Abstract
Human myeloid-derived growth factor (MYDGF; also known as C19orf10) is named based on its identification as a secreted monocyte/macrophage–derived mediator of cardiac repair following myocardial infarction in mice. Homologs of MYDGF, however, are present in organisms throughout and outside of the animal kingdom, some of which lack hematopoietic and circulatory systems. Moreover, the UPF0556 protein domain, which defines these homologs, lacks a known structure. As a result, the functions and properties of MYDGF are unclear. Our current work was initiated to test whether MYDGF is present in secretory vesicles of eosinophils as it was recently reported to be abundant in these cells. However, we could not demonstrate secretion and unexpectedly discovered that MYDGF colocalizes with P4HB in the nuclear envelope, which comprises the bulk of endoplasmic reticulum (ER) in eosinophils, and with P4HB and RCAS1 in Golgi. We noted a ubiquitous C-terminal sequence, BXEL (B, basic; X, variable residue; E, Glu; L, Leu), that has the potential to retain human MYDGF and its homologs in the ER. To test the functionality of this sequence, we expressed full-length human MYDGF or MYDGF lacking the C-terminal Glu-Leu residues in monolayers of human embryonic kidney 293 (HEK293) cells. Full-length MYDGF accumulated in cells, whereas truncated MYDGF appeared in the medium. These observations reveal that MYDGF resides in the ER and Golgi and provide a new framework for investigating and understanding this intriguing protein.
Keywords: endoplasmic reticulum (ER), protein trafficking (Golgi), protein secretion, protein sequence, eosinophil, interleukin, cell polarity, C19orf10, MYDGF, UPF0556
Introduction
The literature on myeloid-derived growth factor (MYDGF)2 is checkered. The protein was identified first in a screen of ST2 murine bone marrow stromal cells for cDNAs encoding proteins that cause proliferation of mutant Ba/F3 murine pre-B cells and was given the name SF20/IL25 (1). The SF20/IL25 paper was retracted when the authors were unable to reproduce the published cell proliferation responses (2). The protein was subsequently identified by two-dimensional electrophoresis separation, in-gel digestion, and MS as SF20/IL25 in murine 3T3-LI adipocytes (3) and as C19orf10 in human fibroblast-like synoviocytes (4), human eosinophils (5), murine macrophages (6), and human cholangiocarcinoma cells (7). Message for C19orf10 was found to be increased in some human hepatocellular carcinomas, and overexpression or addition of recombinant protein to medium resulted in phosphorylation of AKT and proliferation of a liver cancer cell line (8).
The protein was named MYDGF after being identified as a paracrine-acting protein secreted from monocytes/macrophages that improves tissue repair and heart function after myocardial infarction (9). MYDGF was selected from among 42 poorly characterized and putatively secreted proteins that were expressed with C-terminal affinity tags in human embryonic kidney 293 (HEK293) cells. The recombinant proteins were tested for cytoprotective effects on serum-starved, cultured neonatal rat ventricular myocytes. Cytoprotection by MYDGF was associated with phosphorylation of AKT and inhibition of apoptosis (9). In addition, exogenously added MYDGF promoted proliferation of human coronary artery endothelial cells (9). Finally, mice with genetic knockout of MYDGF, although lacking developmental defects, had larger infarct scars and decreased cell proliferation and angiogenesis at the infarct border zone compared with WT mice; prior transplantation of normal bone marrow or administration of recombinant MYDGF after ischemia and reperfusion reversed this effect (9).
Our interest in MYDGF stemmed from a quantitative proteomics study in which C19orf10 was reported as the 559th most abundant protein of human blood eosinophils, present at 1–3% of the molar abundances of the major eosinophil granule proteins RNASE2, RNASE3, PRG2, and PRG3 (10). When we realized that C19orf10 had been characterized functionally as MYDGF (9), we asked whether eosinophils represent a second myeloid source of releasable MYDGF. As described herein, however, upon immunostaining of eosinophils we found that MYDGF did not localize to eosinophil granules but rather to the Golgi and endoplasmic reticulum (ER), which in eosinophils is restricted to the nuclear envelope (11).
A search of various databases revealed homologs of MYDGF throughout and outside of the animal kingdom. Virtually all homologs, which consist of and define the UPF0556 protein domain, have a C-terminal KDEL-like, putative ER retention sequence, BXEL (B, Arg, His, or Lys; X, variable residue; E, Glu; L, Leu). We analyzed the C-terminal RTEL sequence of human MYDGF by expressing the full-length protein or MYDGF lacking the last two residues in HEK293 cells. We found that the truncated protein was extensively secreted. These observations indicate that the RTEL sequence functions to retain MYDGF in the ER and Golgi alongside known modulators of classical protein secretion.
Results
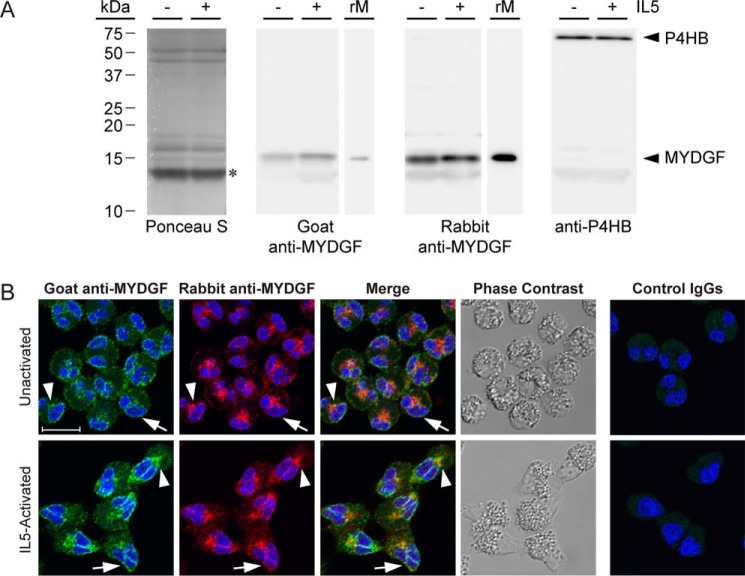
To assess eosinophils as a potential source of releasable MYDGF, we immunoblotted and immunofluorescently stained human blood eosinophils that had been left unactivated or activated by interleukin 5 (IL5) for 10 min (Fig. 1). The assays were performed using either commercial goat or in-house rabbit polyclonal antibodies produced against and affinity-purified with recombinant, human MYDGF.
Figure 1.
Eosinophil MYDGF. A, immunoblots of lysate from 2 × 105 unactivated (−) or IL5-activated (+) eosinophils per lane and 10 ng of recombinant, human MYDGF (rM) produced and purified from E. coli. Ponceau S stain is representative of total lysate protein; the most intense band centered at 13.4 kDa (*) corresponded to a band found in all immunoblot lanes regardless of whether goat anti-MYDGF, rabbit anti-MYDGF, or anti-P4HB primary antibodies were used. Mature MYDGF and P4HB were both detected in close proximity to their theoretical molecular masses of 15.8 and 55.3 kDa, respectively. All immunoblot images were taken with a 3-min exposure and the same image settings. B, human eosinophils, unactivated or activated with IL5, triple-stained with DAPI (blue), goat anti-human MYDGF or control IgGs (green), and rabbit anti-human MYDGF or control IgGs (pseudocolored red). Cells were imaged by confocal fluorescence and transmission microscopy. Images are of single 0.3-μm slices. Arrowheads, patch between nuclear lobes; arrows, periphery of nucleus; scale bar, 10 μm. Immunostaining and immunoblots are representative of experiments conducted on three or more separate occasions.
Both antibodies detected a 15.7-kDa band in eosinophil lysate that migrated in accordance with the theoretical mass of mature, human MYDGF (15.8 kDa) and comigrated with recombinant MYDGF standard (Fig. 1A). Faint detection of a lower apparent molecular mass band (13.4 kDa) was observed that corresponded to the diffuse, intense band stained by Ponceau S (presumably highly abundant histones, Charcot-Leyden crystal protein, and major granular proteins (10)); this band was also detected in blots performed with antibodies to the ER marker P4HB (loading control; Fig. 1A) or with nonimmune control antibodies (data not shown). ImageJ immunoblot quantification of the relative band intensities for MYDGF in eosinophil lysates compared with 10 ng of recombinant protein (Fig. 1B) indicated that ∼2 ng of MYDGF is present in the lysate of 2 × 105 eosinophils. This figure is within 10-fold of the 17 ng of MYDGF per 2 × 105 eosinophils that can be estimated by relating intensity-based absolute quantification values reported for MYDGF to those of several eosinophil granule proteins of known abundance in a global proteomics study of eosinophils (10, 12).
For immunofluorescence, eosinophils were fixed with paraformaldehyde, collected by cytospinning, and extracted with detergent. Cells were then incubated with goat anti-MYDGF, rabbit anti-MYDGF, or nonimmune control antibodies before being incubated with Alexa Fluor 488–conjugated anti-goat IgG or Alexa Fluor 647–conjugated anti-rabbit IgG and imaged for fluorescence and phase (Fig. 1B). We had hypothesized that MYDGF would be packaged for secretion in the large phase–dense granules that fill the cytoplasm of unactivated eosinophils and move to the granular pole when eosinophils are induced to polarize with IL5 (13). We detected no signal in granules above the autofluorescence (due to a high content of FAD (14)) found in the specificity control. Instead, both goat and rabbit anti-MYDGF localized specifically at the periphery of the bilobed nucleus (arrow) and in a centralized patch (arrowhead) located in the cleft between the nuclear lobes (Fig. 1B). The perinuclear staining was most striking as a sharp strip between the nuclear lobes in IL5-treated cells. Colocalization of the two antibodies was quantified as a quotient of the overlapping area stained by both antibodies divided by the area stained by one of the antibodies. Both antibodies had above 85% colocalization in unactivated and IL5-activated eosinophils (Table 1). Despite the overlap and recognition of the same band in immunoblots, visually the rabbit anti-MYDGF antibodies reproducibly stained the central patch more intensely than the goat antibodies, whereas the opposite was true for the perinuclear staining (Fig. 1B). We believe that the discrepancy stems from the two antibody populations differentially recognizing epitopes of formaldehyde-fixed MYDGF depending on the two cellular locations.
Table 1.
Analysis of MYDGF colocalization with organelle markers
Colocalization was quantified as the area of overlapping signal from goat anti-MYDGF (Alexa Fluor 488–conjugated secondary antibody) and rabbit anti-MYDGF or rabbit antibodies to organelle markers (Alexa Fluor 647–conjugated secondary antibody) divided by the signal area from one antibody alone. Values are reported as averages from 9–16 0.3-μm slices from fields of 21–41 eosinophils that were overlaid to create the maximal intensity projections from which the cells shown in Fig. 2 were chosen. Standard deviations represent variation among the slices.
| Alexa Fluor 488 (green) | Alexa Fluor 647 (pseudocolored red) | IL5 | Green having red |
Red having green |
|---|---|---|---|---|
| Green | Red | |||
| Goat anti-MYDGF | Rabbit anti-MYDGF | − | 0.86 ± 0.04 | 0.87 ± 0.12 |
| + | 0.89 ± 0.08 | 0.97 ± 0.03 | ||
| Goat anti-MYDGF | Rabbit anti-P4HB (ER) | − | 0.92 ± 0.06 | 0.91 ± 0.12 |
| + | 0.91 ± 0.06 | 0.99 ± 0.01 | ||
| Goat anti-MYDGF | Rabbit anti-RCAS1 (Golgi) | − | 0.28 ± 0.20 | 0.89 ± 0.27 |
| + | 0.41 ± 0.26 | 1.00 ± 0.01 | ||
| Goat anti-MYDGF | Rabbit anti-AIF (mitochondria) | − | 0.53 ± 0.20 | 0.81 ± 0.06 |
| + | 0.40 ± 0.23 | 0.54 ± 0.12 | ||
| Goat anti-MYDGF | Rabbit anti-EEA1 (early endosomes) | − | 0.28 ± 0.09 | 0.49 ± 0.08 |
| + | 0.26 ± 0.10 | 0.46 ± 0.19 | ||
| Goat anti-MYDGF | Rabbit anti-LAMP1 (lysosome) | − | 0.68 ± 0.18 | 0.84 ± 0.16 |
| + | 0.85 ± 0.05 | 0.93 ± 0.03 |
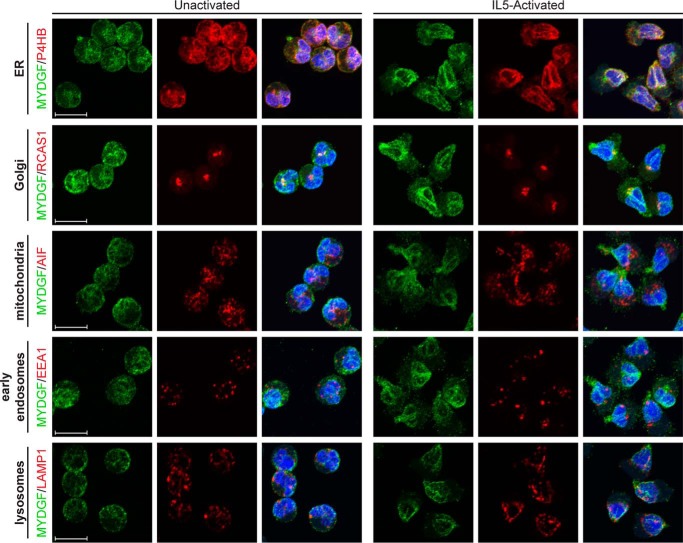
The identities of MYDGF-containing structures in unactivated and IL5-activated eosinophils were investigated by colocalization of goat anti-MYDGF (Alexa Fluor 488; green) with monoclonal rabbit antibodies (Alexa Fluor 647; pseudocolored red) from a commercial organelle-staining kit (Fig. 2 and Table 1). Images shown are maximum intensity projections of multiple slices. MYDGF colocalized most extensively with P4HB (ER marker) around the periphery of the nucleus and in the centralized patch between nuclear lobes with overlapping immunostaining accounting for over 90% of staining relative to either antibody alone in unactivated or IL5-activated eosinophils (Table 1). The immunofluorescence pattern of P4HB is consistent with previous immunogold electron microscopic studies of eosinophils that localized P4HB to the nuclear envelope (11). MYDGF in the patch colocalized with RCAS1, a Golgi marker (Fig. 2). Although total RCAS1 staining almost completely overlapped MYDGF staining, this was not the case for total MYDGF staining overlapping with RCAS1 staining (Table 1); such a result is compatible with the presence of MYDGF in the ER as well as the Golgi. There was poor colocalization of MYDGF with AIF (mitochondria marker) and EEA1 (early endosome marker). MYDGF colocalized more extensively with LAMP1 (lysosome marker) based on quantitation (Table 1). However, the punctate staining pattern of LAMP1 is visually distinct from the perinuclear staining of MYDGF (Fig. 2), and the colocalization observed is in the ER, through which LAMP1 must transit while being processed and trafficked to the lysosome.
Figure 2.
Colocalization of MYDGF and organelle markers. Staining of unactivated and IL5-activated eosinophils with DAPI (blue), goat anti-human MYDGF (green), and rabbit antibodies specific for organelle markers (pseudocolored red) is shown. Images are maximal intensity projections and representative of experiments conducted on three or more separate occasions. Scale bars, 10 μm.
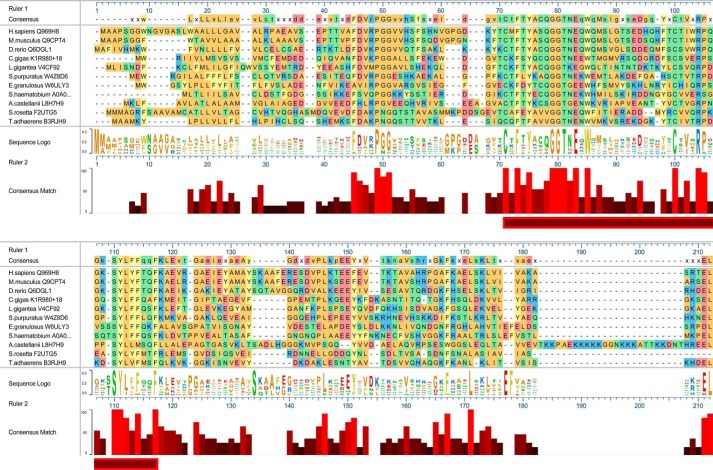
Intracellular staining of C19orf10 in cultured human synoviocytes was interpreted previously as having an ER/Golgi pattern (4). Also, human and murine MYDGF end in variations of the BXEL sequence (Fig. 3) where nearly all BXEL sequences direct intracellular retention and ER–Golgi recycling of proteins via engagement of KDEL receptors (15, 16). We therefore asked whether the BXEL sequence is a general feature of MYDGF homologs. In addition to the homologs present throughout vertebrates as described previously (4), a Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)3 search identified homologs in animals distant from humans, mice, and zebrafish that included the Pacific oyster (Crassostrea gigas), owl limpet (Lottia gigantea), purple sea urchin (Strongylocentrotus purpuratus), hydatid tapeworm (Echinococcus granulosus), and blood fluke (Schistosoma hematobium) (Fig. 3). The homologs belong to the UPF0556 family of proteins (Pfam database; http://pfam.xfam.org/family/upf0556)3 that contain variable signal and N-terminal sequences followed by a core sequence of 47 residues with sequence identity of 25%, which includes a pair of cysteines and a tryptophan (Fig. 3, horizontal bar). A BLAST search using the conserved core sequence revealed homologs in organisms that exist as single cells (Acanthamoeba castellanii), single cells or cellular aggregates (Salpingoeca rosetta), and simple cellular aggregates (Trichoplax adhaerens) (Tree of Life Web Project (38); http://tolweb.org/tree/).3 The relationships of these latter organisms to animals and to one another are obscure (17, 18). The MYDGF homologs shown, except that of S. rosetta, have a BXEL or BXDL sequence at the C terminus (Fig. 3). Six of the nine unique BXEL or BXDL sequences from these MYDGF homologs have been analyzed previously and found to support localization of a model protein to the ER in HeLa cells (16). In the case of the A. castellanii homolog, the BXEL sequence is at the end of a lysine-rich tether. In the rest, the BXEL sequence directly follows a conserved sequence block.
Figure 3.
Sequence alignment of MYDGF homologs. Sequences, based on the UniProt entries (listed after species), are shown for 11 representative MYDGF homologs. The horizontal bar highlights a portion of the sequence with 25% identity. The C. gigas sequence is the UniProt-predicted sequence with addition of an 18-residue N-terminal signal sequence that is predicted by inspection of C. gigas genomic sequence. The figure was generated in MegAlign Pro of the DNASTAR suite.
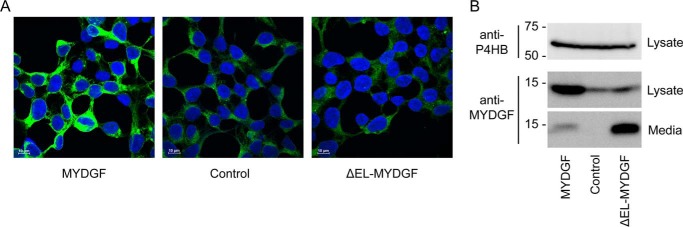
We sought to determine whether human MYDGF in ER and Golgi represents processing of the protein for secretion, recycling of the protein between ER and Golgi due to engagement of KDEL receptors by its RTEL sequence, or interactions of the protein with other ER-resident proteins as is the case for FKBP2, a peptidylprolyl cis-trans isomerase with a naturally occurring C-terminal RTEL sequence (16). In previous studies reporting efficient secretion of recombinantly expressed MYDGF (8, 9), the C terminus had been occluded with an affinity tag. We tested the importance of the C-terminal sequence as a determinant of ER retention versus secretion by comparing the fates of recombinant full-length human MYDGF and MYDGF lacking the final two residues (ΔEL-MYDGF) after transfection into HEK293 cells (Fig. 4). In accord with the variety of cells known to make MYDGF as described in the Introduction, nontransfected HEK293 cells contained endogenous MYDGF as assessed by immunoblotting and immunofluorescence. Antigen had a coarse perinuclear distribution. No MYDGF was detected in the medium of nontransfected cells. Intensities of intracellular staining and MYDGF immunoblotting of cell extracts were much greater in HEK293 cells transfected with full-length MYDGF with some MYDGF also being detected in medium. In contrast, staining and blotting intensities of MYDGF in HEK293 cells transfected with ΔEL-MYDGF were increased minimally compared with nontransfected cells, whereas accumulation in medium was greatly increased. These findings indicate that, unlike FKBP2 (16), RTEL in human MYDGF is necessary for retaining this protein in the ER.
Figure 4.
Endogenous and recombinantly expressed full-length and ΔEL-human MYDGF in HEK293 cells. A, adherent HEK293 cells transfected with MYDGF or ΔEL-MYDGF double stained with DAPI (blue) and goat anti-MYDGF (green) compared with nontransfected cells (Control). B, goat anti-MYDGF or anti-P4HB (loading control) immunoblots of adherent cells or conditioned medium, lysed or collected after 48 h of culture in serum-free medium. Lanes contain cell lysate (10% of the cells plated; ∼2 × 105 HEK293 cells) or 5-fold concentrated conditioned medium (4% of total medium). A and B are both representative of experiments conducted on three separate occasions. Immunoblotting with rabbit anti-MYDGF antibody, also conducted in triplicate, yielded equivalent results (data not shown). Scale bars, 10 μm.
Discussion
These analyses indicate that the name “myeloid-derived growth factor” does not fully capture the functions of human MYDGF and its homologs. First, MYDGF is not specific to hematopoietic cells. Our demonstration of endogenous MYDGF in HEK293 cells is in accord with the presence of MYDGF in several other nonhematopoietic cell types as described in the Introduction and in ProteomicsDB under entry Q969H8 (https://www.proteomicsdb.org/proteomicsdb/#human/proteinDetails/75767/summary),3 which organizes quantitative data on the human proteome (19, 20). ProteomicsDB describes MYDGF (referred to as C19orf10) in almost 140 human tissues and cell lines with the greatest abundance in oral epithelium and skin. Second, the preservation of the MYDGF amino acid sequence in homologs across a wide range of organisms, some of which lack a hematopoietic system, suggests a well-defined global fold subject to evolutionary constraints to fulfill unknown functions. Third, MYDGF has the characteristics of a resident ER protein.
Although we show that the C-terminal RTEL sequence of human MYDGF is necessary for its retention in the ER, we have not provided evidence that this motif is sufficient for retaining a protein in the ER. However, experiments by Raykhel et al. (16) demonstrated that RTEL is among the variants of the canonical KDEL sequence capable of retaining an otherwise secreted reporter protein in the ER of HeLa cells when added to the protein's C terminus. The reporter protein construct to which the KDEL-like sequences were appended comprised a CALR signal sequence, hemagglutinin tag, and P4HB b domain. P4HB folds in the ER, and its b domain was chosen based on structural stability and absence of free thiol groups, cis-proline peptide bonds, disulfide bonds, or post-translational modifications that might contribute to ER retention. The reporter construct colocalized with giantin (Golgi marker) when lacking a C-terminal motif and colocalized with CALR in ER when KDEL, RTEL, or any of 44 other motifs was appended (of 152 motifs examined). Experiments to validate the results with HeLa cells were carried out in COS7 cells using both fluorescence microscopy and secretion into medium as readouts. Finally, colocalization of reporter constructs with the three human KDEL receptors (ERD21, ERD22, and ERD23) responsible for retrieval from the Golgi was examined by a bimolecular fluorescence complementation assay in which the receptors were expressed as fusion proteins containing the N-terminal half of YFP, and the reporter proteins were expressed with the C-terminal half of YFP (16). The reporter protein with the C-terminal RTEL motif interacted with all three human KDEL receptors and had over 80% reporter-receptor localization with ERD21 and ERD22 when compared with the reporter protein bearing the canonical KDEL motif.
Knockout of MYDGF in mice did not result in a describable phenotype other than greater infarct size after experimental coronary artery occlusion (Mouse Genome Informatics under entry MGI:2156020; http://www.informatics.jax.org/marker/key/78906)3 (9). Despite its wide distribution in animals and beyond, we failed to find homologs of MYDGF encoded by Caenorhabditis and Drosophila in directed BLAST searches. Present information regarding function in model organisms, therefore, is based solely on the mouse knockout and suggests roles for MYDGF in responding to cellular stress. These roles likely include modulation of ER stress in addition to serving as a stress-induced autocrine/paracrine factor. Interactions between MYDGF and several quality control proteins have been identified in yeast two-hybrid screens. These proteins include BAG6 (21), SGTA (22), and UBQLN4 (23), all of which play roles in the delivery of improperly folded proteins to the proteasome (24, 25). PDIA5, an ER protein that aids in proper disulfide bond formation and plays a part in initiating the unfolded protein response (26), and EMC10, a subunit of the ER membrane complex that has been implicated as being involved in ER-associated protein degradation (27), have also been identified as interactors with MYDGF using an affinity capture–MS approach (28 (as updated in the Biological General Repository for Interaction Datasets; https://thebiogrid.org/121028/summary/homosapiens/c19orf10.html).3 Finally, a global proteomics survey of ubiquitination sites identified modification of Lys137 of C19orf10 in myeloma cells treated with proteasome inhibitor (29). Collectively, these findings suggest that MYDGF may help in one or more of the steps that deliver proteins from the ER to the proteasome in ER-associated protein degradation (30, 31).
The identification of MYDGF as a resident ER protein does not negate its proposed role as a factor acting from outside cells. There are ample precedents for ER proteins such as P4HB (32), PDIA4 (33), and CALR (34) having dual roles inside and outside cells. Although nontransfected HEK293 cells did not secrete detectable endogenous MYDGF, overexpressed full-length MYDGF appeared in the medium, albeit at lesser amounts than overexpressed ΔEL-MYDGF. Similarly, MYDGF (referred to as C19orf10) was detected in medium of murine bone marrow–derived macrophages stimulated with granulocyte/macrophage colony–stimulating factor and dexamethasone (6), cultured cholangiocarcinoma cells in a hollow fiber bioreactor (7), and human bone marrow cells derived from myocardial infarction patients (9). Finally, the MYDGF homolog of the colonial choanoflagellate S. rosetta lacks a C-terminal BXEL sequence and presumably is secreted constitutively and functions principally outside the cell.
Given its widespread distribution in the biome, MYDGF should be understood better. What is the fold of the UPF0556 domain? With what molecules does intracellular MYDGF interact in MYDGF-centric screens? What receptors allow MYDGF to influence cells from without? Are MYDGF homologs from distant species active on human or murine cells? The latter question may be relevant to diseases caused by E. granulosus (tapeworm), S. hematobium (liver fluke), and A. castellanii (free-living protist widely distributed in the environment that is able to cause keratitis, encephalitis, and skin lesions in humans and animals). Does MYDGF from these organisms modulate the inflammatory or immune response and pathogen–host interactions?
Experimental procedures
Materials
Goat anti-human MYDGF, sold as anti-human SF20 (AF1147), and IL5 (205-IL) were from R&D Systems (Bio-Techne Corp., Minneapolis, MN). The anti-MYDGF antibody had been elicited with recombinant protein produced by Escherichia coli and affinity-purified using the same antigen. Rabbit mAb to P4HB (137110) was from Abcam (Cambridge, MA). Other organelle antibodies were purchased as an organelle staining set (8653) from Cell Signaling Technology (Danvers, MA) composed of rabbit monoclonal antibodies to P4HB (3501), RCAS1 (12290), AIF (5318), EEA1 (3288), and LAMP1 (9091). Nonimmune goat IgG and irrelevant rabbit monoclonal antibodies were used as specificity controls. The following secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA): peroxidase-conjugated bovine anti-goat (805-0350180) and donkey anti-rabbit IgG (711-035-152) for immunoblotting and Alexa Fluor 488–conjugated donkey anti-goat (705-546-147) and Alexa Fluor 647–conjugated donkey anti-rabbit IgG (711-606-152) for immunostaining.
Cloning of MYDGF
MYDGF constructs were amplified from eosinophil RNA by RT-PCR and cloned into pAcGP67.coco (35), pet-ELMER (36), and pcDNA3.1+ by standard molecular biology techniques. Briefly, eosinophil RNA was isolated, and cDNA was generated using random primers and SuperScript III first-strand synthesis system (18080051, Thermo Fisher Scientific–Invitrogen). The cDNA was used as template for PCR to amplify MYDGF and ΔEL-MYDGF inserts, which were then cloned into pcDNA3.1+(V79020, Thermo Fisher Scientific–Invitrogen), pAcGP67.coco (35), and pet-ELMER (36). The inserts for mammalian expression using pcDNA3.1+ included DNA encoding for the MYDGF signal sequence (residues Met1–Ala31). MYDGF inserts for baculoviral (pAcGP67.coco) and bacterial (pet-ELMER) expression start with DNA encoding for residue Val32, the beginning of the mature MYDGF sequence. Full-length MYDGF constructs ended at residue Leu173, and truncated ΔEL-MYDGF ended at residue Thr171. Correct insert sequences were verified before expression.
Expression of MYDGF
For expression in mammalian cells, pcDNA3.1+ plasmids were transfected into HEK293 cells obtained from ATCC (CRL-1573) using FuGENE 6 transfection reagent (E2693, Promega, Madison, WI). Stable transfectants were grown in DMEM supplemented with 10% fetal bovine serum and the selection antibiotic G418 (450 μg/ml; HyClone Laboratories, Logan, UT). For serum-free expression, 2 × 106 cells were plated in serum-containing medium and grown to 80% confluence in 60-mm dishes, after which the cells were gently washed twice with PBS, and 5 ml of serum-free DMEM/F-12 50:50 with l-glutamine (Mediatech, Manassas, VA) medium supplemented with G418 (450 μg/ml) was added to the plates. After 48 h, conditioned medium was carefully collected leaving the cell layer intact, any cells in the collected medium were spun out by centrifugation, and benzamidine was added to 10 mm. The cells were removed from the plate by gentle sloughing with three washes of 0.5 ml of PBS. The cells were pelleted at 750 relative centrifugal force, and the PBS was removed. The pelleted cells were lysed with 50 μl of SDS-PAGE sample buffer (62.5 mm Tris, 4% SDS, 4 m urea, 5% glycerol, pH 6.8) containing 10% β-mercaptoethanol, boiled for 5 min, and vortexed to shear the DNA. With the exception of the addition of G418, the conditions for plating of nontransfected HEK293 cells and subsequent collection of conditioned medium and cell lysates were the same as for the transfected cells.
MYDGF in pAcGP67.coco was expressed using recombinant baculovirus in insect High Five cells and purified via its C-terminal polyhistidine tag by immobilized metal affinity chromatography as described previously (35). Mature, human MYDGF (residues Val32–Leu173) produced in insect cells included N-terminal residues ADP and C-terminal residues LELVPRGSAAGHHHHHH.
MYDGF in pet-ELMER was expressed in Rosetta 2(DE3) competent cells (71400, MilliporeSigma, Burlington, MA), extracted with 8 m urea, and purified by immobilized metal affinity chromatography (36). After elution from the column, the bacterial protein was dialyzed against 100 mm acetic acid, then 1 mm acetic acid, and finally PBS, pH 6, which maximized the yield of soluble, monomeric MYDGF. The N-terminal polyhistidine tag was cleaved by thrombin (HT 1002a, Enzyme Research Laboratories) in a 13-h reaction at room temperature in PBS, pH 6, using 0.05 unit of thrombin/1 μg of MYDGF. Monomeric MYDGF was purified by size-exclusion chromatography (column 28-9893-33, GE Healthcare) immediately after the thrombin cleavage reaction. The final sequence of bacterially produced, mature MYDGF included N-terminal residues GSKGT as a result of the cloning strategy.
Production and purification of rabbit anti-MYDGF
Rabbit anti-MYDGF polyclonal antibody WI552 was produced through Covance Custom Antibody Development Services. The rabbit was immunized against 250 μg of recombinant, baculovirally produced MYDGF followed by two 125-μg booster injections. Antibodies were affinity-purified from the immune serum in the following manner. Recombinant, bacterially produced MYDGF was coupled to cyanogen bromide–activated Sepharose 4B (C9142, Sigma-Aldrich) using the protocol listed on the Sigma-Aldrich product information sheet. Immune serum was diluted 1:2 with PBS, pH 7.4, before loading onto a disposable column containing MYDGF resin. The column was washed with PBS, and bound antibody was eluted with 200 mm glycine, pH 2.8. Elution fractions were immediately neutralized with 3 m Tris-HCl, 3 m KCl, pH 8.8. Pooled fractions containing antibody were dialyzed against PBS, pH 7.4. Affinity-purified rabbit anti-MYDGF was 15-fold more active than commercial goat anti-MYDGF as measured by ELISA in wells coated with 3 μg/ml bacterial MYDGF.
Eosinophils
Human blood eosinophils were purified to >98% purity by Percoll centrifugation and negative selection for neutrophils and monocytes as described previously (13). The cells were received without identifying information in accord with a protocol approved (2013-1570) by the University of Wisconsin-Madison Center for Health Sciences Institutional Review Board. Signed informed consent from the donor was obtained for the use of each sample in research.
Microscopy of eosinophils
Purified eosinophils in RPMI 1640 medium with 0.1% human serum albumin were incubated for 1 h at 37 °C and an additional 10 min with 50 ng/ml IL5 or medium alone. Cells were fixed with 3.7% paraformaldehyde for 10 min, quenched with 0.1 m glycine for 10 min, resuspended in PBS, and cytospun onto poly-l-lysine–coated (12-mm-diameter) glass coverslips. Eosinophils were permeabilized using 0.5% SDS in PBS for 15 min except in the case of LAMP1 for which methanol at −20 °C was used according to the supplier's instructions. After removal of SDS, coverslips were washed, incubated with 10% BSA for 1 h, and incubated for 12–15 h at 4 °C in primary antibodies diluted in 2% BSA, 0.1% SDS in PBS. The primary antibodies were used at the following dilutions: 1 μg/ml for goat anti-MYDGF, 1 μg/ml for rabbit anti-MYDGF, 1:250 for anti-P4HB (137110, Abcam), 1:200 for anti-RCAS1, 1:400 for anti-AIF, 1:200 for anti-EEA1, and 1:100 for anti-LAMP1. Coverslips were washed in PBS and incubated for 1 h at room temperature with Alexa Fluor 488–conjugated donkey anti-goat IgG and Alexa Fluor 647–conjugated donkey anti-rabbit IgG secondary antibodies diluted 1:400 in 2% BSA, 0.1% SDS in PBS. After incubation with DAPI, coverslips were mounted on slides, and sequential 0.3-μm z-step images were acquired using a confocal microscope (Nikon A1R-Si+ Confocal; 60× oil objective) at excitation/emission wavelengths of 409/450, 488/525, and 639/700 nm. Laser power settings and conversion gains were kept constant for activated and unactivated cells and for control IgG staining. Images were reconstructed postacquisition and are displayed as both single slices and maximum projections (signal from all slices).
Colocalization of goat anti-MYDGF with rabbit antibodies was quantified using the General Analysis module of NIS Elements AR version 4.30 software. An intensity threshold was first selected to define a mask of signal for both the 488 and 647 channels that was applied to each 0.3-μm slice of a confocal z-series (21–41 eosinophils in the image plane). The area of the mask was then measured for each slice. The area of signal overlap from the two single-channel masks was measured using the expressions “488 having 647” and “647 having 488.” The fraction of signal overlap was determined using the following quotients: overlapping mask area (488 having 647) divided by 488 mask area and overlapping mask area (647 having 488) divided by 647 mask area. The percent overlap was reported as an average from nine to 16 slices with the standard deviation representing variation among slices.
Immunoblotting of eosinophils
Purified eosinophils in RPMI 1640 medium with 0.02% human serum albumin were incubated for 1 h at 37 °C and an additional 10 min with 50 ng/ml IL5 or medium alone. After the 10-min incubation, protease inhibitor mixture (P8340, MilliporeSigma) was added to the two samples before cells were centrifuged and medium was removed. Hot SDS-PAGE sample buffer containing 5% β-mercaptoethanol was added to the cell pellet to yield cell lysate from 2 × 104 eosinophils/μl of SDS-PAGE sample buffer.
Lysate (2 × 105 eosinophils/well) and recombinant, human MYDGF (bacterially derived; 10 ng/well) were resolved by SDS-PAGE under reducing conditions and transferred to a PVDF membrane. Primary antibodies were used at 2 μg/ml for goat anti-MYDGF, 0.2 μg/ml for rabbit anti-MYDGF, and 1:1000 dilution for rabbit anti-P4HB (3501, Cell Signaling Technology). Peroxidase-conjugated bovine anti-goat and donkey anti-rabbit IgG secondary antibodies were used at 1:20,000 dilutions, and bands were detected using Thermo ScientificTM SuperSignalTM West Pico Chemiluminescent Substrate (34080, Thermo Fisher Scientific). Specificity of the secondary antibodies was assessed by omitting the primary antibodies. Observed molecular weights of protein bands were calculated using a standard curve plotted as the log of known molecular weight standards as a function of their relative migration distance. Analysis of band intensities was quantified using ImageJ software.
Analysis of MYDGF in different species
The Pfam database for the UPF0556 protein domain (http://pfam.xfam.org/family/upf0556)3 describes homologs of MYDGF containing the domain in 104 different species and the presence of the domain in only three other proteins. To visualize the conservation of the MYDGF sequence, we compared homologs of 11 representative species based on their biological interest and distances in phylogenetic trees. The protein sequences are as curated by UniProt (http://www.uniprot.org/;3 entry numbers in parentheses): Homo sapiens (Q969H8), Mus musculus (Q9CPT4), Danio rerio (Q6DGL1), C. gigas (K1R980), Lottia gigantean (V4CF92), S. purpuratus (W4Z8D6), E. granulosus (W6ULY3), S. hematobium (A0A095ACH5), A. castellanii (L8H7H9), S. rosetta (F2UTG5), and T. adhaerens (B3RJH9). Sequences were aligned using the MUSCLE (MUltiple Sequence Comparison by Log-Expectation) alignment tool (37) of MegAlign Pro of the DNASTAR suite (version 12.2.0; Madison, WI), and a sequence logo based on conserved residues was generated. Genomic sequences encoding the C-terminal residues of the S. rosetta (Ensembl gene ID PTSG_11444) and T. adhaerens (Ensembl gene ID TriadG52632) were inspected to confirm the UniProt sequences. The genomic sequence upstream of the homolog in C. gigas (Ensembl gene ID CGI_10024800) was interrogated to determine whether the UniProt-predicted annotation could be missing N-terminal residues. Using Ensembl Genomes (PMID 29092050), we evaluated C. gigas SuperContig scaffold70:891144–891199 and found that a putative 18-residue sequence extends the N-terminal of the UniProt-predicted proteoform. Phobius (39) (http://phobius.sbc.su.se)3 and SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/)3 predict residues Met1–Cys16 from this region to be a signal peptide. The 18-residue extension was added to the UniProt sequence.
Microscopy and immunoblotting of HEK293 cells
HEK293 cells were prepared for microscopy by plating them on acid-washed, 12-mm-diameter glass coverslips in 12-well cluster dishes. After 24 h, cells washed once with PBS and fixed in 3.7% paraformaldehyde for 10 min followed by quenching with 0.1 m glycine for 10 min. Fixed cells were rendered permeable using 0.5% SDS in PBS for 15 min, washed four times with PBS, and blocked in 10% albumin for 1 h. The cells were incubated overnight at 4 °C in 1 μg/ml goat anti-MYDGF antibody diluted in 0.2% BSA, 0.1% SDS in PBS. The following day, coverslips were washed three times with PBS and incubated for 1 h at room temperature with an Alexa Fluor 488–conjugated donkey anti-goat secondary antibody diluted 1:400 in 0.2% BSA, 0.1% SDS in PBS. After DAPI staining, coverslips were mounted on slides and sealed. Images were acquired using a confocal microscope (Nikon A1R-Si+ Confocal). Equal laser power settings and conversion gain were used for nontransfected and transfected cells as well as for control IgG staining.
Immunoblotting of HEK293 conditioned medium and cell lysate samples (see “Expression of MYDGF”) was conducted as described for eosinophils aside from the following differences. Serum-free conditioned medium was concentrated 5-fold using Amicon Ultra-4 centrifugal filters (UFC801024, MilliporeSigma). Lysate (representing 10% of the cells; ∼2 × 105 HEK293 cells/well) and conditioned medium (representing 4% of the medium in the culture dish) were resolved by SDS-PAGE under reducing conditions and transferred to a PVDF membrane. Goat anti-MYDGF was used at 1 μg/ml, rabbit anti-MYDGF was used at 0.2 μg/ml, and rabbit anti-P4HB (3501, Cell Signaling Technology) was used at 1:1000 dilution.
Author contributions
V. B., D. S. A., F. J. F., K. T. B., K. B. T., and D. F. M. conceptualization; V. B., D. S. A., F. J. F., K. T. B., and K. B. T. data curation; V. B., D. S. A., F. J. F., K. T. B., K. B. T., and D. F. M. formal analysis; V. B., D. S. A., F. J. F., K. T. B., K. B. T., and D. F. M. validation; V. B., D. S. A., F. J. F., K. T. B., K. B. T., and D. F. M. visualization; V. B., D. S. A., F. J. F., K. T. B., and K. B. T. methodology; V. B. and D. F. M. writing-original draft; V. B., D. S. A., F. J. F., K. T. B., K. B. T., and D. F. M. writing-review and editing; D. F. M. supervision; D. F. M. funding acquisition; D. F. M. investigation; D. F. M. project administration; V. B. designed and executed the strategies for production, purification, and analysis of recombinant proteins and antibodies; D. S. A. designed and executed the strategies for production, purification, and analysis of recombinant proteins; F. J. F. and K. T. B. designed, performed, and analyzed the microscopic experiments; K. B. T. carried out the bioinformatics investigations; D. F. M. conceived and coordinated the study and organized the paper.
Acknowledgments
We thank Elle Grevstad for help in obtaining and processing microscopy data. Microscopy was performed at the University of Wisconsin-Madison Biochemistry Optical Core, which was established with support from the University of Wisconsin-Madison Department of Biochemistry Endowment.
This work was supported by National Institutes of Health Grants HL088594 and AI125390. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- MYDGF
- myeloid-derived growth factor
- ER
- endoplasmic reticulum
- HEK
- human embryonic kidney
- IL
- interleukin
- AIF
- apoptosis-inducing factor
- BLAST
- Basic Local Alignment Search Tool
- CALR
- calreticulin
- YFP
- yellow fluorescent protein
- DMEM
- Dulbecco's modified Eagle's medium
- DAPI
- 4′,6-diamidino-2-phenylindole.
References
- 1. Tulin E. E., Onoda N., Nakata Y., Maeda M., Hasegawa M., Nomura H., and Kitamura T. (2001) SF20/IL-25, a novel bone marrow stroma-derived growth factor that binds to mouse thymic shared antigen-1 and supports lymphoid cell proliferation. J. Immunol. 167, 6338–6347 10.4049/jimmunol.167.11.6338 [DOI] [PubMed] [Google Scholar]
- 2. Tulin E. E., Onoda N., Nakata Y., Maeda M., Hasegawa M., Nomura H., and Kitamura T. (2003) SF20/IL-25, a novel bone marrow stroma-derived growth factor that binds to mouse thymic shared antigen-1 and supports lymphoid cell proliferation. J. Immunol. 170, 1593 10.4049/jimmunol.170.3.1593 [DOI] [PubMed] [Google Scholar]
- 3. Wang P., Mariman E., Keijer J., Bouwman F., Noben J. P., Robben J., and Renes J. (2004) Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell. Mol. Life Sci. 61, 2405–2417 10.1007/s00018-004-4256-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weiler T., Du Q., Krokhin O., Ens W., Standing K., El-Gabalawy H., and Wilkins J. A. (2007) The identification and characterization of a novel protein, c19orf10, in the synovium. Arthritis Res. Ther. 9, R30 10.1186/ar2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Straub C., Pazdrak K., Young T. W., Stafford S. J., Wu Z., Wiktorowicz J. E., Haag A. M., English R. D., Soman K. V., and Kurosky A. (2009) Toward the proteome of the human peripheral blood eosinophil. Proteomics Clin. Appl. 3, 1151–1173 10.1002/prca.200900043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailey M. J., Lacey D. C., de Kok B. V., Veith P. D., Reynolds E. C., and Hamilton J. A. (2011) Extracellular proteomes of M-CSF (CSF-1) and GM-CSF-dependent macrophages. Immunol. Cell Biol. 89, 283–293 10.1038/icb.2010.92 [DOI] [PubMed] [Google Scholar]
- 7. Weeraphan C., Diskul-Na-Ayudthaya P., Chiablaem K., Khongmanee A., Chokchaichamnankit D., Subhasitanont P., Svasti J., and Srisomsap C. (2012) Effective enrichment of cholangiocarcinoma secretomes using the hollow fiber bioreactor culture system. Talanta 99, 294–301 10.1016/j.talanta.2012.05.054 [DOI] [PubMed] [Google Scholar]
- 8. Sunagozaka H., Honda M., Yamashita T., Nishino R., Takatori H., Arai K., Yamashita T., Sakai Y., and Kaneko S. (2011) Identification of a secretory protein c19orf10 activated in hepatocellular carcinoma. Int. J. Cancer 129, 1576–1585 10.1002/ijc.25830 [DOI] [PubMed] [Google Scholar]
- 9. Korf-Klingebiel M., Reboll M. R., Klede S., Brod T., Pich A., Polten F., Napp L. C., Bauersachs J., Ganser A., Brinkmann E., Reimann I., Kempf T., Niessen H. W., Mizrahi J., Schönfeld H. J., et al. (2015) Myeloid-derived growth factor (C19orf10) mediates cardiac repair following myocardial infarction. Nat. Med. 21, 140–149 10.1038/nm.3778 [DOI] [PubMed] [Google Scholar]
- 10. Wilkerson E. M., Johansson M. W., Hebert A. S., Westphall M. S., Mathur S. K., Jarjour N. N., Schwantes E. A., Mosher D. F., and Coon J. J. (2016) The peripheral blood eosinophil proteome. J. Proteome Res. 15, 1524–1533 10.1021/acs.jproteome.6b00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dias F. F., Amaral K. B., Carmo L. A., Shamri R., Dvorak A. M., Weller P. F., and Melo R. C. (2014) Human eosinophil leukocytes express protein disulfide isomerase in secretory granules and vesicles: ultrastructural studies. J. Histochem. Cytochem. 62, 450–459 10.1369/0022155414531437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mosher D. F., Wilkerson E. M., Turton K. B., Hebert A. S., and Coon J. J. (2017) Proteomics of eosinophil activation. Front. Med. 4, 159 10.3389/fmed.2017.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han S. T., and Mosher D. F. (2014) IL-5 induces suspended eosinophils to undergo unique global reorganization associated with priming. Am. J. Respir. Cell Mol. Biol. 50, 654–664 10.1165/rcmb.2013-0181OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mayeno A. N., Hamann K. J., and Gleich G. J. (1992) Granule-associated flavin adenine dinucleotide (FAD) is responsible for eosinophil autofluorescence. J. Leukoc. Biol. 51, 172–175 10.1002/jlb.51.2.172 [DOI] [PubMed] [Google Scholar]
- 15. Andres D. A., Rhodes J. D., Meisel R. L., and Dixon J. E. (1991) Characterization of the carboxyl-terminal sequences responsible for protein retention in the endoplasmic reticulum. J. Biol. Chem. 266, 14277–14282 [PubMed] [Google Scholar]
- 16. Raykhel I., Alanen H., Salo K., Jurvansuu J., Nguyen V. D., Latva-Ranta M., and Ruddock L. (2007) A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179, 1193–1204 10.1083/jcb.200705180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cavalier-Smith T., Fiore-Donno A. M., Chao E., Kudryavtsev A., Berney C., Snell E. A., and Lewis R. (2015) Multigene phylogeny resolves deep branching of Amoebozoa. Mol. Phylogenet. Evol. 83, 293–304 10.1016/j.ympev.2014.08.011 [DOI] [PubMed] [Google Scholar]
- 18. Cavalier-Smith T., Chao E. E., Snell E. A., Berney C., Fiore-Donno A. M., and Lewis R. (2014) Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol. Phylogenet. Evol. 81, 71–85 10.1016/j.ympev.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 19. Kim M. S., Pinto S. M., Getnet D., Nirujogi R. S., Manda S. S., Chaerkady R., Madugundu A. K., Kelkar D. S., Isserlin R., Jain S., Thomas J. K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N. A., et al. (2014) A draft map of the human proteome. Nature 509, 575–581 10.1038/nature13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilhelm M., Schlegl J., Hahne H., Gholami A. M., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., et al. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 10.1038/nature13319 [DOI] [PubMed] [Google Scholar]
- 21. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., et al. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 10.1016/j.cell.2005.08.029 [DOI] [PubMed] [Google Scholar]
- 22. Rolland T., Taşan M., Charloteaux B., Pevzner S. J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., Kamburov A., Ghiassian S. D., Yang X., Ghamsari L., Balcha D., et al. (2014) A proteome-scale map of the human interactome network. Cell 159, 1212–1226 10.1016/j.cell.2014.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim J., Hao T., Shaw C., Patel A. J., Szabó G., Rual J. F., Fisk C. J., Li N., Smolyar A., Hill D. E., Barabási A. L., Vidal M., and Zoghbi H. Y. (2006) A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125, 801–814 10.1016/j.cell.2006.03.032 [DOI] [PubMed] [Google Scholar]
- 24. Shao S., Rodrigo-Brenni M. C., Kivlen M. H., and Hegde R. S. (2017) Mechanistic basis for a molecular triage reaction. Science 355, 298–302 10.1126/science.aah6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riley B. E., Xu Y., Zoghbi H. Y., and Orr H. T. (2004) The effects of the polyglutamine repeat protein ataxin-1 on the UbL-UBA protein A1Up. J. Biol. Chem. 279, 42290–42301 10.1074/jbc.M406284200 [DOI] [PubMed] [Google Scholar]
- 26. Higa A., Taouji S., Lhomond S., Jensen D., Fernandez-Zapico M. E., Simpson J. C., Pasquet J. M., Schekman R., and Chevet E. (2014) Endoplasmic reticulum stress-activated transcription factor ATF6α requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol. Cell. Biol. 34, 1839–1849 10.1128/MCB.01484-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Harper J. W., and Kopito R. R. (2011) Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 10.1038/ncb2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., et al. (2015) The BioPlex Network: a systematic exploration of the human interactome. Cell 162, 425–440 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Preston G. M., and Brodsky J. L. (2017) The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem. J. 474, 445–469 10.1042/BCJ20160582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olzmann J. A., Kopito R. R., and Christianson J. C. (2013) The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5, a013185 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bowley S. R., Fang C., Merrill-Skoloff G., Furie B. C., and Furie B. (2017) Protein disulfide isomerase secretion following vascular injury initiates a regulatory pathway for thrombus formation. Nat. Commun. 8, 14151 10.1038/ncomms14151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou J., Wu Y., Chen F., Wang L., Rauova L., Hayes V. M., Poncz M., Li H., Liu T., Liu J., and Essex D. W. (2017) The disulfide isomerase ERp72 supports arterial thrombosis in mice. Blood 130, 817–828 10.1182/blood-2016-12-755587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Owusu B. Y., Zimmerman K. A., and Murphy-Ullrich J. E. (2018) The role of the endoplasmic reticulum protein calreticulin in mediating TGF-β-stimulated extracellular matrix production in fibrotic disease. J. Cell Commun. Signal. 12, 289–299 10.1007/s12079-017-0426-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mosher D. F., Huwiler K. G., Misenheimer T. M., and Annis D. S. (2002) Expression of recombinant matrix components using baculoviruses. Methods Cell Biol. 69, 69–81 10.1016/S0091-679X(02)69008-9 [DOI] [PubMed] [Google Scholar]
- 36. Maurer L. M., Tomasini-Johansson B. R., Ma W., Annis D. S., Eickstaedt N. L., Ensenberger M. G., Satyshur K. A., and Mosher D. F. (2010) Extended binding site on fibronectin for the functional upstream domain of protein F1 of Streptococcus pyogenes. J. Biol. Chem. 285, 41087–41099 10.1074/jbc.M110.153692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maddison D. R., Schulz K.-S., and Maddison W. P. (2007) The Tree of Life web project in Linnaeus Tercentenary: Progress in Invertebrate Taxonomy (Zhang Z.-Q., and Shear W.A., eds) Zootaxa 1668, 1–766 [Google Scholar]
- 39. Käll L., Krogh A., and Sonnhammer E. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 35, W429–W432 10.1093/nar/gkm256 [DOI] [PMC free article] [PubMed] [Google Scholar]