Abstract
During mitosis, replicated chromosomes are separated to daughter cells by the microtubule-based mitotic spindle. Chromosomes attach to the mitotic spindle through specialized DNA/protein structures called kinetochores, but the mechanism of attachment is not well understood. We show here that the yeast microtubule-binding protein, Dam1p, associates physically and functionally with kinetochores, suggesting a role in kinetochore attachment to the spindle. An epitope-tagged version of Dam1p colocalizes with the integral kinetochore component Ndc10p/Cbf2p in immunofluorescence analysis of chromosome spreads. In addition, Dam1p is associated preferentially with centromeric DNA as shown by chromatin immunoprecipitation experiments, and this association depends on Ndc10p/Cbf2p. We also demonstrate genetic interactions between DAM1 and CTF19 or SLK19 genes encoding kinetochore proteins. Although the defect caused by the dam1-1 mutation leads to activation of the spindle checkpoint surveillance system and consequent persistence of sister chromatid cohesion, the metaphase arrest spindle abnormally elongates, resulting in virtually complete chromosome missegregation. Execution point experiments indicate that Dam1p has a role in formation of a metaphase spindle and in anaphase spindle elongation. Finally, we have observed that the protein encoded by the dam1-1 allele becomes delocalized at the nonpermissive temperature, correlating with the subsequent onset of the mutant phenotype. Our studies are consistent with a role for Dam1p in attachment of sister chromatids through the kinetochore to the mitotic spindle before chromosome segregation.
The accurate segregation of replicated chromosomes to progeny cells during mitosis is essential for survival in unicellular organisms and for proper development and fitness in multicellular organisms. During each cell cycle, eukaryotic cells build an elaborate structure called the mitotic spindle apparatus to carry out chromosome segregation (reviewed in ref. 1). This apparatus is composed of a microtubule-based spindle anchored at both poles to centrosomes (called spindle pole bodies, SPBs, in yeast) and attached to chromosomes at the centromeric region through a DNA–protein complex called the kinetochore. A surveillance system known as the spindle checkpoint monitors for errors in formation of the mitotic spindle apparatus and arrests cells in metaphase until damage is repaired (2, 3). Many proteins have been identified in the mitotic spindle and the checkpoint pathway (1, 2, 4). However, the precise mechanisms for chromosome attachment to the spindle, chromosome segregation, detection of defects, and signal transduction through the checkpoint pathway are not understood.
It has been known for some time that depolymerization of microtubules as well as certain kinetochore and centrosome/SPB defects lead to activation of the checkpoint in a variety of eukaryotes (2). There is evidence for at least two potential signals recognized by the checkpoint machinery in vivo: (i) the presence of kinetochores improperly attached or unattached to the spindle (2, 5, 6); and (ii) lack of tension in the spindle microtubules (7). Some studies have suggested unattached kinetochores are the more important signal detected by the spindle checkpoint in budding yeast (5, 6). The unattached kinetochore is thought to provide a site for recruitment of a number of checkpoint factors, eventually leading to Mad2p-mediated inhibition of Cdc20p. The role of Cdc20p at metaphase is to direct the action of the cyclosome (also called the APC for anaphase promoting complex) to ubiquitinate and degrade Pds1p (securin) so that its binding partner Esp1p (separin) is free to promote anaphase through the separation of sister chromatids.
The kinetochore must be at least partially intact to be recognized by the checkpoint machinery, because mutations in NDC10/CBF2, a gene encoding an integral kinetochore component, appear unable to activate the checkpoint (8). Ndc10p is part of a complex called CBF3 (which also includes Cep3p, Ctf13p, and Skp1p) that is known to bind to a conserved DNA sequence in the centromeric DNA (CDE 111) (reviewed in ref. 9). A second complex containing Ctf19p, Mcm21p, and Okp1 is thought to link the Cbf3/CDE 111 complex with other kinetochore proteins bound at the centromere such as Mif2p, Cbf1p, and the centrosome-specific histone-like protein Cse4p. A third complex contains Ndc80p, Nuf2p, Spc24p, and Spc25p and also appears to be required for checkpoint activation (10, 11). Less is known about the proteins that provide the link between the kinetochore and spindle; although Slk19p and Mtw1p are possible candidates because they (or their fission yeast homologs) have genetic interactions with microtubule-associated proteins (9, 12, 13).
Dam1p is a member of an essential microtubule-binding protein complex also containing Duo1p (14). Previously, we isolated the dam1-1 mutation in a genetic screen for interactors with MPS1, a gene-encoding a protein kinase involved in both SPB duplication and the spindle checkpoint (15). We found that the dam1-1 mutation has genetic interactions with STU1, CIN8, and KAR3, other genes involved in spindle function. We also showed that although the spindle checkpoint is activated at the nonpermissive temperature, as ascertained by molecular and cytological markers, the cells are unable to hold a traditional arrest. The DNA [by 4′,6-diamidino-2-phenylindole (DAPI) staining] appears to separate, and the spindle elongates and breaks, suggesting either that later steps of the checkpoint pathway are not functioning or a structural defect prevents achievement of a proper arrest. Our immunofluorescence (IF) experiments using an epitope-tagged version of Dam1p showed potential kinetochore or kinetochore microtubule localization. Similar to recent studies by Cheeseman et al. (16), our current work suggests a role for Dam1p at the kinetochore. We demonstrate that Dam1p has a physical and functional association with the kinetochore, and that DAM1 shows genetic interactions with genes encoding kinetochore components. Furthermore, delocalization of the dam1-1 protein correlates with subsequent onset of the mutant phenotype. Finally, our work suggests a function for Dam1p in assembly and elongation of a bipolar spindle, consistent with a kinetochore role.
Materials and Methods
Yeast Genetic Techniques.
General yeast techniques were as in ref. 17. Cell synchronization, budding index, flow cytometry, and nocodazole treatment (15 μg/ml nocodazole and 30 μg/ml benomyl, confirmed by IF of microtubules) were as in Jones et al. (15). Strains (Table 1) were constructed as follows: To visualize chromosome V with green fluorescent protein (GFP) (35 kb from centromere, “arm” location) strain KN-7479 (18) was crossed to 371 (15) and 906 and haploids were chosen. To analyze colocalization with Ndc10p, the DAM1-Myc strain (see below) was crossed to SBy169 and haploids were chosen. To check for genetic interactions, we crossed strain yKH35 (same genotype as yPH1315 in ref. 19 or strain WSY837 (12) to strain 906 and isolated double mutants. To analyze Gal-Mps1p arrest in the presence of dam1-1, we crossed strain 2023 with strain 906 and used haploids 2900 and 2902, and we crossed strain 2023 with strain 2946 to monitor localization. To perform the execution point experiment, these strains were synchronized at 23°C in G1 by using α-factor in raffinose and then shifted to galactose (or glucose) with α-factor at 23°C (or shifted to 33°C). The cells were then washed and released into the appropriate medium (galactose or glucose) and temperature (23°C or 33°C), and samples were collected for IF, fluorescence-activated cell sorting analysis, and budding index. The cultures were released from Mps1p arrest by shift to glucose at 37°C plus α-factor so that cells would stop at G1 of the next cell cycle.
Table 1.
Yeast strains
| Strain | Relevant genotype |
|---|---|
| 906* | a dam1-1 |
| 2453† | a ura3∷URA3 tetOs leu2∷LEU2 tetRGFP PDS1myc∷TRP |
| 2461† | a ura3∷URA3 tetOs leu2∷LEU2 tetRGFP PDS1myc∷TRP dam1-1 |
| 2023†‡ | α Gal-MPS1myc∷LEU2 |
| 2902† | a ura3-1∷dam1-1∷URA3, Gal-MPS1myc∷LEU2 |
| 2900† | a ura3-1∷dam1-1∷URA3, Gal-MPS1myc∷LEU2, DAM1∷HIS3 |
| 2768* | α DAM1-GFP∷kanmx |
| 2946* | a dam1-1-GFP∷kanmx |
| 2911† | a ctf19∷HIS3, dam1-1 |
| 2984† | α slk19∷HIS3, dam1-1 |
| XHY329† | a DAM1∷DAM1-GFP∷URA3 |
| XHY330† | α DAM1∷DAM1-GFP∷URA3, ndc10-1 |
| SBy169†§ | a NDC10HA∷URA3 |
| 2814† | a NDC10HA∷URA3, DAM1-Myc∷TRP1 |
All strains are from this study unless indicated otherwise.
Derived from S288C.
Derived from W303.
From B. Huneycutt, Univ. of Colorado, Boulder, CO.
From S. Biggins, Fred Hutchinson Cancer Research Institute, Seattle.
dam1-1 Mutation.
We prepared DNA from dam1-1 strain 906 for sequencing (using an Applied Biosystems automated sequencer) and used two sets of primers flanking the gene (sequence information for all primers used in this study available on request). We used primer dam1–1Rl and primer MOE-CLA in PCR to generate a fragment that was cloned into pRS306 (20) containing the DAM1 gene from DraI–NheI. This was integrated at the URA3 gene of both a W303- and a S288C-derived (21) diploid strain heterozygous for a replacement of DAM1 with HIS3. The strains were sporulated and URA3 HIS3 spores were chosen.
Epitope Tagging.
We used plasmids for epitope tagging from Knop et al. (22). pYM6 (9xMyc) and pYM12 (GFP) were used in PCRs with primers DAM1-STWO and DAM1-S3. PCR products were transformed into W303- and S288C-derived diploid strains, and TRP1 or G418-resistant transformants were picked and sporulated to obtain the desired haploid strain (strains confirmed by PCR). The epitope-tagged proteins were the only copies of Dam1p in the cell and were fully functional at all temperatures.
Cytological Techniques.
Chromsome spread experiments were performed as in Klein et al. (23), but using 1% Lipsol. Flow cytometric analysis was performed as in ref. 15 by using a Becton Dickinson FACScan flow cytometer and cell quest software. For IF experiments, we used a Leica DMRXA/RF4/V automated microscope with a Cooke Sensicam digital camera and slidebook software (Intelligent Imaging Innovations, Denver, CO). Fixation techniques involved a 15- to 60-min treatment with 4% formaldehyde followed by Zymolyase treatment as described (24). The following Abs were used: for visualizing microtubules, primary mAb YOL1/34 and secondary Texas red- or FITC-conjugated anti-rat Abs (Accurate Chemical and Scientific, Westbury, NY); and for visualizing Pds1-myc, Dam1p-myc, and dam1–1p-myc, we used polyclonal anti-myc Ab (kind gift of J. Yucel, Univ. of Colorado, Boulder, CO) and secondary Cy3-conjugated anti-rabbit Abs. Immunoelectron microscopy was performed as in Chial et al. (25) with strain 2768.
Molecular Techniques.
For the chromatin immunoprecipitation (ChIP), 50 ml of OD600 = 0.5 wild-type (W303) or ndc10-1 (8) cells that carry Dam1-GFP, YXH329, and YXH330 were shifted from 25°C to 37°C for 2.5 h and subjected to ChIP procedure as described in ref. 26. Anti-GFP Abs (CLONTECH 8372-2) were used for immunoprecipitation. Two pairs of oligonucleotides, KOL34/KOL38 and CEOL153/CEOL155, were used in PCR to detect DNA fragments that cover CEN4 or URA3.
Results
Mutation Responsible for the dam1-1 Phenotype.
DAM1 encodes a protein that is part of a microtubule-binding complex (14). To learn more about the dam1-1 phenotype, we sequenced the dam1-1 gene and found a single change resulting in a tyrosine replacing a cysteine at residue 111, confirming that Cheeseman et al. (16) independently isolated the dam1-1 mutation. We made this change in a cloned DAM1 gene, integrated the mutated gene into the genome, and found the phenotype to be identical to the original dam1-1 mutant strain. The changed residue lies 14 residues before a domain highly likely to contain dimer and trimer coiled-coils [using the multicoil program (27)], in a region shown recently to be conserved between Saccharomyces cerevisiae and Candida albicans (16). Thus the mutation may affect a conserved protein–protein interaction.
The dam1-1 Mutation Leads to a Chromosome Segregation Defect.
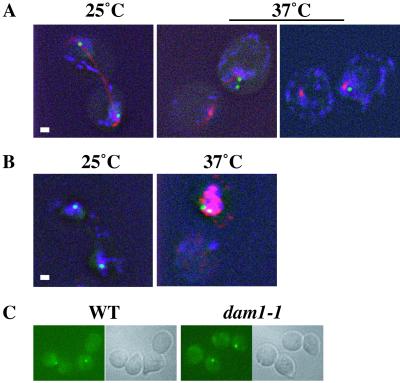
Although the spindle checkpoint appears to be activated in dam1-1 cells at the nonpermissive temperature (i.e., cells show a mitotic delay that depends on the checkpoint gene, MAD1), a typical metaphase arrest is not achieved (15). First, the DNA appears to separate; second, the spindle appears to elongate and then lose integrity; and finally, the cells show rapid loss in viability. We were interested whether sister chromatids separate properly to opposite poles in dam1-1 strains at the nonpermissive temperature. Recently, Cheeseman et al. (16) demonstrated that dam1 mutant strains show a chromosome segregation defect at the nonpermissive temperature. We made the same observation in our dam1-1 strain by using a system designed to visualize chromosome movement by introducing a gene encoding a GFP-TetR fusion protein and TetR-binding sites on chromosome V (18). The chromosome-marked dam1-1 strain was released from a synchronization in G1 to the permissive (25°C) and nonpermissive (37°C) temperatures. At the permissive temperature, 95% of large budded cells (Fig. 1A) showed two separated GFP-marked chromosomes, colocalizing with DNA, flanking an elongated spindle. At the nonpermissive temperature, no cells were observed that showed two separated GFP dots; instead there was either one dot or two closely spaced dots associated with one of the two spindle fragments and a portion of the DAPI staining (Fig. 1A). Thus we, too, find that sister chromatids in dam1-1 strains at the nonpermissive temperature do not achieve proper segregation to opposite poles but rather remain together at one pole.
Figure 1.
The dam1-1 mutation leads to chromosome missegregation at the nonpermissive temperature. Large-budded dam1-1 cells at permissive temperature (25°C) and nonpermissive temperature (37°C). Green shows chromosome V-GFP; blue shows DAPI (DNA). (A) At 25°C, two separated dots in different buds, 95% (n = 206) and at 37°C, two separated dots, 0%, two close dots in the same bud, 32%, one dot, 68% (n = 205). Red shows primary rat anti-α tubulin plus secondary Texas red anti-rat Abs. (Bar = 1 μm.) (B) Pds1p-myc = Red shows primary rabbit anti-Myc plus secondary Cy3-conjugated anti-rabbit Abs. (Bar = 1 μm.) (C) Wild-type and dam1-1 cells at 37°C after nocodazole treatment. Cells with one dot: wild-type = 93% (n = 200); dam1-1 = 95% (n = 200). (Bar = 1.5 μm.)
We used two criteria to determine that the two GFP dots seen in the latter category represented sister chromatids that maintained cohesion but were far enough to be resolvable as two dots rather than sister chromatids that had lost cohesion but not segregated. First, we looked for Pds1 protein stabilization, which is a hallmark of checkpoint activation and leads to persistent sister chromatid cohesion. We used a Myc-epitope-marked Pds1p (18) to demonstrate by Western analysis (data not shown) and by whole cell IF (Fig. 1B) that Pds1p is stabilized at the nonpermissive temperature (37°C) but not at the permissive temperature (25°C). Second, we ruled out a cohesion defect by analyzing the dam1-1 phenotype in the presence of the microtubule-depolymerizing agent nocodazole. Under these conditions, in strains with cohesion defects sister chromatids have been shown to drift apart (18); however, we observed that the majority of dam1-1 cells contained one dot in both wild-type and dam1-1 strains (Fig. 1C). Together, these observations suggest that sister chromatids maintain cohesion in dam1-1 strains and also show that the small separation observed is microtubule dependent.
Thus, the dam1-1 mutation leads to normal activation of the spindle checkpoint resulting in persistent cohesion of sister chromatids. However, unlike a normal checkpoint arrest, the spindle appears to elongate, leading to chromosome missegregation, consistent with our observed decrease in cell viability.
The Protein Encoded by dam1-1 Becomes Delocalized at the Restrictive Temperature.
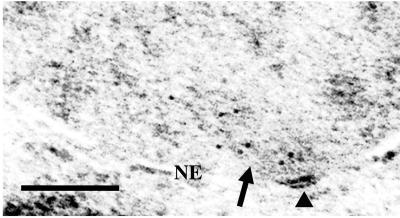
Previously we have localized Dam1p tagged with a C-terminal Myc epitope by IF to a region overlapping the ends of the spindle but peripheral to the SPB (15). We suggested this localization may correspond to kinetochores because kinetochores are thought to cluster on the spindle side of the SPB. To confirm that the localization is not at SPBs per se, we used chromosomal integration to place a GFP tag at the C terminus of the Dam1p and used anti-GFP Abs in conjunction with gold-labeled secondary Abs in immunoelectron microscopy analysis. As expected, the gold particles were not on the SPB but clustered on the mitotic spindle (Fig. 2). Analysis of the immunoelectron microscopy signal of Dam1-GFP was limited to mitotic cells wherein spindle microtubules and/or SPBs could be identified. In such cells, the SPB and associated microtubules occupy a small fraction of the area of the nucleus examined; however, 80% of the gold particles (n = 191 in 19 cells) were within 300 nm of the SPB and generally associated with microtubules (Fig. 2). This distance from the SPB is consistent with the position of kinetochores based on electron microscopy analysis of spindle structure (28). Furthermore, in seven of the 19 cells, some of which contained clear anaphase spindles (>2 μm), 84% of the gold particles (n = 69) are within 150 nm of the SPB, consistent with the anaphase position of the kinetochore because of anaphase A shortening of the kinetochore microtubules (28).
Figure 2.
Dam1p-GFP is clustered on the mitotic spindle near the SPB. Immunoelectron microscopy was performed on 19 whole cells from strain 2768. Arrows denote spindle microtubules and the arrowhead shows the SPB. NE, nuclear envelope. (Bar = 0.2 μm.)
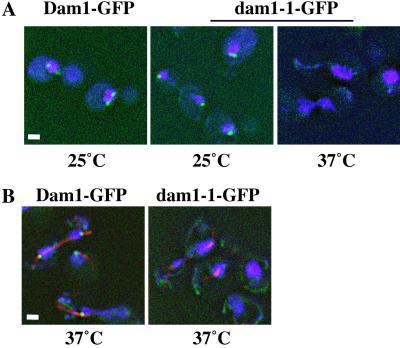
Next, we were interested whether localization of the mutant Dam1p changed at the nonpermissive temperature. We placed a GFP tag on the dam1–1p and analyzed the autofluorescence of wild-type Dam1p-GFP and dam1–1p-GFP at permissive (25°C) and nonpermissive (37°C) temperatures. Although the localization of dam1–1p-GFP at 25°C looks similar to wild-type Dam1p-GFP (Fig. 3A), and to Dam1p-myc (15), the mutant but not the wild-type protein becomes delocalized at 37°C after 1 or 2 h (Fig. 3 A and B, respectively). The delocalization of dam1–1p-GFP at the nonpermissive temperature correlates with the spindle losing integrity after longer incubation (Fig. 3B; ref. 15). This result suggests that the observed localization is related to the function of Dam1p and its delocalization at the nonpermissive temperature leads to activation of the checkpoint, eventual spindle elongation and breakage, and chromosome missegregation.
Figure 3.
The dam1–1pGFP becomes delocalized at the nonpermissive temperature. (A) Asynchronous cells containing GFP-tagged DAM1 or dam1-1 were grown at 25°C or shifted to 37°C for 1 h. (B) Cells as described in A but shifted to 37°C for 2 h and treated with rat anti-tubulin Abs plus Texas red anti-rat secondary Ab. (Bar = 1.5 μm.)
dam1-1 Strains Cannot Assemble a Spindle at the Nonpermissive Temperature.
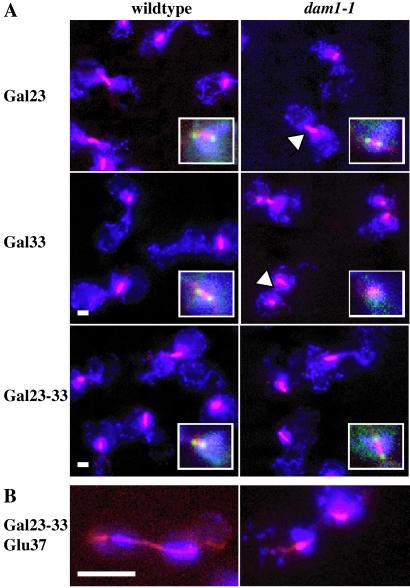
The broken spindle phenotype in dam1-1 mutant cells suggests that the protein is required for spindle formation or integrity. We wanted to distinguish whether Dam1p is required to: (i) assemble/maintain a metaphase spindle and/or (ii) elongate the spindle properly during anaphase. Recent experiments by Cheeseman et al. (16) have suggested that Dam1p is not required for an arrest caused by treatment with a replication inhibitor (“S phase spindle”) but is required for arrest because of inhibition of the APC (“metaphase spindle”). We examined this question by using a different, reversible system to achieve a metaphase spindle. Overexpression of the checkpoint protein kinase Mps1p activates the spindle checkpoint, leading to a mitotic arrest of cells with replicated, unseparated DNA and a short metaphase spindle (29). We analyzed the mitotic arrest state of wild-type and dam1-1 cells, using IF microscopy, after induction of MPS1 from a galactose-inducible promoter. First, cells were arrested in G1 (with α factor) at the permissive temperature. Then, MPS1 transcription was induced at permissive or nonpermissive temperature (33°C in this strain background). To determine whether Dam1p is required to assemble a metaphase spindle, cells were released from G1 into the cell cycle (still under inducing conditions). At the permissive temperature, both the wild-type and dam1-1 cells show a typical checkpoint arrest (Fig. 4A Top, Gal23). The cells contained a short (1–2 μm, see arrowhead in dam1-1 panel) metaphase spindle, a single mass of DAPI stain, sometimes in the bud neck, large-budded morphology, and 2C DNA content by FACS analysis (data not shown). In addition, the Dam1GFP protein localizes normally, overlapping the ends of the short spindle (see Insets). At the nonpermissive temperature (Fig. 4A Middle, Gal33), the wild-type cells arrest normally with a short intact spindle retaining Dam1GFPp; the dam1-1 cells instead contain two spindle fragments (see arrowhead) that lack dam1–1GFPp.
Figure 4.
Dam1p is required for metaphase spindle assembly and anaphase spindle elongation. Blue shows DAPI (DNA), red shows microtubules detected with rat anti-tubulin Abs plus FITC anti-rat secondary Ab (pseudocolored red), and green (Insets) are Dam1GFPp. Wild-type and dam1-1 cells after induction of GAL-MPS1 and release from α factor in four different conditions. (A) Gal23, galactose medium at 23°C; Gal33, galactose medium at 33°C; and Gal23–33, galactose medium at 23°C then shifted to 33°C. (Bar = 1.5 μm.) (B) Gal23–33-Glu37, galactose medium at 23°C then shifted to 33°C and then shifted to glucose medium at 37°C plus α factor. (Bar = 5 μm.)
However, when the cells were allowed to form a checkpoint-arrest spindle at 23°C, before shift to 33°C [Fig. 4A Bottom (Inset), Gal23–33] the dam1–1GFPp appears to be at least partially stabilized because it does not become delocalized. Therefore, although dam1-1 cells maintain the short spindle arrest at 33°C, we cannot evaluate the requirement for Dam1p in spindle maintenance because we have no other assay besides delocalization for protein inactivation. Next, we were interested in whether Dam1p is needed for spindle elongation. Although proper galactose-mediated induction does not occur at temperatures above 33°C, we were able to use a more stringent restrictive temperature of 37°C when we released the cells from the Mps1p-induced arrest by shift to glucose-containing media. Under these conditions, we observed that many of the dam1-1 cells contained broken or aberrant spindles (41%, n = 49), whereas virtually all of the wild-type cells contained intact elongating spindles (95%, n = 38, Fig. 4B).
Overall, this experiment suggests that Dam1p plays a role in metaphase spindle assembly, in agreement with Cheeseman et al. (16), and localization of Dam1p flanking the spindle is correlated with its function. In fact, it is likely that Dam1p has a continuous role in spindle integrity, as it is also required for proper spindle elongation.
Dam1p Localizes to the Kinetochore.
Our localization studies have shown a pattern of Dam1p staining consistent with kinetochore localization (ref. 15; Figs. 2–4) and the phenotypic experiments suggest that dam1-1 strains may have a kinetochore defect. This idea was strengthened by our finding that the dam1-1 mutation shows synthetic growth defects with deletion of two different genes encoding the kinetochore components, Ctf19p and Slk19p (data not shown). Ctf19p is thought to be involved in linking the Ndc10p-containing CBF3/CDE 111 complex with other kinetochore proteins, such as Mif2p (reviewed in ref. 9). Slk19p is thought to have a role in kinetochore attachment to the microtubule. These interactions appear specific because dam1-1 does not show synthetic defects with ndc10-1, ndc10-42, or mif2-3 (15).
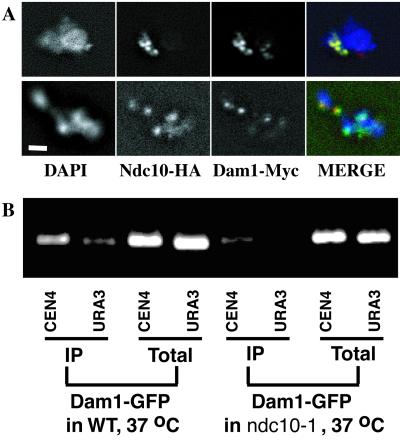
Cheeseman et al. (16) recently showed Dam1p localization to kinetochores in chromosome spreads. We made the same observation by using a strain containing Dam1p-Myc and a kinetochore component, Ndc10p, tagged with a hemagglutinin (HA) epitope. Ndc10p-HA can be visualized in a punctate pattern of four to five dots overlapping the DNA (DAPI) (Fig. 5A). In general, the Dam1p-Myc staining is coincident with Ndc10p-HA, although occasionally (6%, n = 130) a strongly Ndc10p-HA-reactive region does not contain Dam1p-Myc (Ndc10-HA, Fig. 5A Lower), which is not surprising because Dam1p has not been shown to interact directly with Ndc10p. Also, faint Dam1p staining sometimes appears (8%, n = 130) in the absence of Ndc10-HA (Dam1p-Myc, Fig. 5A Upper), consistent with our previous observation of weak punctate nuclear Dam1p staining in whole cells (15).
Figure 5.
Dam1p is at the kinetochore. (A) IF of chromosome spreads from cells containing Ndc10pHA and Dam1p-myc. Blue shows DAPI (DNA), green shows Ndc10pHA detected with anti-HA mAb plus FITC anti-mouse secondary Ab, and red shows Dam1pMyc detected with polyclonal anti-Myc Ab plus Cy3 anti-rabbit secondary Ab. (Bar = 1.5 μm.) (B) Dam1-GFP is associated with centromeric DNA in vivo, in an NDC10-dependent manner. CEN4 association was tested in wild-type or ndc10-1 cells at 37°C. Background signal was detected by using URA3-specific oligonucleotides. Total input, DNA extract of total lysate before immunoprecipitation; IP, DNA extract after immunoprecipitation.
To test for a physical association of Dam1p with centromeres, the kinetochore-associated chromosome regions, we performed ChIP experiments, crosslinking cells containing Dam1p-GFP by using formaldehyde and then performing immunoprecipitation experiments using anti-GFP Abs (Fig. 5B). The identity of the DNA region that is enriched in co-immunoprecipitates with Dam1p-GFP was determined by using PCR, using primers either to a centromere-specific region (CEN4) or to a noncentromeric region (URA3). The centromere-specific region is enriched in the anti-GFP ChIP, suggesting Dam1p-GFP has a functional association with the kinetochore; furthermore, when the kinetochore is disrupted, in an ndc10-1 mutant strain, the enrichment of centromeric DNA is lost. Interestingly, in whole cells, Dam1GFP localization does not appear to change in ndc10-1 cells under conditions in which the kinetochore protein Ndc80p becomes delocalized (data not shown). These data suggest that Dam1p may be associated primarily with the plus end of the kinetochore microtubule, linking it to the core kinetochore.
Our localization experiments in whole cells and in chromosome spreads together with the ChIP experiment suggest that Dam1p is associated with the kinetochore, perhaps through its interaction with the end of the kinetochore microtubule and this association is lost in a dam1-1 mutant strain. From this position, Dam1p probably interacts indirectly with centromeric DNA, an association lost in an ndc10-1 mutant strain.
Discussion
The data we present here together with recent data from Cheeseman et al. (16) support a role for Dam1p at the kinetochore, perhaps in kinetochore attachment to the mitotic spindle. We show by several criteria that Dam1p associates both physically and functionally with kinetochores. First, a Myc-epitope tagged Dam1p colocalizes with an integral kinetochore component, Ndc10p/Cbf2p by IF microscopy of chromosome spreads. Second, Dam1p-GFP preferentially associates with centromeric DNA and this association depends on Ndc10p. Third, immunoelectron microscopy of Dam1p-GFP is consistent with kinetochore localization. Finally, DAM1 shows genetic interaction with CTF19 and SLK19, genes encoding kinetochore components. Cheeseman et al. (16) also have shown that Dam1p as well as its binding partner Duo1p localize with Ndc10p by IF in chromosome spreads.
Our further characterization of the dam1-1 mutation is consistent with a role in attachment of the kinetochore to the mitotic spindle. At the nonpermissive temperature, the dam1-1 mutation leads to activation of the spindle checkpoint as evidenced by a MAD1-dependent mitotic delay and Mad1p hyperphosphorylation (15), as well as Pds1p stabilization and maintenance of sister chromatid cohesion. The abnormal elongation of the spindle despite persistent chromatid cohesion leads to virtually complete chromosome missegregation, as shown by using a GFP-tagged chromosome system, and suggests a defect in the connection of the sister chromatids to the spindle. Our experiments do not distinguish whether lack of proper kinetochore attachment per se or a resulting defect in spindle tension is the signal to activate the checkpoint. This attachment defect appears to be partial, as the paired sister chromatids move to one or the other pole, as opposed to the defect in ndc10-1 mutants in which the DNA is completely unattached from the spindle and remains in the mother cell (8).
Dam1p appears to have a continuous role in kinetochore-microtubule attachment during mitosis, which is needed for spindle integrity. Cheeseman et al. (16) showed that Dam1p is not required for an arrest caused by a replication inhibitor, in which kinetochore microtubules are attached to only a single kinetochore. In contrast, we, here, and Cheeseman et al. (16) have demonstrated a requirement for Dam1p in establishing a bipolar metaphase spindle, and we have shown, additionally, that Dam1p is required for proper anaphase spindle elongation.
By using two different C-terminal epitope tags, we have observed localization of Dam1p to be primarily at the poles, on the nuclear side of the SPB, consistent with our localization to kinetochores in chromosome spreads. At the nonpermissive temperature, dam1–1p becomes delocalized, correlating with the eventual mutant phenotype of dam1-1 cells. Using anti-Dam1p Abs, Cheeseman et al. (16) also observed kinetochore localization in chromosome spreads; however, in whole cells they observe uniform staining along the spindle, which is disrupted in dam1 mutants even at the permissive temperature. It is possible that Dam1p has distinct domains for binding to the lateral region of microtubules and for binding to the kinetochore, by interaction with the plus ends of microtubules or with another kinetochore protein. If the lateral microtubule-binding domain is at the C terminus, it may be somewhat affected in our epitope-tagged versions, despite the fact that they are fully functional by biological phenotype. This could potentially enhance the kinetochore binding, which may explain why we observe localization at the permissive temperature and delocalization at the nonpermissive temperature whereas Cheeseman et al. (16) observed delocalization at both temperatures.
Although it is not clear which proteins in the kinetochore directly mediate attachment to the spindle, dephosphorylation of Ndc10p is likely to play a role; mutations in the phosphatase gene GLC7 result in Ndc10p hyperphosphorylation in vivo and reduced kinetochore-microtubule binding in vitro (30, 31). Kinetochore attachment appears to require a balance between the Glc7p phosphatase and the spindle-associated Ipl1p protein kinase (30, 31). It is possible that Dam1p is involved in this pathway, because of both its inherent microtubule-binding activity and its genetic and physical interactions with the Ipl1p kinase (see discussion in ref. 16).
A detailed study monitoring localization of various kinetochore proteins in the presence of other kinetochore mutations as well as further development of functional kinetochore assays (32) will be necessary to unravel the complex architecture of proteins in the kinetochore.
Acknowledgments
We thank members of the Winey lab for stimulating discussion; A. Castillo, S. Jasperson, A. Stemm-Wolf, and T. Su for critical reading of the manuscript; and I. Cheeseman and G. Barnes for sharing results before publication and for helpful discussion. We thank S. Biggins, K. Nasmyth, K. Hyland, B. Huneycutt, W. Saunders, and J. Yucel for reagents; G. Morgan for help with immunoelectron microscopy; and G. Sluder, S. Biggins, and the reviewers for helpful suggestions. This work was supported by a grant from the National Institutes of Health (GM-51312 to M.W.). Deconvolution microscopy was made possible in part by a gift from Virginia and Mel Clark.
Abbreviations
- SPB
spindle pole body
- GFP
green fluorescent protein
- APC
anaphase promoting complex
- IF
immunofluorescence
- DAPI
4′,6-diamidino-2-phenylindole
- ChIP
chromatin immunoprecipitation
- HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Saunders W S. Curr Opin Cell Biol. 1999;11:129–133. doi: 10.1016/s0955-0674(99)80016-7. [DOI] [PubMed] [Google Scholar]
- 2.Amon A. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 3.Gardner R D, Burke D J. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- 4.Wigge P A, Jensen O N, Holmes S, Soues S, Mann M, Kilmartin J V. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters J C, Chen R H, Murray A W, Salmon E D. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnan R, Pangilinan F, Lee C, Spencer F. Genetics. 2000;156:489–500. doi: 10.1093/genetics/156.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Nicklas R B. Nature (London) 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 8.Goh P, Kilmartin J. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pidoux A L, Allshire R C. Curr Opin Cell Biol. 2000;12:308–319. doi: 10.1016/s0955-0674(00)00094-6. [DOI] [PubMed] [Google Scholar]
- 10.Wigge P A, Kilmartin J V. J Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera M M, Schramm C, Schiebel E. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng X, Kahana J A, Silver P A, Morphew M K, McIntosh J R, Fitch I T, Carbon J, Saunders W S. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goshima G, Saitoh S, Yanagida M. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann C, Cheeseman I M, Goode B L, Mcdonald K L, Barnes G, Drubin D G. J Cell Biol. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones M H, Bachant J B, Castillo A R, Giddings T H, Jr, Winey M. Mol Biol Cell. 1999;10:2377–2391. doi: 10.1091/mbc.10.7.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheeseman I M, Enquist-Newman M, Muller-Reichert T, Drubin D G, Barnes G. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthrie C, Fink G, editors. Guide to Yeast Genetics and Molecular Biology. 1991. Methods in Enzymology (Academic, New York), Vol. 194. [PubMed] [Google Scholar]
- 18.Michaelis C, Ciosk R, Nasmyth K. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 19.Hyland K, Kingsbury J, Koshland D, Hieter P. Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 23.Klein F, Laroche T, Cardenas M E, Hofmann J F, Schweizer D, Gasser S M. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pringle J A, Adams A E, Drubin D G, Haarer B K. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 25.Chial H. Ph.D. thesis. Boulder, CO: Univ. of Colorado; 1998. [Google Scholar]
- 26.Hecht A, Grunstein M. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 27.Wolf E, Kim P S, Berger B. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winey M, O'Toole E T. Nat Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- 29.Hardwick K, Weiss E, Luca F C, Winey M, Murray A. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 30.Biggins S, Severin F F, Bhalla N, Sassoon I, Hyman A A, Murray A W. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sassoon I, Severin F F, Andrews P D, Taba M R, Kaplan K B, Ashford A J, Stark M J, Sorger P K, Hyman A A. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Severin F, Kaplan K, Sorger P, Hyman T. Methods Cell Biol. 1999;61:145–153. doi: 10.1016/s0091-679x(08)61979-2. [DOI] [PubMed] [Google Scholar]