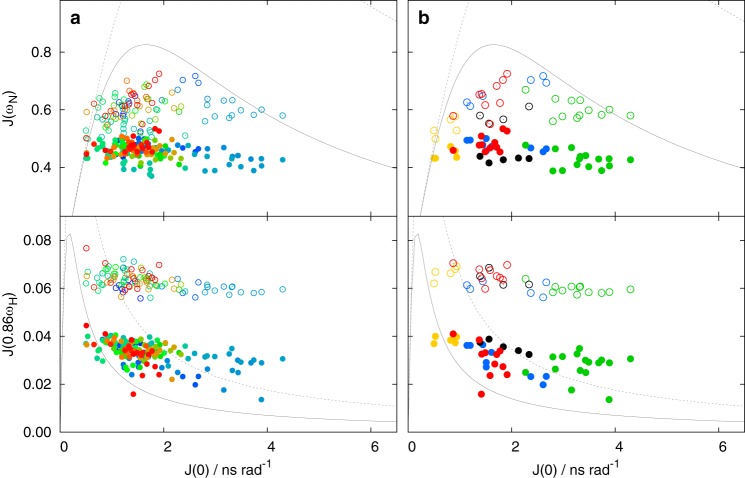
Values of spectral density function of unphosphorylated MAP2c. The J(0) values in all plots were calculated from R1, Γx, and steady-state NOE measured at 950 MHz. The J(ωN) and J(0.86ωH) values calculated from R1 and steady-state NOE for ωN = 61 MHz and ωH = 600 MHz are displayed as open circles. The J(ωN) and J(0.86ωH) values calculated from R1 and steady-state NOE for ωN = 96 MHz and ωH = 950 MHz are displayed as filled circles. a, all available values are displayed in a as circles color-coded in a rainbow manner according to the residue number (N terminus is shown in blue, C terminus in red). b, values for selected regions are shown as circles colored according to the regions: vicinity of Trp14 and Glu51 in blue, RII site in green, MTBR3 in black, C-terminal α helix in red, and vicinity of Ser157 in gold (see Fig. 1). The limits corresponding to mono-exponential TCF for 600- and 950-MHz spectrometers are plotted as dotted and solid gray curves, respectively.
Figure 8.
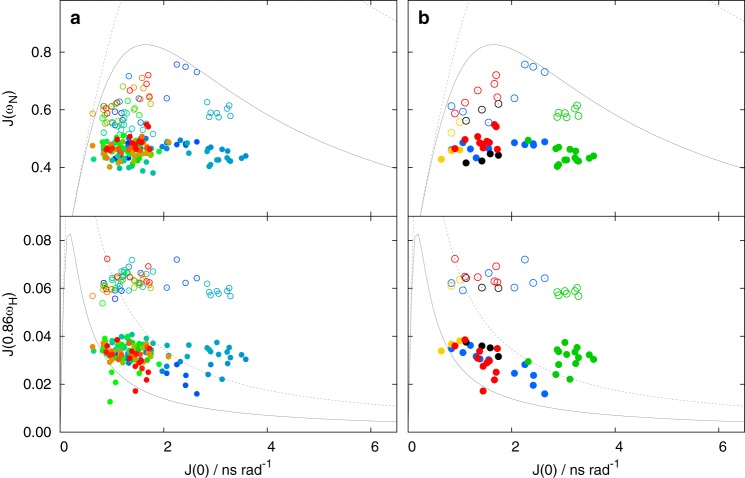
Values of spectral density function of PKA-phosphorylated MAP2c. The J(0) values in all plots were calculated from R1, Γx and steady-state NOE measured at 950 MHz. The J(ωN) and J(0.86ωH) values calculated from R1 and steady-state NOE for ωN = 61 MHz and ωH = 600 MHz are displayed as open circles. The J(ωN) and J(0.86ωH) values calculated from R1 and steady-state NOE for ωN = 96 MHz and ωH = 950 MHz are displayed as filled circles. a, all available values are displayed in a as circles color-coded in a rainbow manner according to the residue number (N terminus is shown in blue, C terminus in red). b, values for selected regions are shown in b as circles colored according to the regions: vicinity of Trp14 and Glu51 in blue, RII site in green, MTBR3 in black, C-terminal α helix in red, and vicinity of Ser157 in gold (see Fig. 1). The limits corresponding to mono-exponential TCF for 600- and 950-MHz spectrometers are plotted as dotted and solid gray curves, respectively.
Graphical analysis of the J(0) versus J(ω) plots showed that J(ω) values of residues in different known or proposed functional sites formed well-defined clusters, indicating distinct types of motions of the individual sites. Several examples are presented in Figs. 7 and 8, and their position in the MAP2c molecule is indicated in Fig. 1. J(0) values of residues of the C-terminal helix 456EDVTAALAK464 (red cluster) and of MTBR3 (black cluster) range between 0.5 and 2 ns. Higher J(0) values (up to 3 ns) were observed for residues in the vicinity of Trp14 and Glu51 (blue cluster). The highest local ordering was observed for residues Asp83–Glu113 (green cluster), where the contribution of longer correlation time(s) is most significant and where J(0) exceeds 4 ns in unphosphorylated MAP2c. The region in the vicinity of Ser157 (gold cluster) is an example of a flexible region with dynamics dominated by the short correlation time.
Interaction with plectin SH3 domain
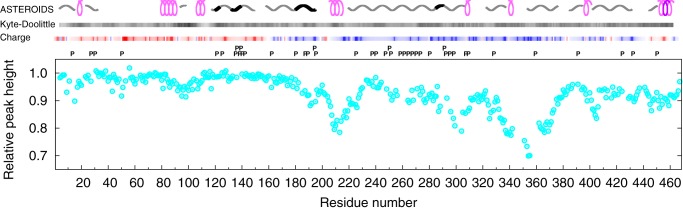
Regions of MAP2c responsible for interactions with several proteins have been characterized in the past. To extend the knowledge of binding sites, we examined interaction of MAP2c with a region of plectin containing a noncanonical SH3 domain. Interactions of disordered MAP2c with a well-folded fragment of plectin, consisting of spectrin repeats SR4 and SR5 and of the SH3 domain (29), were monitored as a decrease of MAP2c peak height in HNCO spectra after the addition of the SR4-SR5-SH3 plectin fragment (Fig. 9). Peak heights were reduced in several regions, most notably in MTBR3 and the region 208SLNSSISSARR218. Canonical SH3 domains are known to interact with PXXP motifs and positively charged region (RXXK) motifs and via tertiary contacts involving hydrophobic residues (52). Comparison of the peak height decrease with distribution of prolines, hydrophobic residues, charged amino acids, and secondary structure motifs (Fig. 9) shows that positively charged residues, but not the PXXP motifs, are often found in the interaction sites. Interestingly, the 208SLNSSISSA216 sequence contains no prolines or positively charged amino acids but tends to form an amphiphilic helical structure.
Figure 9.
Relative decrease of peak height in HNCO NMR spectra of 70 μm MAP2c upon the addition of 70 μm SR4-SR5-SH3 plectin fragment. Secondary structure motifs determined by the ASTEROIDS analysis, hydropathicity, and charge distribution are shown above the plot. The symbols used to describe the secondary structure motifs are explained in the legend to Fig. 4. The hydrophaticity index according to Kyte and Doolittle (85) is shown as darkness of the top bar above the plot (white and black correspond to values of −4.5 and +4.5, respectively). The charge distribution is represented by the color in the bottom bar above the plot, corresponding to a relative electrostatic potential approximated by ΣjCQi/(d0 + d1|ni − nj|), where Qi and ni are charge and sequential number of the ith residue, C is a constant including the electric permitivity, and dk are distance constants. The ratio d1/d0 was set to 2.0, and the colors were chosen so that red and blue correspond to the highest negative and positive potential, respectively, which makes the color code independent of C/d0 (13). Letters P above the plot indicate positions of prolines.
Discussion
Intrinsic conformational behavior of MAP2c is an important factor in a complex network of intermolecular interactions controlling microtubule dynamics essential for neuron development. Therefore, we examined how the observed regions of distinctive conformations and dynamics correlate with known and proposed functional motifs of MAP2c. We also compared the properties of MAP2c with its close homologue Tau (clone htau40, splicing variant 2N4R), exhibiting different expression and phosphorylation patterns and cellular localization and playing an important role in neurodegenerative processes associated with Alzheimer's disease. The determined populations of conformations and dynamics did not differ substantially between MAP2c and Tau in regions of high sequence homology (boxes m–r in Fig. 1), including MTBD. Therefore, we can conclude that the structural basis of interactions with microtubules and their competitors (such as 14-3-3 proteins) is similar for MAP2c and Tau. Results providing new insight into the binding of MAP2c to its important interaction partners were obtained for regions of lower sequence similarity with Tau and are discussed below.
Site of interaction with canonical Fyn SH3 domain
Fyn is an important kinase containing an SH3 domain and phosphorylating single tyrosine residues in different regions of the compared proteins: Tyr18 in Tau (12) and Tyr67 in MAP2c (53). In general, SH3 domains bind to proteins with the sequential motif PXXP, preferably with positively charged amino acids in its vicinity. Tau and MAP2c contain 7 and 13 PXXP motifs, respectively, most of them in the proline-rich region P2 (box j in Fig. 1) with a high sequence homology. P2 of Tau contains two Fyn-binding sites, the classical class II motif 216PTTPTR221 (box w in Fig. 1) and the classical Class I motif 230RTPPKSP236 (box k). Only the latter site is present in MAP2c (288RTPPKSP294). Binding assays performed with synthetic biotinylated peptides showed that the Fyn SH3 domain binds preferentially to the Class II site of Tau (28) and Class I site of MAP2c (27). Our analysis (Fig. 4) revealed that the selected ensembles contained 6% MAP2c structures with all residues of the 288RTPPKSP294 motif in the polyproline II conformation, optimal for binding to the Fyn SH3 domain. This number is 25 times higher than the corresponding population in the statistical coil pool. Only two other sites outside the proline-rich regions P1 and P2 have populations of seven-residue polyproline II stretches higher than 5% (one of them being 134PPSPPPSP141 with the discussed conformation highly populated already in the statistical coil pool). This shows that (i) the 288RTPPKSP294 sequence preferentially adopts the conformation optimal for binding already in the free form of MAP2c, and (ii) the 288RTPPKSP294 site differs from the other PXXP motifs not only in charge distribution, but also by well-defined conformation. The relaxation data (Fig. 6) also show somewhat higher ordering for the 288RTPPKSP294 site.
Site of interaction with the noncanonical plectin SH3 domain
Interactions with plectin where proposed to regulate the microtubule-stabilizing activity of MAP2c. Binding of MAP2c to the plectin SH3 domain and longer plectin fragments was observed experimentally (30). X-ray crystallography revealed that the plectin SH3 domain adopts the standard fold, but it lacks some aromatic side chains involved in canonical interactions with PXXP motifs, and its PXXP-binding site is partially occluded (29). It was therefore proposed that the plectin SH3 domain does not recognize the classical PXXP motifs. Here, we tested this hypothesis for intrinsically disordered MAP2c by solution NMR spectroscopy. Decrease in NMR peak height, indicative for MAP2c interactions with the well-folded SR4-SR5-SH3 plectin fragment, was not observed for the PXXP motifs, but in other regions (Fig. 9). Some of the observed decrease can be explained by electrostatic interactions of positively charged MAP2c residues in MTBD (e.g. of 298KQLR301 in MTBR1 and 352KNIR355 in MTBR3, resembling the alternative SH3-binding motifs RXXK (52)). However, significant binding was observed also in the region 208SLNSSISSARR218, preceding a positively charged sequence, 217RRTTRSEPIRRAGK230. We used the conformational analysis to examine whether this region can be distinguished by its structure. The analysis revealed (Fig. 4) that the sequence 206SFSLNSSISSARR218 is one of three regions with the highest population of α-helical stretches (together with the C-terminal helix and with the RII site). Based on this finding, we propose that the SR4-SR5-SH3 plectin fragment recognizes also an amphiphilic helix consisting of residues 206SFSLNSSISSARR218 and present in free MAP2c.
Site of interaction with the PKA regulatory domain
The sequence between Asp83 and Glu113 of MAP2c (box e in Fig. 1) aligns with a motif binding both the RI- and RII-regulatory subunit of PKA (54, 55), and the binding to the RII subunit is well documented (5, 31, 38). Unlike the canonical SH3 domains, dimers of the regulatory subunits of PKA form X-type helical bundles, recognizing relatively long amphiphilic α-helices (at least five turns) of well-folded protein kinase A–anchoring proteins (55). Therefore, the obvious question was whether such a helix is also significantly populated among conformers of free MAP2c. Increased population of α-helical conformation was observed in the selected ensembles for residues Thr80–Ser91 and Lys107–Gln110. In the middle part of the motif, the helical population in the selected ensembles did not exceed the statistical coil values, whereas the polyproline II conformation was more populated. A particularly low population of α-helix was observed for Gln96 and Val98. It shows that the RII site does not form a highly populated, uniform secondary structure in free MAP2c, as observed for the SH3-binding sites. It should be emphasized that the lack of a uniform secondary structure does not imply higher flexibility of the RII site. On the contrary, relaxation data revealed that the whole RII site is significantly more ordered than other parts of MAP2c. Contacts observed in PRE data suggest that hydrophobic interactions with aromatic residues in the N-terminal region may be involved. Taken together, our results indicate that the RII site of MAP2c represents a relatively complex local structure, qualitatively different from the SH3-binding sites. Further investigation will be needed to find out whether a regular helical structure is formed in complex with the regulatory subunit of PKA. In any case, it is interesting that the α-helical conformation is more populated in terminal regions of the motif, whereas the polyproline II conformation prevails in the interior part, which is supposed to form the amphiphilic helix critical for the interaction.
Global conformation
The obtained experimental data not only describe local conformational motifs, discussed above, but also provide information about global structural features of MAP2c. The overall shape of the MAP2c molecule is mostly given by electrostatic interactions between acidic N-terminal and positively charged C-terminal domains. The structural effect of the intramolecular electrostatic interactions is formation of bent “paper clip” conformations (56). At higher concentration, formation of antiparallel dimers is expected based on the charge distribution. Both types of structures have been observed earlier by Wille et al. (33). The antiparallel nature of the dimers is important for the interpretation of the PRE data. Whereas PRE observed between the N-terminal and C-terminal regions can be due to intramolecular and/or intermolecular interactions, the observed PRE inside the N-terminal region can be reliably attributed to the intramolecular contacts, discussed above. Regardless of the possible contribution of intermolecular contacts to the PRE data, it is clear that the actual interacting regions of MAP2c and htau40 differ. The contacts of MTSL-labeled E52C with the vicinity of Trp14 and the RII site are in agreement with the proposed model of the hydrophobic neurosteroid binding site and its interactions with the RII site (57). Most of the other differences can be explained by different charge distributions in htau40 and MAP2c (13). In the cellular environment, the intramolecular contacts contribute to the delicate equilibrium of interactions related to microtubule dynamics (58), aggregation of Tau (23), and interactions with other partners. The biological relevance of the intermolecular interactions is less clear. Quantitative data (53, 59–63) show that prenatal cytosolic concentrations of MAP2c are comparable with the concentrations of Tau in adult neurons (5–10 μm) (63), but formation of dimers can be expected in regions with locally increased MAP2c concentration (64). Interestingly, intermolecular interactions in MAP2c are reduced by PKA phosphorylation. Notably, PKA phosphorylation sites differ between MAP2c and Tau. For example, the major PKA phosphorylation site in MAP2c, Ser435, is localized close to the C terminus in a slightly positively charged region, and its phosphorylation makes interactions with the acidic N-terminal region in the antiparallel dimer less favorable.
In summary, high-resolution NMR experiments allowed us to study structural features and motions of a vast majority of MAP2c residues. Despite the high complexity of the conformational ensembles of MAP2c, the obtained data could be converted into parameters describing populations of conformations, long-range contacts, and average dynamics of individual amino acids using calibrated analytical procedures (37) to derive representative ensemble descriptions of the conformational space sampled by AP2c. Results of the analysis and NMR relaxation data revealed lower flexibility and increased populations of specific conformations in regions of MAP2c interacting with other proteins. They indicate that the local conformational propensities may play an important role in the protein partner recognition. As tested here for MAP2c, these conformational propensities cannot be reliably predicted for IDPs by currently available bioinformatics tools. Therefore, the presented type of structural ensemble analysis based on a set of experimental (NMR) data is essential for discovering yet unexplored binding motifs.
Experimental procedures
Sample preparation
The C348S and E52C/C348S MAP2c mutants were obtained previously (13). The same protocol was used to obtain the double mutants (Q110C/C348S, T153C/C348S, E203C/C348S, K282C/C348S, and L402C/C348S), using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies), following the manufacturer's protocol, using C348S MAP2c in pET3a as a template. The result of mutations was confirmed by sequencing. MAP2c expression, purification, and phosphorylation by PKA were performed as described earlier (13, 25, 65). The SR4-SR5-SH3 plectin fragment was expressed in Escherichia coli using a DNA construct including exons 16–21 (30).
Spin labeling
After purification, the cysteines were reduced by incubating MAP2c with 0.5 mm TCEP. For spin labeling, TCEP was removed by size-exclusion chromatography using the HiTrap desalting column (GE Healthcare), and the protein was incubated in 20 mm potassium phosphate, 150 mm potassium chloride, pH 7.4. A 10-fold molar excess of MTSL (Toronto Research Chemicals) dissolved in DMSO was added, and MTSL was allowed to bind overnight at 4 °C. The protein was then dialyzed into NMR buffer (50 mm MOPS, 150 mm sodium chloride, pH 6.9) before measurement. Paramagnetic MTSL label was converted into diamagnetic MTS by adding a 3-fold molar excess of ascorbic acid to the sample and incubating for 15 min at room temperature. 3D HNCO spectra were measured for the paramagnetic and diamagnetic MAP2c mutants. The PRE profiles were obtained by calculating the ratio of peak intensities of the paramagnetic and diamagnetic samples.
NMR spectroscopy
NMR experiments were acquired using 600-, 850-, and 950-MHz Bruker Avance III spectrometers equipped with TCI cryogenic probe heads with z axis gradients and a 700-MHz Bruker Avance III spectrometer equipped with a 1H/13C/15N TXO cryogenic probe head with z axis gradients. All experiments were performed at 27 °C with the temperature calibrated according to the chemical shift differences of pure methanol peaks. The indirect dimensions in 3D and 5D experiments were acquired in a nonuniformly sampled manner. On-grid Poisson disk sampling with a Gaussian probability distribution (66) was applied.
The 3D (CACO)NCACO spectrum and the 5D CACONCACO spectrum (25) were acquired as described earlier (13) to confirm assignment of mutants used in the PRE study. The 3D HNCO (67) spectra were acquired with spectral widths set to 18,939 (direct) × 2000 (15N) × 2000 (13C′) Hz and maximal evolution times of 120 ms for 15N and 80 ms for 13C′ indirectly detected dimensions. The overall number of 2048 complex points was acquired in the acquisition dimension, and 2000 hypercomplex points were randomly distributed over the indirectly detected dimensions. Interactions with the plectin SH3 domain were observed by comparing nonuniformly sampled HNCO spectra of samples containing 70 μm MAP2c with and without 70 μm SR4-SR5-SH3 plectin fragment in 50 mm MOPS, pH 6.9, 150 mm NaCl, and 0.7 mm TCEP. The spectra were recorded at 850 MHz and 10 °C (due to the limited thermal stability of the SR4-SR5-SH3 plectin fragment). Peaks with low intensity in the spectrum of free MAP2c were excluded from the analysis. 1D(NH) RDCs in PKA-phosphorylated MAP2c were measured at 600 MHz using a nonuniformly sampled 3D in-phase anti-phase HNCO experiment (68) with spectral widths of 2000 (15N) × 2000 (13C′) Hz in the indirect dimensions, with maximal evolution times of 180 ms for 15N and 120 ms for 13C′, and with 600 hypercomplex points randomly distributed over the indirectly detected dimensions. The 1D(NH) values were calculated as the difference between the splitting in spectra of phosphorylated MAP2c in the isotropic buffer and in a stretched 5% polyacrylamide gel. The relaxation rates of 0.9 mm unphosphorylated and phosphorylated MAP2c were measured using 3D HNCO-based relaxation experiments as described recently (39). The spectral density values J(ωN) and J(0.86ωH) (i.e. J(61 MHz) and J(515 MHz), respectively, on the 600-MHz spectrometer and J(96 MHz) and J(815 MHz), respectively, on the 950 MHz spectrometer) were calculated from R1 and steady-state NOE using reduced spectral density mapping (47, 48). The J(0) values were calculated from R1, Γx, and steady-state NOE was measured at 950 MHz using the LNX protocol described previously (46). The graphical spectral density analysis was performed as described earlier (46, 49, 51).
SAXS measurement
SAXS data were collected on the beamline BM29 BioSAXS ESFR in Grenoble, France. The highest concentrations of MAP2c in the dilution series were 12.0 and 13.6 mg/ml for unphosphorylated and phosphorylated sample, respectively. The data were recorded using the pixel 1M PILATUS detector at a sample–detector distance of 2.867 m, and a wavelength (λ) of 0.099 nm, covering the range of momentum transfer 0.025 nm−1 < q < 5 nm−1 (q = 4πsin(θ)/λ, where 2θ is the scattering angle). 50 μl of the MAP2c protein solution was loaded into a flow-through quartz capillary cell at 20.12 °C. The total exposure time was 10 s/sample. Several successive frames (10 for MAP2c and 20 for the buffer) were recorded for both the sample and the buffer. Each frame was inspected to check for the possible presence of protein damage before calculating average intensities. Solvent contributions (buffer backgrounds collected before and after the protein sample) were averaged and subtracted from the associated protein sample. No radiation damage was observed during the data collection. The data were processed using standard procedures with PRIMUS (69).
Conformational analysis
The flexible-meccano program (70, 71) was used to generate a large ensemble of conformers of the protein. For each conformer in the ensemble, the side-chain and hydrogen atoms were added to the model using SCCOMP (72). The random-coil torsion angle database was used by flexible-meccano to create an ensemble of 10,000 conformers. Moreover, a database of torsion angles filtered to be in agreement with experimental chemical shifts of 15N, 13C′, 13Cα, and 13Cβ was prepared to enhance convergence of ASTEROIDS using flexible-meccano/ASTEROIDS selection and used by flexible-meccano to create another ensemble of 74,000 conformers. Both ensembles were joined together to define a starting pool of 84,000 conformers, and experimental observables were calculated for each conformer. Chemical shifts were predicted using SPARTA (36), SAXS using CRYSOL (73–75), and PRE as described previously (76). Local alignment windows of 15 amino acids in length were used to calculate averaged RDCs (77). These were then combined with a generic baseline to account for long-range effects (34, 76). The starting pool was used for selection of a sub-ensemble representing the disorder protein by ASTEROIDS (34). The optimal number of structures in the selection was estimated from cross-validation procedures to be ∼600. To achieve convergence, 200,000 iterations of the genetic algorithm were required. The ASTEROIDS selection was repeated five times with the same set of data to confirm reproducibility of torsion angle distribution in the selected ensembles and of the back-calculated chemical shift, PRE, and SAXS data. Differences between statistical coil (COIL) sampling and selected ensembles (AST) were characterized as described previously (26) with the metric ΔRama defined for each residue as follows,
| (Eq. 1) |
where pCOIL and pAST are statistical coil and selected populations, respectively, and k covers four regions of the Ramachandran diagram (α-helix, β-sheet, polyproline II, and the remaining region including the left-handed α-helical conformation).
Author contributions
K. M., V. Z., S. J., L. Z., J. H., and M. B. conceived and designed the research and wrote the manuscript; S. J., J. N., and K. M. prepared the samples; K. M., V. Z., S. J., E. N., and M. Z. performed the experiment and analyzed the data; V. Z., M. R.-J., and M. B. performed the conformational analysis; M. Z. provided the htau40 data; and all authors reviewed the manuscript.
Acknowledgments
This work used the platforms of the Grenoble Instruct Center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). The CIISB research infrastructure project LM2015043 funded by the Ministry of Education, Youth, and Sports of the Czech Republic (MEYS CR) is gratefully acknowledged for the partial financial support of the measurements at the Josef Dadok National NMR Centre and Proteomics Core Facilities, CEITEC–Masaryk University. Computational resources were provided by CESNET (LM2015042) and the CERIT Scientific Cloud (LM2015085) under the program “Projects of Large Research, Development, and Innovations Infrastructures” funded by MEYS CR, and by the IT4Innovations National Supercomputing Center (LM2015070) under the program “Large Infrastructures for Research, Experimental Development, and Innovations” funded by MEYS CR. We thank G. Wiche for providing plasmids for protein expression, and M. Z. thanks E. Mandelkow and J. Biernat for useful discussions.
This work was supported in part by Czech Science Foundation Grant GA15-14974S. The authors declare that they have no conflicts of interest with the contents of this article.
The small angle scattering data and models reported in this paper have been submitted to the Small Angle Scattering Biological Data Bank under SASBD accession numbers SASDD62, SASDD72, SASDD82, SASDD92, SASDDA2, SASDDB2, and SASDDC2.
The data reported in this paper have been submitted to the Biological Magnetic Resonance Bank under BMRB accession number 27350.
- MAP
- microtubule-associated protein
- MTBD
- microtubule-binding domain
- IDP
- intrinsically disordered protein
- SH3
- Src-homology 3
- PKA
- cAMP-dependent protein kinase
- SAXS
- small-angle X-ray scattering
- PRE
- paramagnetic relaxation enhancement
- MTSL
- (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl)-methanethiosulfonate
- 2D
- 3D, and 5D, two-, three-, and five-dimensional, respectively
- RDC
- residual dipolar coupling
- TCF
- time-correlation function
- TCEP
- tris(2-carboxyethyl)phosphine.
References
- 1. Jalava N. S., Lopez-Picon F. R., Kukko-Lukjanov T. K., and Holopainen I. E. (2007) Changes in microtubule-associated protein-2 (MAP2) expression during development and after status epilepticus in the immature rat hippocampus, Int. J. Dev. Neurosci. 25, 121–131 10.1016/j.ijdevneu.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 2. Wang Y., and Mandelkow E. (2016) Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 10.1038/nrg.2015.11 [DOI] [PubMed] [Google Scholar]
- 3. Sündermann F., Fernandez M. P., and Morgan R. O. (2016) An evolutionary roadmap to the microtubule-associated protein MAP tau. BMC Genomics 17, 264 10.1186/s12864-016-2590-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Viereck C., Tucker R. P., and Matus A. (1989) The adult rat olfactory system expresses microtubule-associated proteins found in the developing brain. J. Neurosci. 9, 3547–3557 10.1523/JNEUROSCI.09-10-03547.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dehmelt L., and Halpain S. (2005) The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 6, 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brugg B., and Matus A. (1991) Phosphorylation determines the binding of microtubule-associated protein 2 (MAP2) to microtubules in living cells. J. Cell Biol. 114, 735–743 10.1083/jcb.114.4.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brandt R., Lee G., Teplow D. B., Shalloway D., and Abdel-Ghany M. (1994) Differential effect of phosphorylation and substrate modulation on tau's ability to promote microtubule growth and nucleation. J. Biol. Chem. 269, 11776–11782 [PubMed] [Google Scholar]
- 8. Illenberger S., Drewes G., Trinczek B., Biernat J., Meyer H. E., Olmsted J. B., Mandelkow E. M., and Mandelkow E. (1996) Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark: phosphorylation sites and regulation of microtubule dynamics. J. Biol. Chem. 271, 10834–10843 10.1074/jbc.271.18.10834 [DOI] [PubMed] [Google Scholar]
- 9. Sánchez C., Díaz-Nido J., and Avila J. (2000) Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 61, 133–168 10.1016/S0301-0082(99)00046-5 [DOI] [PubMed] [Google Scholar]
- 10. Alexa A., Schmidt G., Tompa P., Ogueta S., Vázquez J., Kulcsár P., Kovács J., Dombrádi V., and Friedrich P. (2002) The phosphorylation state of threonine-220, a uniquely phosphatase-sensitive protein kinase A site in microtubule-associated protein MAP2c, regulates microtubule binding and stability. Biochemistry 41, 12427–12435 10.1021/bi025916s [DOI] [PubMed] [Google Scholar]
- 11. Landrieu I., Lacosse L., Leroy A., Wieruszeski J. M., Trivelli X., Sillen A., Sibille N., Schwalbe H., Saxena K., Langer T., and Lippens G. (2006) NMR analysis of a Tau phosphorylation pattern. J. Am. Chem. Soc. 128, 3575–3583 10.1021/ja054656+ [DOI] [PubMed] [Google Scholar]
- 12. Martin L., Latypova X., Wilson C. M., Magnaudeix A., Perrin M.-L., Yardin C., and Terro F. (2013) Tau protein kinases: involvement in Alzheimer's disease. Ageing Res. Rev. 12, 289–309 10.1016/j.arr.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 13. Jansen S., Melková K., Trošanová Z., Hanáková K., Zachrdla M., Nováček J., Župa E., Zdráhal Z., Hritz J., and Žídek L. (2017) Quantitative mapping of microtubule-associated protein 2c (MAP2c) phosphorylation and regulatory protein 14-3-3ζ-binding sites reveals key differences between MAP2c and its homolog Tau. J. Biol. Chem. 292, 6715–6727 10.1074/jbc.M116.771097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie C., Soeda Y., Shinzaki Y., In Y., Tomoo K., Ihara Y., and Miyasaka T. (2015) Identification of key amino acids responsible for the distinct aggregation properties of microtubule-associated protein 2 and tau. J. Neurochem. 135, 19–26 10.1111/jnc.13228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bibow S., Mukrasch M. D., Chinnathambi S., Biernat J., Griesinger C., Mandelkow E., and Zweckstetter M. (2011) The dynamic structure of filamentous Tau. Angew. Chem. Int. Ed. Engl. 50, 11520–11524 10.1002/anie.201105493 [DOI] [PubMed] [Google Scholar]
- 16. Dunker A. K., Obradovic Z., Romero P., Garner E. C., and Brown C. J. (2000) Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 11, 161–171 [PubMed] [Google Scholar]
- 17. Dunker A. K., Oldfield C. J., Meng J., Romero P., Yang J. Y., Chen J. W., Vacic V., Obradovic Z., and Uversky V. N. (2008) The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 9, Suppl. 2, S1 10.1186/1471-2164-9-S2-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dyson H. J., and Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- 19. Tompa P. (2005) The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579, 3346–3354 10.1016/j.febslet.2005.03.072 [DOI] [PubMed] [Google Scholar]
- 20. Fink A. L. (2005) Natively unfolded proteins. Curr. Opin. Struct. Biol. 15, 35–41 10.1016/j.sbi.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 21. Mukrasch M. D., Biernat J., von Bergen M., Griesinger C., Mandelkow E., and Zweckstetter M. (2005) Sites of tau important for aggregation populate β-structure and bind to microtubules and polyanions. J. Biol. Chem. 280, 24978–24986 10.1074/jbc.M501565200 [DOI] [PubMed] [Google Scholar]
- 22. Mukrasch M. D., von Bergen M., Biernat J., Fischer D., Griesinger C., Mandelkow E., and Zweckstetter M. (2007) The “jaws” of the tau-microtubule interaction. J. Biol. Chem. 282, 12230–12239 10.1074/jbc.M607159200 [DOI] [PubMed] [Google Scholar]
- 23. Mukrasch M. D., Bibow S., Korukottu J., Jeganathan S., Biernat J., Griesinger C., Mandelkow E., and Zweckstetter M. (2009) Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 7, e34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sibille N., Huvent I., Fauquant C., Verdegem D., Amniai L., Leroy A., Wieruszeski J.-M., Lippens G., and Landrieu I. (2012) Structural characterization by nuclear magnetic resonance of the impact of phosphorylation in the proline-rich region of the disordered tau protein. Proteins 80, 454–462 10.1002/prot.23210 [DOI] [PubMed] [Google Scholar]
- 25. Nováček J., Janda L., Dopitová R., Žídek L., and Sklenář V. (2013) Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: transient secondary structure analysis of 49.2 kDa microtubule associated protein 2c. J. Biomol. NMR 56, 291–301 10.1007/s10858-013-9761-7 [DOI] [PubMed] [Google Scholar]
- 26. Schwalbe M., Ozenne V., Bibow S., Jaremko M., Jaremko L., Gajda M., Jensen M. R., Biernat J., Becker S., Mandelkow E., Zweckstetter M., and Blackledge M. (2014) Predictive atomic resolution descriptions of intrinsically disordered hTau40 and α-synuclein in solution from NMR and small angle scattering. Structure 22, 238–249 10.1016/j.str.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 27. Zamora-Leon S. P., Lee G., Davies P., and Shafit-Zagardo B. (2001) Binding of Fyn to MAP-2c through an SH3 binding domain. J. Biol. Chem. 276, 39950–39958 10.1074/jbc.M107807200 [DOI] [PubMed] [Google Scholar]
- 28. Reynolds J. G., McCalmon S. A., Donaghey J. A., and Naya F. (2008) Deregulated protein kinase A signaling and myospryn expression in muscular dystrophy. J. Biol. Chem. 283, 8070–8074 10.1074/jbc.C700221200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ortega E., Buey R. M., Sonnenberg A., and de Pereda J. M. (2011) The structure of the plakin domain of plectin reveals non-canonical SH3 domain interacting with its fourth spectrin repeat. J. Biol. Chem. 286, 12429–12438 10.1074/jbc.M110.197467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valencia R. G., Walko G., Janda L., Novacek J., Mihailovska E., Reipert S., Andrä-Marobela K., and Wiche G. (2013) Intermediate filament-associated cytolinker plectin 1c destabilizes microtubules in keratinocytes. Mol. Biol. Cell 24, 768–784 10.1091/mbc.e12-06-0488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malmendal A., Halpain S., and Chazin W. J. (2003) Nascent structure in the kinase anchoring domain of microtubule-associated protein 2. Biochem. Biophys. Res. Commun. 301, 136–142 10.1016/S0006-291X(02)02989-3 [DOI] [PubMed] [Google Scholar]
- 32. Gillespie J. R., and Shortle D. (1997) Characterization of long-range structure in the denatured state of staphylococcal nuclease. I. Paramagnetic relaxation enhancement by nitroxide spin labels. J. Mol. Biol. 268, 158–169 10.1006/jmbi.1997.0954 [DOI] [PubMed] [Google Scholar]
- 33. Wille H., Mandelkow E. M., and Mandelkow E. (1992) The juvenile microtubule-associated protein MAP2c is a rod-like molecule that forms antiparallel dimer. J. Biol. Chem. 267, 10737–10742 [PubMed] [Google Scholar]
- 34. Nodet G., Salmon L., Ozenne V., Meier S., Jensen M. R., and Blackledge M. (2009) Quantitative description of backbone conformational sampling of unfolded proteins at amino acid resolution from NMR residual dipolar couplings. J. Am. Chem. Soc. 131, 17908–17918 10.1021/ja9069024 [DOI] [PubMed] [Google Scholar]
- 35. Marsh J. A., Singh V. K., Jia Z., and Forman-Kay J. D. (2006) Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: implications for fibrillation. Protein Sci. 15, 2795–2804 10.1110/ps.062465306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen Y., and Bax A. (2007) Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J. Biomol. NMR 38, 289–302 10.1007/s10858-007-9166-6 [DOI] [PubMed] [Google Scholar]
- 37. Ozenne V., Schneider R., Yao M., Huang J.-R., Salmon L., Zweckstetter M., Jensen M. R., and Blackledge M. (2012) Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J. Am. Chem. Soc. 134, 15138–15148 10.1021/ja306905s [DOI] [PubMed] [Google Scholar]
- 38. Obar R. A., Dingus J., Bayley H., and Vallee R. B. (1989) The RII subunit of cAMP-dependent protein kinase binds to a common amino-terminal domain in microtubule-associated proteins 2A, 2B, and 2C. Neuron 3, 639–645 10.1016/0896-6273(89)90274-2 [DOI] [PubMed] [Google Scholar]
- 39. Srb P., Nováček J., Kadeřávek P., Rabatinová A., Krásný L., Žídková, Bobálová J. J., Sklenář V., and Žídek L. (2017) Triple resonance 15N NMR relaxation experiments for studies of intrinsically disordered proteins. J. Biomol. NMR 69, 133–146 10.1007/s10858-017-0138-1 [DOI] [PubMed] [Google Scholar]
- 40. Ferrage F. (2012) Protein dynamics by 15N nuclear magnetic relaxation. Methods Mol. Biol. 831, 141–163 10.1007/978-1-61779-480-3_9 [DOI] [PubMed] [Google Scholar]
- 41. Wangsness R., and Bloch F. (1953) The dynamical theory of nuclear induction. Phys. Rev. Lett. 89, 728–739 [Google Scholar]
- 42. Abragam A. (1961) The Principles of Nuclear Magnetism, pp. 264–300, Clarendon Press, Oxford [Google Scholar]
- 43. Redfield A. G. (1965) The theory of relaxation processes. Adv. Magn. Reson. 1, 1–32 10.1016/B978-1-4832-3114-3.50007-6 [DOI] [Google Scholar]
- 44. Cavanagh J., Faibrother W., Palmer A. 3rd, Rance M., and Skelton N. (2007) Protein NMR Spectroscopy: Principles and Practice, 2nd Ed., pp. 334–404, Elsevier Academic Press, Burlington, MA [Google Scholar]
- 45. Kowalewski A., and Mäler L. (2006) Nuclear Spin Relaxation in Liquids: Theory, Experiments, and Applications, pp. 127–157, Taylor & Francis, New York [Google Scholar]
- 46. Kadeřávek P., Zapletal V., Rabatinová A., Krásný L., Sklenář V., and Žídek L. (2014) Spectral density mapping protocols for analysis of molecular motions in disordered proteins. J. Biomol. NMR 58, 193–207 10.1007/s10858-014-9816-4 [DOI] [PubMed] [Google Scholar]
- 47. Ishima R., and Nagayama K. (1995) Quasi-spectral-density function analysis for nitrogen-15 nuclei in proteins. J. Magn. Reson. Ser. B 108, 73–76 10.1006/jmrb.1995.1104 [DOI] [Google Scholar]
- 48. Ishima R., and Nagayama K. (1995) Protein backbone dynamics revealed by quasi spectral density function analysis of amide N-15 nuclei. Biochemistry 34, 3162–3171 10.1021/bi00010a005 [DOI] [PubMed] [Google Scholar]
- 49. Lefevre J. F., Dayie K. T., Peng J. W., and Wagner G. (1996) Internal mobility in the partially folded DNA binding and dimerization domains of GAL4: NMR analysis of the N-H spectral density functions. Biochemistry 35, 2674–2686 10.1021/bi9526802 [DOI] [PubMed] [Google Scholar]
- 50. Barthe P., Chiche L., Declerck N., Delsuc M. A., Lefèvre J. F., Malliavin T., Mispelter J., Stern M. H., Lhoste J. M., and Roumestand C. (1999) Refined solution structure and backbone dynamics of 15N-labeled C12A-p8MTCP1 studied by NMR relaxation. J. Biomol. NMR 15, 271–288 10.1023/A:1008336418418 [DOI] [PubMed] [Google Scholar]
- 51. Krízová H., Zídek L., Stone M. J., Novotny M. V., and Sklenár V. (2004) Temperature-dependent spectral density analysis applied to monitoring backbone dynamics of major urinary protein-I complexed with the pheromone 2-sec-butyl-4,5-dihydrothiazole. J. Biomol. NMR 28, 369–384 10.1023/B:JNMR.0000015404.61574.65 [DOI] [PubMed] [Google Scholar]
- 52. Kaneko T., Li L., and Li S. S. (2008) The SH3 domain-A family of versatile peptide- and protein-recognition module, Front. Biosci. 13, 4938–4952 [DOI] [PubMed] [Google Scholar]
- 53. Zamora-Leon S. P., Bresnick A., Backer J. M., and Shafit-Zagardo B. (2005) Fyn phosphorylates human MAP-2c on tyrosine 67. J. Biol. Chem. 280, 1962–1970 10.1074/jbc.M411380200 [DOI] [PubMed] [Google Scholar]
- 54. Newlon M. G., Roy M., Morikis D., Carr D. W., Westphal R., Scott J. D., and Jennings P. A. (2001) A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 20, 1651–1662 10.1093/emboj/20.7.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Götz F., Roske Y., Schulz M. S., Autenrieth K., Bertinetti D., Faelber K., Zühlke K., Kreuchwig A., Kennedy E. J., Krause G., Daumke O., Herberg F. W., Heinemann U., and Klussmann E. (2016) AKAP18:PKA-RIIα structure reveals crucial anchor points for recognition of regulatory subunits of PKA. Biochem. J. 473, 1881–1894 10.1042/BCJ20160242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jeganathan S., von Bergen M., Brutlach H., Steinhoff H. J., and Mandelkow E. (2006) Global hairpin folding of tau in solution. Biochemistry 45, 2283–2293 10.1021/bi0521543 [DOI] [PubMed] [Google Scholar]
- 57. Laurine E., Lafitte D., Grégoire C., Sérée E., Loret E., Douillard S., Michel B., Briand C., and Verdier J. M. (2003) Specific binding of dehydroepiandrosterone to the N terminus of the microtubule-associated protein MAP2. J. Biol. Chem. 278, 29979–29986 10.1074/jbc.M303242200 [DOI] [PubMed] [Google Scholar]
- 58. Goode B. L., Denis P. E., Panda D., Radeke M. J., Miller H. P., Wilson L., and Feinstein S. C. (1997) Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol. Biol. Cell 8, 353–365 10.1091/mbc.8.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guo Y., Gong H. S., Zhang J., Xie W. L., Tian C., Chen C., Shi Q., Wang S. B., Xu Y., Zhang B. Y., and Dong X. P. (2012) Remarkable reduction of MAP2 in the brains of scrapie-infected rodents and human prion disease possibly correlated with the increase of calpain, PLoS One 7, e30163 10.1371/journal.pone.0030163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tie L., Zhang J. Z., Lin Y. H., Su T. H., Li Y. H., Wu H. L., Zhang Y. Y., Yu H. M., and Li X. J. (2008) Epinephrine increases phosphorylation of MAP-2c in rat pheochromocytoma cells (PC12 cells) via a protein kinase C- and mitogen activated protein kinase-dependent mechanism. J. Proteome Res. 7, 1704–1711 10.1021/pr700711s [DOI] [PubMed] [Google Scholar]
- 61. Ackmann M., Wiech H., and Mandelkow E. (2000) Nonsaturable binding indicates clustering of tau on the microtubule surface in a paired helical filament-like conformation. J. Biol. Chem. 275, 30335–30343 10.1074/jbc.M002590200 [DOI] [PubMed] [Google Scholar]
- 62. Meixner A., Haverkamp S., Wässle H., Führer S., Thalhammer J., Kropf N., Bittner R. E., Lassmann H., Wiche G., and Propst F. (2000) MAP1B is required for axon guidance and is involved in the development of the central and peripheral nervous system. J. Cell Biol. 151, 1169–1178 10.1083/jcb.151.6.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu F., Iqbal K., Grundke-Iqbal I., Rossie S., and Gong C. X. (2005) Dephosphorylation of tau by protein phosphatase 5: impairment in Alzheimer's disease. J. Biol. Chem. 280, 1790–1796 10.1074/jbc.M410775200 [DOI] [PubMed] [Google Scholar]
- 64. Ozer R. S., and Halpain S. (2000) Phosphorylation-dependent localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol. Biol. Cell 11, 3573–3587 10.1091/mbc.11.10.3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gamblin T. C., Nachmanoff K., Halpain S., and Williams R. C. (1996) Recombinant microtubule-associated protein 2c reduces the dynamic instability of individual microtubules. Biochemistry 35, 12576–12586 10.1021/bi961135d [DOI] [PubMed] [Google Scholar]
- 66. Kazimierczuk K., Zawadzka A., and Koźmiński W. (2008) Optimization of random time domain sampling in multidimensional NMR. J. Magn. Reson. 192, 123–130 10.1016/j.jmr.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 67. Kay L. E., Ikura M., Tschudin R., and Bax A. (1990) Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins, J. Magn. Reson. 89, 496–514 [DOI] [PubMed] [Google Scholar]
- 68. Nováček J., Žídek L., and Sklenář V. (2014) Toward optimal-resolution NMR of intrinsically disordered proteins. J. Magn. Reson. 241, 41–52 10.1016/j.jmr.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 69. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 10.1107/S0021889803012779 [DOI] [Google Scholar]
- 70. Bernadó P., Blanchard L., Timmins P., Marion D., Ruigrok R. W., and Blackledge M. (2005) A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc. Natl. Acad. Sci. U.S.A. 102, 17002–17007 10.1073/pnas.0506202102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ozenne V., Bauer F., Salmon L., Huang J. R., Jensen M. R., Segard S., Bernadó P., Charavay C., and Blackledge M. (2012) Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 28, 1463–1470 10.1093/bioinformatics/bts172 [DOI] [PubMed] [Google Scholar]
- 72. Eyal E., Najmanovich R., McConkey B. J., Edelman M., and Sobolev V. (2004) Importance of solvent accessibility and contact surfaces in modeling side-chain conformations in proteins, J. Comput. Chem. 25, 712–724 10.1002/jcc.10420 [DOI] [PubMed] [Google Scholar]
- 73. Bernadó P., and Blackledge M. (2009) A self-consistent description of the conformational behavior of chemically denatured proteins from NMR and small angle scattering. Biophys. J. 97, 2839–2845 10.1016/j.bpj.2009.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28, 768–773 10.1107/S0021889895007047 [DOI] [Google Scholar]
- 75. Wells M., Tidow H., Rutherford T. J., Markwick P., Jensen M. R., Mylonas E., Svergun D. I., Blackledge M., and Fersht A. R. (2008) Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 105, 5762–5767 10.1073/pnas.0801353105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Salmon L., Nodet G., Ozenne V., Yin G., Jensen M. R., Zweckstetter M., and Blackledge M. (2010) NMR characterization of long-range order in intrinsically disordered proteins. J. Am. Chem. Soc. 132, 8407–8418 10.1021/ja101645g [DOI] [PubMed] [Google Scholar]
- 77. Marsh J. A., Baker J. M. R., Tollinger M., and Forman-Kay J. D. (2008) Calculation of residual dipolar couplings from disordered state ensembles using local alignment. J. Am. Chem. Soc. 130, 7804–7805 10.1021/ja802220c [DOI] [PubMed] [Google Scholar]
- 78. Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., and Jones D. T. (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20, 2138–2139 10.1093/bioinformatics/bth195 [DOI] [PubMed] [Google Scholar]
- 79. Medina M. W., Gao F., Naidoo D., Rudel L. L., Temel R. E., McDaniel A. L., Marshall S. M., and Krauss R. M. (2011) Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PLoS One 6, e19420 10.1371/journal.pone.0019420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dosztányi Z., Csizmok V., Tompa P., and Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- 81. Kozlowski L. P., and Bujnicki J. M. (2012) MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 13, 111 10.1186/1471-2105-13-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ishida T., and Kinoshita K. (2007) PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–W464 10.1093/nar/gkm363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yang Z. R., Thomson R., McNeil P., and Esnouf R. M. (2005) RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21, 3369–3376 10.1093/bioinformatics/bti534 [DOI] [PubMed] [Google Scholar]
- 84. Vullo A., Bortolami O., Pollastri G., and Tosatto S. (2006) Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 34, W164–W168 10.1093/nar/gkl166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kyte J., and Doolittle R. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]