Abstract
Therapeutic monoclonal antibodies (mAbs) are an important class of drugs for a wide spectrum of human diseases. Liquid chromatography (LC) coupled to mass spectrometry (MS) is one of techniques in the forefront for comprehensive characterization of analytical attributes of mAbs. Among various protein chromatography modes, hydrophobic interaction chromatography (HIC) is a popular offline non-denaturing separation technique utilized to purify and analyze mAbs, typically with the use of non-MS-compatible mobile phases. Herein we demonstrate for the first time, the application of direct HIC-MS and HIC-tandem MS (MS/MS) with electron capture dissociation (ECD) for analyzing intact mAbs on Q-TOF and FT-ICR mass spectrometers, respectively. Our method allows for rapid determination of relative hydrophobicity, intact masses, and glycosylation profiles of mAbs, as well as sequence and structural characterization of the complementarity-determining regions in an online configuration.
TOC

Since the first approval of monoclonal antibody drug in 1986, therapeutic monoclonal antibodies (mAbs) and their derivatives have arguably become the most important and promising class of therapeutics for many human diseases such as cancer, autoimmunity, metabolic disorders, infection.1–3 The unique pharmacological advantages of mAbs (e.g. target specificity, selectivity, long half-life, and excellent safety profile) and the evolving protein engineering (bispecific antibody, fusion protein, antibody-drug conjugate and nanobody) continuously propel the development of new mAb-based therapeutics, resulting in hundreds of candidates in clinical trials.4 However, with complex conformational and structural dynamics, large molecular sizes, and micro-heterogeneity due to various post-translational modifications, mAbs and their derivatives are among the most complex biologics ever produced, imposing tremendous analytical challenges for comprehensive characterization and control to ensure product quality.5,6
Liquid chromatography (LC) and mass spectrometry (MS) play essential roles, and are the dominant techniques for characterizing mAb-based therapeutics.6,7 Reversed-phase chromatography (RPC), size exclusion chromatography (SEC), hydrophobic interaction chromatography (HIC), and affinity chromatography are often employed for assessing heterogeneity and impurity; top-down, middle-down, bottom-up, and native MS have been used to identify and characterize primary sequences and study higher order structures.6–8 Coupling LC and MS in an online fashion (LC-MS) not only integrates the advantages from separation and detection of these techniques,8–10 but also simplifies the workflow, eliminating offline-handling steps.
Recently we reported the first online coupling of HIC to MS by using columns of greater hydrophobic character and ammonium acetate with some organic solvent in the eluting mobile phase for top-down proteomics.11–14 Importantly, HIC remains the only non-denaturing separation method that exploits hydrophobicity of the native analytes in their native structures, and has been adopted as a powerful approach to analyze mAbs.9,15 Therefore, online coupling of HIC to native MS holds great promise for determining relative hydrophobicity, assessing heterogeneity, and sequence and structural characterization of mAbs when applying tandem MS (MS/MS) techniques. Herein, for the first time, we demonstrate online HIC-MS analysis of intact mAbs using Q-TOF and FT-ICR mass spectrometers, revealing variation in hydrophobicity and glycosylation of proteoforms of the mAbs. Non-covalent interactions that produce higher order dimers and trimers were preserved and detected upon elution condition. Moreover, we have demonstrated that online HIC-MS with electron capture dissociation (ECD) enabled sequence and structural characterization of mAbs, particularly on the complementarity-determining regions (CDRs) outside of the disulfide bridges.
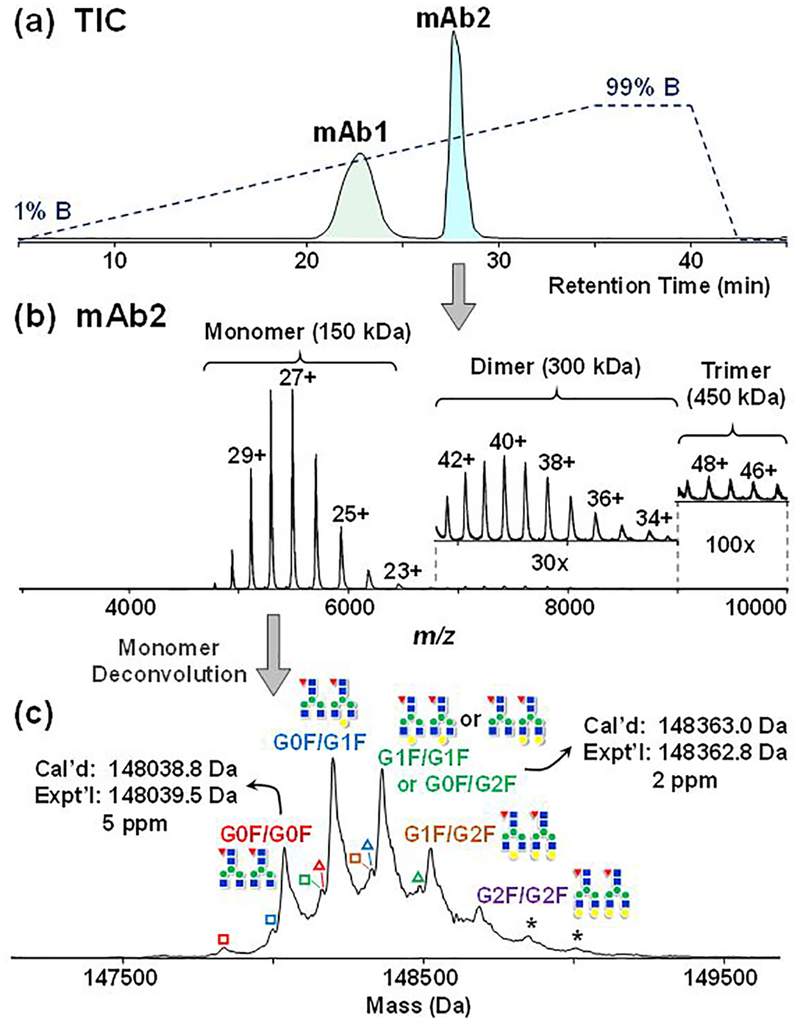
We first applied HIC-MS on a Q-TOF mass spectrometer that has extended m/z range. A mixture of equal amounts of two different immunoglobulin G 1’s (IgG1), namely mAb1 and mAb2 (refer to Experimental procedures in SI), are well separated in the HIC-MS experiments, in which a linear gradient from 99% MPA (1M ammonium acetate) to 99% MPB (20 mM ammonium acetate in 50% ACN) was applied. mAb2 appears to be relatively more hydrophobic than mAb1, and elutes in a sharper peak in the total ion chromatogram (Figure 1a). The two mAb peaks were calculated to have a roughly 1:1 area under the curve (AUC), indicating that they had similar ionization efficiency between the two mAbs (Table S1). Additionally, using mAb2 as an example, the AUCs showed good linear response as a function of injection amounts (Figure S1). mAb2 can be detected with the injection amount as low as 50 ng on a 10 cm, 200 μm i.d. PolyPENTYL A capillary column (Figure S1).
Figure 1.
Online HIC-MS of mAb mixtures on a maXis II Q-TOF mass spectrometer. (a) Total ion chromatogram demonstrating the separation of mAb1 and mAb2. The dash line indicates the gradient changes of mobile phase B. (b) Mass spectrum of mAb2 showing the detection of monomers, dimers (30× zoom-in), and trimers (100× zoom-in). (c) Deconvoluted mass spectrum of mAb2 monomer with annotated glycosylation forms (red triangle: Fucose; blue square: GlcNAc; green circle: Mannose; yellow circle: Galactose); hollow square represents the loss of one GlcNAc (−204 Da), hollow triangle represents the preservation of C-terminal Lys on heavy chain (+128 Da) and asterisk * represents the addition of a hexose (+162 Da). GxF indicates Fc-oligosaccharides terminated by x number of galactoses.
The average mass spectrum from the Q-TOF mass spectrometer further demonstrates that the low-charge-state envelopes of both mAbs fell into the 5,000 to 7,000 m/z range (Figure 1b and Figure S2), which accords with the characteristics of native MS as shown in the previous studies 16,17. The deconvoluted spectra further reveal heterogeneity due to different glycosylations (Figure 1c and Figure S2).18 For instance, multiple addition of 162 Da were observed on mAb2, corresponding to mAb2 with glycan structure G0F/G0F, G0F/G1F, G1F/G1F or G0F/G2F, G1F/G2F, G2F/G2F, with G0F/G1F being the most abundant proteoform (Figure 1c), consistent with previous studies.18,19 The average masses are in good agreement with the theoretical values within 5 ppm of the identified proteoforms, except for the low abundance G2F/G2F proteoform (Figure 1c and Table S2). Minor proteoforms with 204 Da truncation and 128 Da addition were also detected, which correspond to the removal of one terminal GlcNAc and the preservation of C-terminal lysine on one of the heavy chains, respectively.18 On the other hand, mAb1 exhibited a significantly different glycosylation profile from those of mAb2 (Figure S2 and Figure 1C), most likely due to different manufacturing procedures. More interestingly, under the HIC-MS conditions, we also observed high-molecular weight aggregates for both mAbs.20 In particular, dimers (300 kDa) and trimers (450 kDa) of mAb2 were detected within the same elution time frame as the monomers (150 kDa) (Figure 1b). The presence of high molecular-weight dimers and trimers is consistent with the SEC-UV analysis from the mAb2 investigation report (~3.2%),18 consisting of about 2.6 % of the total amount from our HIC-MS result based on the peak areas of the deconvoluted spectrum. Dimers and trimers of mAb2 were eluted in about 33% ACN and 370 mM ammonium acetate, under which the rather strong non-covalent interaction was preserved. We speculated that the kinetics of the chromatography was faster than the kinetics of the denaturation under the HIC-MS conditions used here, and the high concentration of ammonium acetate salt seemed to prevent excessive exposure to ACN, which is consistent with our previous observation.11
To further apply the HIC-MS method to gain more sequence and structural information, we coupled HIC online to a 12T FT-ICR mass spectrometer to analyze mAb2 with a faster gradient. Deglycosylation was performed with an endoglycosidase IgGZERO to reduce sample heterogeneity. Removal of glycosylation also increased mAb2 hydrophobicity, leading to slightly longer retention time and the revelation of preserved heavy chain C-terminal Lysine (+128 Da) and glycated (+162 Da) proteoforms and their relative intensities (Figure S3).18 Without inducing dissociation, the signal of mAb2 can be greatly improved by more than 10-fold after increasing the direct current (DC) bias between the quadrupole and the collision cell in the FT-ICR mass spectrometer (Figure S4).17 This observation could result from further desolvation of the small charged droplets during ion transfer by applying additional collisional activation.
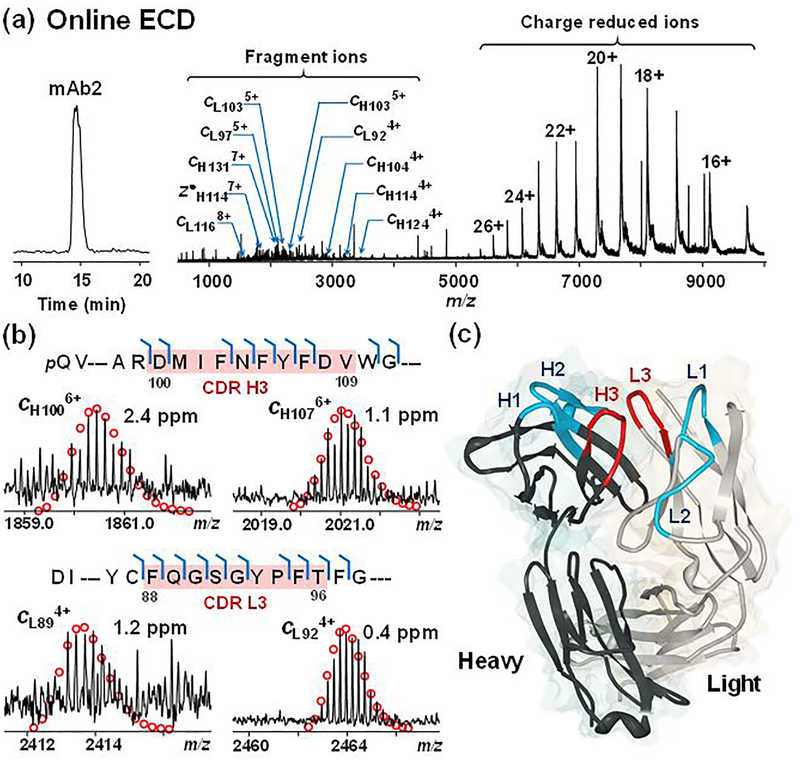
We then performed online broadband ECD on mAb2 in the Para cell with the improved signals, resulting in around 80 ECD scans on the LC time scale (Figure 2a). Low electron energy yielded higher signal to noise ratio (S/N), as higher electron energy is likely to cause secondary dissociation leading to low S/N (Figure S5). The averaged spectrum from a single experiment with 0.6 eV electron energy and 50 ms irradiation time shows charge-reduced species from 5,000 to 10,000 m/z. Fragment ions are readily distinguishable from 1,000 to 3,000 m/z (Figure 2a). The residues of the fragment ions identified within a short 30 min LC-MS/MS run represent 66% of the sequence for the light chain, and 57% for the heavy chain (Figure S6). However, ECD was inefficient in cleaving disulfide bonds of the 150 kDa mAb2, yielding no fragment ions within disulfide bridges, except c141 from the heavy chain.21,22 Similarly, because of the disulfide linkage between light chain and heavy chain, no z• ions were observed from the C-terminus of the light chain. Therefore, only part of the CDRs, namely DMIFNFYFDV from the heavy chain (H3) and FQGSGYPFT from the light chain (L3), had cleavages. For the heavy chain, c100, c103, c104, c105, c106, and c107 consisted of the 6 out of 10 possible N-Cα bonds fragmented from H3 (Figure 2b). The CDR L3 from light chain were well fragmented, where the 8 possible N-Cα bonds were all cleaved (c88, c89, c90, c91, c92, c94, c95, and c96), except the N-terminal side of proline (Figure 2b). Representative fragment ions around 10 kDa were detected in low charge states with mass accuracy below 3 ppm (Figure 2b). ECD has previously been shown to provide tertiary and even quaternary information of native protein and complexes.23,24 We then compared the ECD fragments obtained from the native-like mAb2 MS spectrum to the known crystal structure of mAb2. The crystal structure of the Fab fragment of mAb2 (PDB 5K8A)25 shows that all CDR loops are facing outward and well-exposed, and therefore susceptible to ECD fragmentation (Figure 2c). Indeed, we observed cleavages around the exposed loops of CDRs (H3 and L3) highlighted in red (Figure 2c). Since ECD preserves non-covalent interactions, antigen binding sites on the H3 and L3 CDRs could potentially be localized. We speculate that ECD also cleaved at other CDRs (H1, H2, L1, and L2), however, because of the disulfide bond linkages, no fragment ions could be observed. To our surprise, ECD yielded no cleavages near the N- and C- termini of both the light chain and heavy chain, which indicated their potential roles in the interaction interface that might have prevented electron migration during the fragmentation event. Overall, 106 bond cleavages were observed from the full mAb2 sequence (Figure S6).
Figure 2.
Online HIC-MS/MS with broadband ECD of deglycosylated mAb2 on a 12T solariX XR FT-ICR mass spectrometer. (a) Chromatogram and ECD spectrum of mAb2 showing charge reduced species and fragment ions. (b) Representative fragment ions from the CDRs H3 and L3 (highlighted in red) of heavy chain (top) and light chain (bottom). (c) Crystal structure of the Fab fragment of mAb2 (PDB 5K8A). The CDRs fragmented by ECD (H3 and L3) are highlighted in red, and the other CDRs are highlighted in blue (H1, H2, L1, and L2).
With a gradient that includes increasing organic solvent concentration, it is reasonable to question the use of the term HIC. In our previous work, we speculate that this combination of stationary and mobile phase obliterates the boundary between HIC and RPC. Here we propose that the term HIC continue to be applied in its traditional usage, because it describes a mode that separates proteins based on their hydrophobic character while preserving their three-dimensional structure. The results we have demonstrated above fit in this circumstance.
To recapitulate, we have demonstrated the use of online HIC-MS for the analysis of mAbs in two different types of mass spectrometers. In a high-throughput online manner, HIC separates mAbs based on their relative hydrophobicity; and MS reveals the intact masses and proteoform heterogeneity. Sequence and structural characterization have been further obtained by applying MS/MS techniques such as ECD. As described above, HIC separates proteins based on hydrophobicity under native conditions. When coupled online with MS, it has high potential and value for efficient separation and comprehensive characterization of complex mAbs. We envision growing interest in this method. Continued improvement of the HIC material will further refine HIC-MS, expanding the toolbox for characterizing mAb-based therapeutics.
Supplementary Material
ACKNOWLEDGEMENT
We would like to acknowledge the financial support from Abbvie. We also would like to acknowledge the high-end instrument grant S10OD018475 (to Y.G.) for the acquisition of the high-resolution mass spectrometers that were used in this study. Furthermore, Y.G. would like to acknowledge NIH R01 grants, GM117058, HL109810, and HL096971.
Footnotes
Supporting Information Available:
This material is available free of charge via the Internet at http://pubs.acs.org. Additional information as noted in the text. Part I: Experimental Procedures. Part II. Retention time and area under curve of mAb1 and mAb2 (Table S1). Experimental vs. theoretical masses of mAb2 glycoprotoeoforms (Table S2). Quantitative HIC-MS analysis of mAb2 (Figure S1). Average mass spectrum of mAb1 (Figure S2). Comparison of HIC-MS analysis of glycosylated and deglycosylated mAb2 (Figure S3). Improvement of mAb2 signals in the FT-ICR mass spectrometer by applying collisional energy (Figure S4). Online broadband ECD fragmentation with different electron energies (Figure S5). Online broadband ECD fragmentation map of intact deglycosylated mAb2 (Figure S6).
REFRENCES
- (1).Leavy O Nat. Rev. Immunol 2010, 10, 297–297. [DOI] [PubMed] [Google Scholar]
- (2).Buss NAPS; Henderson SJ; McFarlane M; Shenton JM; de Haan L Curr. Opin. Pharm 2012, 12, 615–622. [DOI] [PubMed] [Google Scholar]
- (3).Weiner GJ Nat. Rev. Cancer 2015, 15, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ecker DM; Jones SD; Levine HL Mabs 2015, 7, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Beck A; Wurch T; Bailly C; Corvaia N Nat. Rev. Immunol 2010, 10, 345–352. [DOI] [PubMed] [Google Scholar]
- (6).Beck A; Wagner-Rousset E; Ayoub D; Van Dorsselaer A; Sanglier-Cianferani S Anal. Chem 2013, 85, 715–736. [DOI] [PubMed] [Google Scholar]
- (7).Fekete S; Guillarme D; Sandra P; Sandra K Anal. Chem 2016, 88, 480–507. [DOI] [PubMed] [Google Scholar]
- (8).Zhang H; Cui WD; Gross ML FEBS Lett 2014, 588, 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Haverick M; Mengisen S; Shameem M; Ambrogelly A mAbs 2014, 6, 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cusumano A; Guillarme D; Beck A; Fekete SJ Pharm. Biomed. Anal 2016, 121, 161–173. [DOI] [PubMed] [Google Scholar]
- (11).Chen BF; Peng Y; Valeja SG; Xiu LC; Alpert AJ; Ge Y Anal. Chem 2016, 88, 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cai WX; Tucholski TM; Gregorich ZR; Ge Y Expert Rev. Proteomics 2016, 13, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Toby TK; Fornelli L; Kelleher NL Annu. Rev. Anal. Chem 2016, 9, 499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chen B; Brown KA; Lin Z; Ge Y Anal. Chem 2018, 90, 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Queiroz JA; Tomaz CT; Cabral JM S. J. Biotechnol 2001, 87, 143–159. [DOI] [PubMed] [Google Scholar]
- (16).Rosati S; Yang Y; Barendregt A; Heck AJ Nat. Protoc 2014, 9, 967–976. [DOI] [PubMed] [Google Scholar]
- (17).Campuzano IDG; Netirojjanakul C; Nshanian M; Lippens JL; Kilgour DPA; Van Orden S; Loo JA Anal. Chem 2018, 90, 745–751. [DOI] [PubMed] [Google Scholar]
- (18).Formolo T; Ly M; Levy M; Kilpatrick L; Lute S; Phinney K; Marzilli L; Brorson K; Boyne M; Davis D; Schiel J State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization, Vol 2: Biopharmaceutical Characterization: The Nistmab Case Study 2015, 1201, 1–62. [Google Scholar]
- (19).Hilliard M; Alley WR; McManus CA; Yu YQ; Hallinan S; Gebler J; Rudd PM mAbs 2017, 9, 1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Alain B; François D; Hélène D; Elsa WR; Olivier C; Van DA; Sarah CJ Mass Spectrom 2015, 50, 285–297. [Google Scholar]
- (21).Ganisl B; Breuker K ChemistryOpen 2012, 1, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang J; Ogorzalek Loo RR; Loo JA Int. J. Mass spectrom 2015, 377, 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang H; Cui WD; Wen JZ; Blankenship RE; Gross ML Anal. Chem 2011, 83, 5598–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Li HL; Nguyen HH; Loo RRO; Campuzano IDG; Loo JA Nat. Chem 2018, 10, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Karageorgos I; Gallagher ES; Galvin C; Gallagher DT; Hudgens JW Biologicals 2017, 50, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.