Abstract
Cancer cells have a very different metabolism from that of normal cells from which they are derived. Their metabolism is elevated, which allows them to sustain higher proliferative rate and resist some cell death signals. This phenomenon, known as the “Warburg effect”, has become the focus of intensive efforts in the discovery of new therapeutic targets and new cancer drugs. Both glycolysis and glutaminolysis pathways are enhanced in cancer cells. While glycolysis is enhanced to satisfy the increasing energy demand of cancer cells, glutaminolysis is enhanced to provide biosynthetic precursors for cancer cells. It was recently discovered that there is a tyrosine phosphorylation of a specific isoform of pyruvate kinase, the M2 isoform, that is preferentially expressed in all cancer cells, which results in the generation of pyruvate through a unique enzymatic mechanism that is uncoupled from ATP production. Pyruvate produced through this unique enzymatic mechanism is converted primarily into lactic acid, rather than acetyl-CoA for the synthesis of citrate, which would normally then enter the citric acid cycle. Inhibition of key enzymes in glycolysis and glutaminolysis pathways with small molecules has provided a novel but emerging area of cancer research and has been proven effective in slowing the proliferation of cancer cells, with several inhibitors being in clinical trials. This review paper will cover recent advances in the development of chemotherapeutic agents against several metabolic targets for cancer therapy, including glucose transporters, hexokinase, pyruvate kinase M2, glutaminase, and isocitrate dehydrogenase.
Keywords: Cancer, glycolysis, glutaminolysis, hexokinase, glucose transporters, pyruvate kinase M2, glutaminase, isocitrate dehydrogenase
1. INTRODUCTION
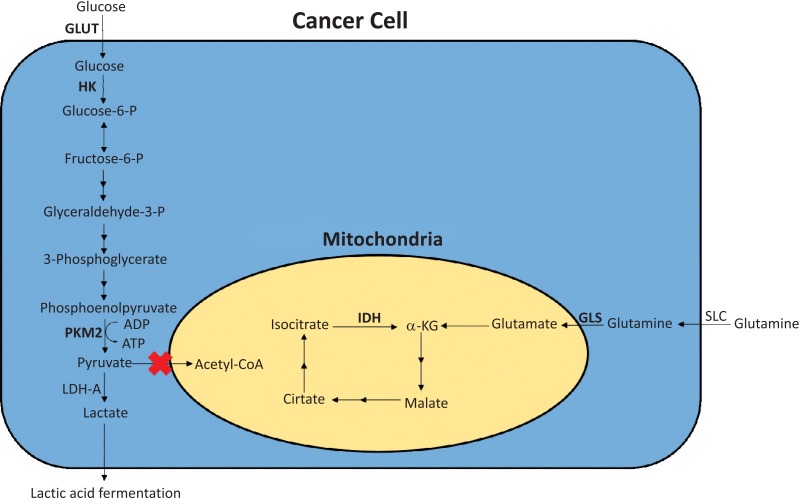
According to the World Cancer Report 2014, cancer accounted for 14.6% of all human deaths in 2012, equal to 8.2 million lives [1]. If considered as a single entity, cancer has become the second biggest cause of mortality worldwide. Costs of cancer treatment are spiraling with increasing demands that are placed on the health-care budgets of all countries. Cancer cells have a very different metabolism from that of normal cells from which they are derived [2]. Their metabolism is elevated, which allows them to sustain higher proliferative rate and resist some cell death signals. The importance of cellular metabolism in the development of cancer was observed early by Warburg when he noticed that tumor cells exhibited enhanced glycolytic activity than normal cells [3]. This phenomenon, now known as the “Warburg effect”, has recently become the focus of intensive efforts in the discovery of new therapeutic targets and new cancer drugs [4,5]. Cancer cells, however, do not enhance the glycolytic activity in their metabolism by just simply upregulating the expression of enzymes in the glycolytic pathway to accelerate many reactions in this pathway. They were observed to predominantly produce energy by a high rate of glycolysis followed by lactic acid fermentation in the cytosol, rather than by a comparatively low rate of glycolysis followed by oxidation of pyruvate in mitochondria as in most normal cells [6]. The former process is anaerobic, which does not use oxygen, and the latter process is aerobic, which uses oxygen. This switch in cancer cell metabolism occurs even if oxygen is plentiful. It was recently discovered by Christofk et al. that this switch in cancer cell metabolism happens because the conversion of phosphoenolpyruvate to pyruvate, which is catalyzed by the enzyme pyruvate kinase, is not accelerated, but rather attenuated in cancer cells [7]. There is a tyrosine phosphorylation of a specific isoform of pyruvate kinase, the M2 isoform, that is preferentially expressed in cancer cells, as well as in embryonic cells, but not in differentiated cells, which results in the generation of pyruvate through a unique enzymatic mechanism that is uncoupled from ATP production. Pyruvate produced through this unique enzymatic mechanism is converted primarily into lactic acid [6], rather than acetyl-CoA for the synthesis of citrate, which would normally enter the citric acid cycle (Fig. 1). Glycolysis, although enhanced in cancer cells, is no longer a source of biosynthetic precursors. To accommodate the alterations in the glycolytic pathway, cancer cells shift to increased rates of glutamine metabolism to maintain the citric acid cycle, particularly given the loss of the input from pyruvate [6]. This shift to increased rates of glutamine metabolism occurs through the acceleration of the conversion of glutamine in the cytosol to glutamate in the mitochondria, catalyzed by glutaminase, a mitochondrial membrane enzyme. Glutamate is subsequently converted to α-ketoglutarate by either glutamate dehydrogenase or glutamate oxaloacetate aminotransferase [8]. The outcome of elevated glutamine metabolism in cancer cells is an enhanced production of α-ketoglutarate for the citric acid cycle [6]. This outcome is critical to cancer cells as it provides carbons for the citric acid cycle to produce glutathione, fatty acids, and nucleotides, and contributes nitrogens to produce hexosamines, nucleotides, and many nonessential amino acids [9-12]. It has been shown that cancer cells are dependent on glutamine metabolism, and this dependency causes cancer cells to be highly sensitive to the exogenous levels of glutamine [13]. Inhibition of key enzymes in glycolysis and glutaminolysis pathways with small molecules has provided a novel but emerging area of cancer research and has been proven effective in slowing the proliferation of cancer cells, with several inhibitors being in clinical trials [10,14-24]. This review paper will cover recent advances in the development of chemotherapeutic agents against several metabolic targets for cancer therapy, including glucose transporters, hexokinase, pyruvate kinase M2, glutaminase, and isocitrate dehydrogenase (Fig. 1).
Fig. (1).
Glycolysis and glutaminolysis in cancer cells. Abbreviations: GLUT = glucose transporters; HK = hexokinase; PKM2 = pyruvate kinase M2; LDH-A = lactate dehydrogenase A; SLC = solute carrier-type transporters; GLS = glutaminase; IDH = isocitrate dehydrogenase; α-KG = α-ketoglutarate.
2. Inhibition of Glucose Transporters (GLUT)
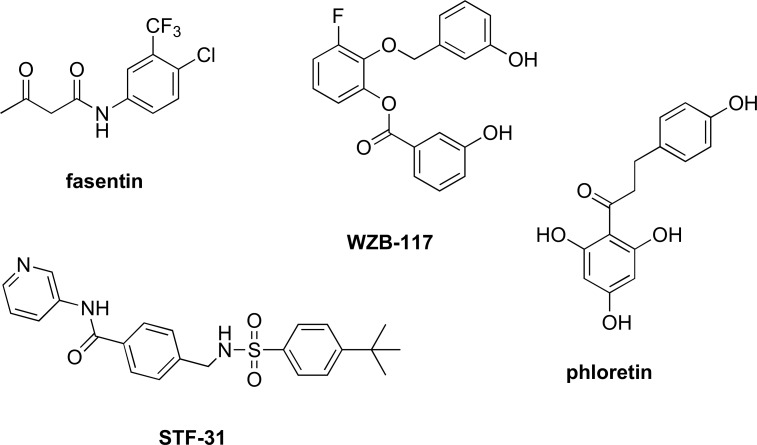
Glucose transport is the first step in the metabolism of glucose, which transports glucose across the plasma membrane. There are 14 glucose transport proteins (GLUT) present in humans, and they are often overexpressed in malignant cells [25]. Small molecule inhibitors of GLUT1, GLUT2, and GLUT4 have already been developed and studied [26-32]. In addition, recent studies have shown multiple myeloma exhibiting dependence on GLUT4, GLUT8, and GLUT11 [28]. Studies on the inhibition of GLUT started with GLUT1 antibodies, but recently, several small molecule inhibitors of GLUT, including fasentin, phloretin, STF-31, and WZB117 (Fig. 2), were discovered and have shown promising results in anticancer therapy [9,26,27,33,34].
Fig. (2).
Structures of known GLUT inhibitors.
Fasentin (Fig. 2) is a small molecule that was previously found leading to cell death. Further studies on the mechanism of action of fasentin have shown that it works as a GLUT1 inhibitor [26]. Studies by Wood et al. showed that fasentin and its analogues not only exhibit partial inhibition of the glucose transportation pathway but also break down the resistance of caspase activation, which is normally seen in malignant cells that are resistant to chemotherapy and other treatments [35,36].
Polyphenol Phloretin (Ph) (Fig. 2), isolated from apple, was recently found to be an antagonist of GLUT2 in triple-negative breast cancer (TNBC), a poorly understood subclass of breast cancer [32]. Ph was shown to suppress TNBC cell growth and metastasis, as well as to possess potential benefits for breast, bladder, liver, and colon cancer chemoprevention [32,37-39]. The benefits of Ph may have come from the antagonistic effects of GLUT1. Cao et al. observed that GLUT1 is significantly overexpressed in hypoxic regions of colon and breast cancers and that Ph showed greater than 60% inhibition of glucose uptake in Daunorubicin (DRN)-resistant breast/colon cancer cell lines [40]. This inhibition enhanced the anticancer effect of DRN by overcoming the hypoxia-conferred drug resistance and induced apoptosis in doxorubicin (a chemotherapy medication)-resistant cancer cells. Lin et al. recently observed that Ph inhibited colorectal cancer cell growth not only via inhibition of GLUT2 but also via activation of p53-mediated signaling, which is a protein that plays an important role in cell cycle control and apoptosis [41]. While other flavonoids similar to Ph have also been shown to inhibit glucose efflux, Ph exhibits the highest inhibitory activity [34]. Further studies would be needed for other flavonoids and antiestrogens.
Compared to other GLUTs, GLUT1 plays a pivotal role in basal glucose uptake, but there is a lack of potent and selective inhibitors of GLUT1. WZB117 (Fig. 2) is one of the few inhibitors that are selective for GLUT1 (IC50 = ~0.6 µM). In addition to inhibition of GLUT1, WZB117 also lowers the amount of intracellular ATP and causes stress on the endoplastic reticulum (ER), which leads to cell cycle arrest [42]. WZB117 alone was shown to have inhibitory effects on cancer cell growth in vitro and in vivo, but it also has synergistic effects with cisplatin and paclitaxel. STF-31 (Fig. 2) is another compound that is a selective inhibitor of GLUT1 (IC50 = ~1.0 µM). Chan et al. used STF-31 to exploit the loss of von Hippel-Lindau (VHL) tumor suppressor genes [27]. STF-31 suppressed renal cell carcinoma’s dependence on glycolysis by GLUT1 inhibition. This small molecule resulted in an inhibition of cancer cell growth and a decrease of tumor size in VHL-dependent models.
The antiretroviral medication ritonavir is a protease inhibitor, but recently it was discovered to have the potential utility as a noncompetitive inhibitor of GLUT4 for myeloma [28-31]. The HIV-protease inhibitor slowed the growth of multiple myeoloma cells and was used in combination with other drugs, such as metformin, to create synergistic effects.
Inhibition of GLUT3 has also been shown to delay the resistance to temozolomide (TMZ) in the treatment of glioblastoma, the most frequent malignant glioma [23,43]. This was elucidated by showing that long-term treatment using concurrent radiation and chemotherapy with the alkylating drug TMZ can lead to overexpression of GLUT3.
3. Inhibition of Hexokinase (HK)
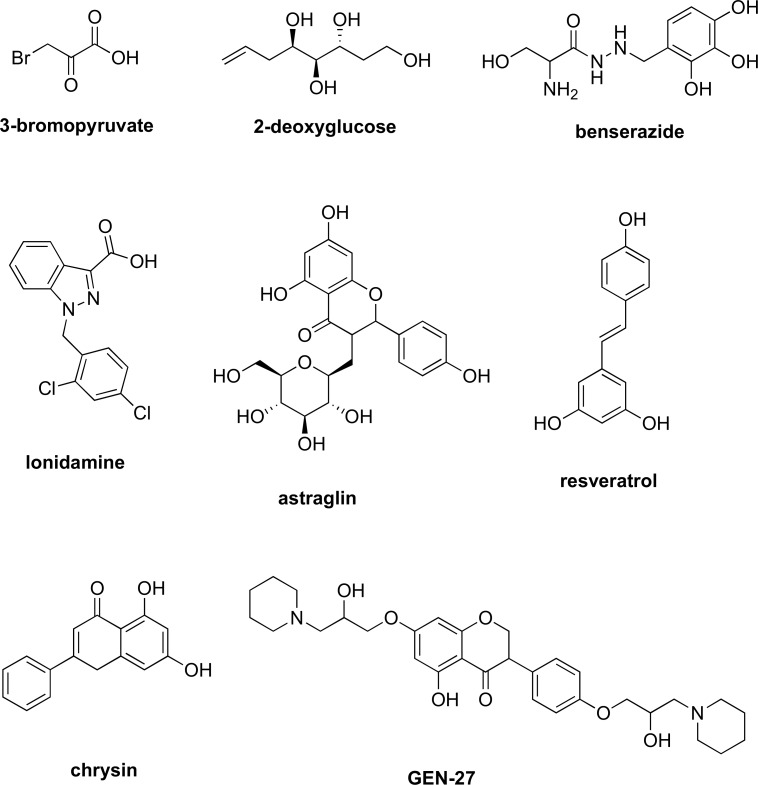
Hexokinase is a tissue-specific isoenzyme that phosphorylates glucose to glucose-6-P (G-6-P). The formation of G-6-P is the first reaction in glycolysis and also the starting point of the pentose phosphate pathway, a metabolic pathway parallel to glycolysis, which generates NADPH, pentoses, and ribose 5-phosphate, a precursor for the synthesis of nucleotides. To date, there are four hexokinase isozymes discovered in mammals [44-46]. Among the four, it has been observed that hexokinase II (HKII) plays a major role in maintaining the high glucose catabolic rate, which is needed for tumor cells to thrive [47-49]. Many inhibitors of HKII, such as 3-bromopyruvate, 2-deoxyglucose, GEN-27, benserazide, and lonidamine (Fig. 3), have been discovered and have shown efficacy in pre-clinical trial testing, with 2-deoxyglucose advancing to phase I/II clinical trials for prostate cancer before stoppage due to insignificant effects on tumor growth in vivo as a single agent [21,50-54]. Several natural products have also been shown to be HKII inhibitors [55-57].
Fig. (3).
Structures of known hexokinase inhibitors.
Since its discovery, 3-bromopyruvate (Fig. 3) has been the subject in multiple studies for its anticancer activity against many cancer types. The compound was found to inhibit HKII, activate the mitochondrial pathway for cell death, and deplete the ATP levels [49-51,58-60]. 3-Bromopyruvate was also shown to induce apoptosis in the breast cancer cell lines MDA-MB-435 and MDA-MB-231 [61]. While these results are very promising, 3-bromopyruvate was observed to trigger autophagy, which increased the breast cancer cell resistance to 3-bromopyruvate treatment [50]. When chloroquine, a large stage autophagy inhibitor, was co-administered with 3-bromopyruvate, a stimulation of Reactive Oxygen Species (ROS) formation, an enhanced 3-bromopyruvate-induced cell death in breast cancer cells was observed. Detailed mechanisms driving ROS generation and autophagy, however, are unclear. 3-Bromopyruvate was also shown to induce cell death in colon cancer [62]. The chemo-resistant cells were depleted of ATP via treatment with 3-bromopyruvate. This treatment rendered chemo-resistant colon cancer cells susceptible to oxaliplatin and 5-fluorouracil, the first-line chemotherapeutic agents for colorectal cancer [62-64]. 3-Bromopyruvate was also observed to suppress the ATP levels in malignant cell lines of multiple myeloma and in leukemic cells [65]. Resistant cell lines were chosen to study the role of glycolysis inhibition in drug resistance due to the aberrant expression of drug-expelling ATP-binding cassette (ABC) transporters. Overexpression of ABC transporters prevents the sufficient accumulation of anticancer drugs within the cells, which eventually leads to drug resistance. The malignant cell lines, when treated with DRN or mitoxantrone and 3-bromopyruvate, were observed to have a larger uptake of the chemotherapeutic drugs. This suggested that the inhibition of glycolysis with 3-bromopyruvate simultaneously led to inactivation of all types of ABC transporters in cancer cells since these transporters were dependent on the ATPs that were generated through enhanced glycolysis. The overall result was a restoration of susceptibility to anticancer drugs in these resistant cells. 3-Bromopyruvate has been hailed by many researchers as a potential breakthrough; however, it has yet to undergo formal clinical trials. It recently made the news for being the suspect after the death of three patients at an alternative medicine clinic where the drug was used without having first conducted the proper clinical studies [66]. Certainly, more human data about its efficacy and safety are much needed.
2-Deoxyglucose (Fig. 3), a glucose molecule that has the 2-hydroxyl group replaced by hydrogen, cannot undergo further glycolysis. In most cells, HK phosphorylates 2-deoxyglucose into 2-deoxyglucose-6-phosphate, which then acts to competitively inhibit the production of G-6-P from glucose. The phosphorylated 2-deoxyglucose cannot be metabolized, and its accumulation leads to ATP depletion and eventual cell death [67,68]. In addition, because of its structural similarity to mannose, 2-deoxyglucose also interferes with N-linked glycosylation, resulting in ER stress [68]. The usage of this glucose analog for cancer treatment was first reported by Jain et al [19,69,70]. In that study, 2-deoxyglucose was administered (four weekly fractions of oral 200 mg/kg body weight) before radiation (5 Gy) to increase the efficacy of radiation in brain cancer. Two anticancer drugs, ABT-263 (navitoclax) and ABT-737, which are antagonists of the Bcl-2 family and induce apoptosis in limited cell types [71], were co-administered with 2-deoxyglucose. The results showed that they were very effective against a tumor xenograft of highly metastasized chemo-resistant human prostate cancer cells. 2-Deoxyglucose has also been shown to restore sensitivity of lymphoma cells to ABT-737-induced apoptosis [71]. As a single agent therapy, 2-deoxyglucose reached phase I/II clinical trials for the treatment of solid tumors and hormone refractory prostate cancer before stoppage due to insignificant effects on tumor growth in vivo [21].
Lonidamine (Fig. 3) is another drug that acts as an inhibitor of HKII. Since its discovery over thirty years ago, lonidamine has gone through a multitude of clinical trials against many types of cancer [18,20,23]. Besides targeting HKII, subsequent studies have uncovered additional pharmacological targets for the drug, including the mitochondrial pyruvate carrier, the plasma membrane monocarboxylate transporters, the electron transport chain, and the mitochondrial permeability transition pore [72,73]. These results have shown that the anti-cancer effects of lonidamine do not occur through a single target but rather at multiple sites. Overall, the drug is capable of sensitizing tumors to chemotherapy, hyperthermia, and radiotherapy [74-76].
Genistein-27 (GEN-27) (Fig. 3), a derivative of genistein, was synthesized and reported by Du et al. to exhibit potent anti-proliferation activity against colon cancer cells as well as prevention of colitis-associated tumorigenesis [54]. This anti-proliferative activity was observed across three human colorectal carcinoma cell lines with IC50’s of less than 31µM. Further studies have shown that GEN-27 works as an inhibitor of glycolysis and induces cell apoptosis via suppression of HKII in human breast cancer cells [53].
Therapies for acute lymphoblastic leukemia have been using several classes of drugs such as glucocorticoids, vinca alkaloids, and anthracyclines [77]. However, cellular resistance to glucocorticoids is common, and therefore, therapies involving these compounds fail at about 20% of the time. Co-administration of glucocorticoids with 3-bromopyruvate, 2-deoxyglucose, or lonidamine was shown to increase in vitro sensitivity of glucocorticoids [78].
Benserazide (Fig. 3) is another HKII inhibitor that binds to HKII specifically and significantly. Screened against a large amount of targets, benserazide was identified to inhibit HKII in colorectal cancer cells, and induce apoptosis and suppress tumor growth in colorectal cancer xenograft models [52]. Benserazide was also tested in breast cancer cell lines and was shown to decrease malignant properties associated with tumorigenesis, but via a different mechanism independent of HKII inhibition [79].
Recent studies have also discovered several natural products, including astragalin, resveratrol, and the flavonoid chrysin (Fig. 3), possessing inhibitory efficacy against HKII and some specific cancer types. Astraglin was shown to reduce the activity of HKII by boosting microRNA-125b, and therefore inhibiting the proliferation of hepatocellular carcinoma (HCC) cells in vitro and in vivo [56,80]. The increase in expression of microRNA-125b was shown to suppress the proliferation of the HCC cells via the microRNA-125b/HKII cascade. Inhibition of HKII by chrysin and resveratrol were also shown to induce apoptosis in HCC cells, and therefore leading to a decrease in their proliferation [55,57].
4. Inhibition of Pyruvate Kinase M2 (PKM2)
Pyruvate kinase (PK) is the enzyme that catalyzes the final step of glycolysis, the transformation of phosphoenolpyruvate to pyruvate. There are four isoforms of PK expressed in mammals, but the M2 isoform (PKM2) has been shown to be predominantly expressed in tumor cells and play a pivotal role in cancer metabolism and tumor growth [81-84]. Studies have shown that suppression of PKM2 leads to mixed responses to some chemotherapy drugs and that resistance to oxaliplatin in many colorectal cell lines was linked to an inverse relationship between PKM2 and p53 protein levels [85-89]. P53 is a protein that plays an important role in cell cycle control and apoptosis. Mutations in p53 could result in proliferation of abnormal cells and contribute to the complex network of molecular events leading to tumor formation [90]. The p53 protein levels are usually low in normal cells, but stresses to the cell such as DNA damage, hypoxia, telomere erosion, nutrient deprivation, and ribosomal stress, can cause an increase in p53 proteins. A large increase of p53 proteins can lead to growth arrest, DNA repair, and apoptosis [91]. It is not known whether high levels of p53 and low levels of PKM2 are a good marker for colorectal cancer, but they result in a poor response to oxaliplatin [86]. Moreover, when PKM2 was studied in human gastric carcinoma cell lines, a low expression of PKM2 was observed leading to resistance to cisplatin [85]. Interestingly, it was seen in colon cancer that upregulation of PKM2 was linked to 5-fluorouracil resistance [87]. In human lung cancer cells, the silencing of PKM2 was observed increasing the efficacy of docetaxel in vitro and in vivo [82].
Few studies have been published on small molecule inhibitors or activators of PKM2, but there is some promising development. First, a class of N,N’-diarylsulfonamides was discovered to have PKM2 activation, but no further studies have been carried out [92]. In addition, resveratrol (Fig. 3), a natural phytoalexin found in a variety of plants, was shown to reduce PKM2 expression in tumor cells, which resulted in an increase in the expression of ER stress and mitochondrial fission proteins but decrease in cell viability and the levels of fusion proteins [93]. Finally, apigenin, a natural product belonging to the flavone class that is found in many plants, was recently found to restrain colon cancer cell proliferation via blocking of PKM2 [94]. These results further highlight the pivotal role of PKM2 in cancer metabolism and tumor growth. In a study led by Hitosugi, it was discovered that oncogenic forms of fibroblast growth factor receptor type 1 inhibit PKM2 by direct phosphorylation of PKM2’s Tyr105, which inhibits the formation of active, tetrameric PKM2 [95]. Thus, tyrosine phosphorylation is suggested to regulate PKM2 to provide a metabolic advantage to tumor cells, thereby promoting tumor growth.
5. Inhibition of Glutaminase (GLS)
Glutaminase is an amidohydrolase that catalyzes the first step in glutaminolysis, the conversion of glutamine in the cytosol to glutamate in the mitochondria. There are 3 isoforms of glutaminase: kidney-type (GLS1), the splice variant of GLS1 (Glutaminase C or GAC), and liver-type (GLS2) [96]. GLS1 is widely distributed throughout extra-hepatic tissues, whereas GLS2 is found primarily in adult liver [97]. While the role of the gene GLS2, a novel p53 target gene, in cancer is controversial with conflicting reports concerning its function as a tumor suppressor [98,99], GLS1, however, has emerged as a critical enzyme in a number of types of cancer cells [9]. siRNA-mediated knockdown of the gene GLS1 was shown to slow the growth of several different cancer types, including human P-493 B lymphoma cells [100], human PC3 prostate cancer cells [100], breast cancer cells [101], non-small cell lung cancer cells [102], and glioma cells [103].
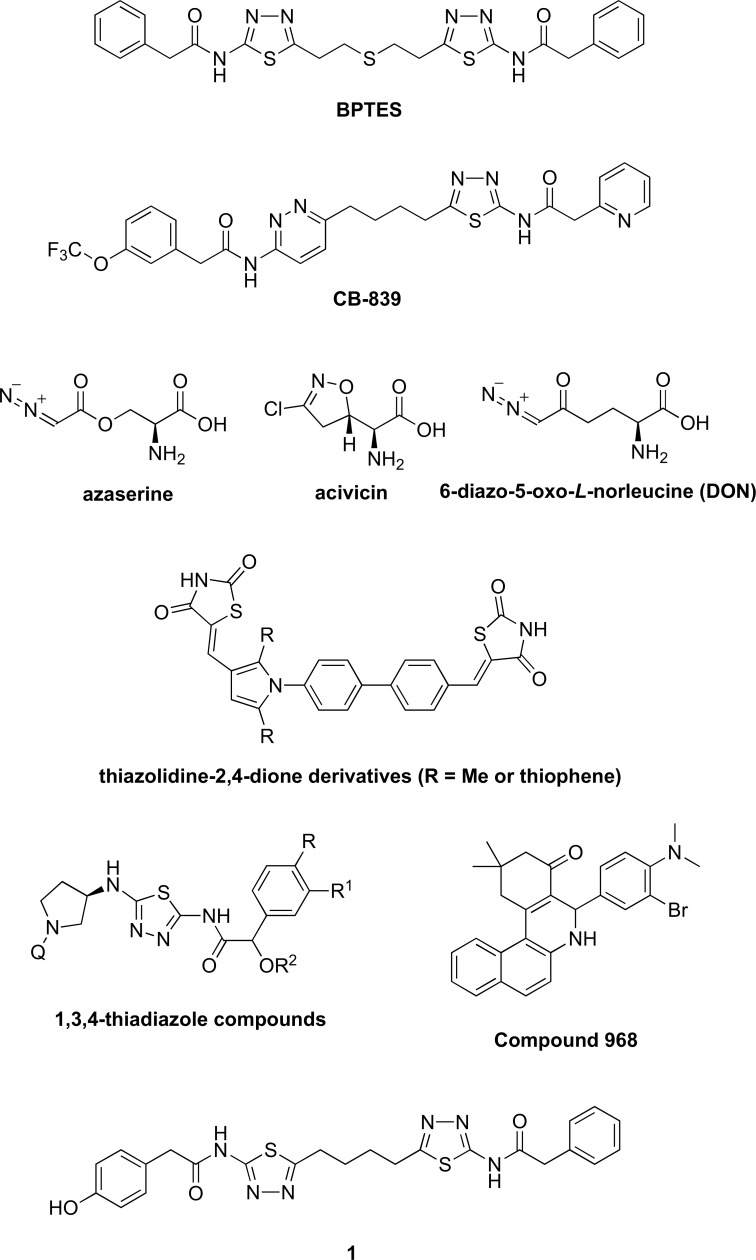
Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES) (Fig. 4) was identified to be the first allosteric inhibitor of GLS1 [97,104]. BPTES has shown to slow the proliferation in several cancer cell types in vitro and in xenograft models [8,9,12,103,105]. Docking studies and crystal structure of BPTES-bound GLS1 showed that the complex is locked into a tetramer, essentially locking GLS1 in an off mode and disabling the enzyme [106-109]. The potency of BPTES, however, is only moderate (IC50 = 3.3 µM) [97]. In addition, its poor metabolic stability and low solubility have limited its potential for clinical development [97]. An attempt to identify more BPTES-like GLS1 inhibitors by Shukla et al. did not lead to significantly better compounds [97]. CB-839 (Fig. 4), structurally related to BPTES [110], is an allosteric and more potent inhibitor of GLS1 than BPTES with a much lower IC50 (IC50 = 30 nM) [111]. CB-839 diminished growth of mouse HCC cells at a concentration (333 nM) at which BPTES had no effect on their growth [9]. CB-839 is currently in phase 1 clinical trial development for treatment of various cancer types [112].
Fig. (4).
Structures of known glutaminase inhibitors.
Azaserine [113], acivicin [113-116], and 6-diazo-5-oxo-L-norleucine (DON) [17,22,117] (Fig. 4) are naturally occurring glutamine analogues that are inactivators of GLS1. They modify the enzyme by forming covalent bonds with Ser286 in the active site. These compounds have shown to suppress the growth of a variety of tumors and have demonstrated their activity in some clinical trials [17,117]. A phase IIa study of PEGylated glutaminase (PEG-PGA) plus DON in patients with advanced refractory solid tumors was conducted, and the results were promising in late stage colorectal and lung cancers and warranted further investigations [22]. One major setback for azaserine, acivicin, and DON is that their selectivities towards GLS1 are extremely low. Besides GLS1, they also inhibit many other glutamine-dependent enzymes [9] and demonstrate variable degrees of gastrointestinal toxicity, myelosuppression, and neurotoxicity [118,119].
A class of thiazolidine-2,4-dione derivatives (Fig. 4) was recently shown to inhibit GLS1 and GLS2 in vitro cell assays and in vivo tumor growth assays [120]. When these compounds or BPTES was co-administered with doxorubicin, a stabilizer of the topoisomerase−DNA complex, a synergistic effect for suppressing AsPC-1 carcinoma cell proliferation was observed. A class of 1,3,4-thiadiazole compounds was also discovered to possess potent activities as GLS1 inhibitors and have been patented as a potential cancer treatment [13]. Another analog (1, Fig. 4) based on the 1,4-di(5-amino-1,3,4-thiadiazol-2-yl)butane scaffold was shown to be a potent GLS1 inhibitor [121]. Anti-proliferative effects of the compound on human breast cancer lines are comparable with those observed with BPTES or CB-839. Compound 968 (Fig. 4) was also discovered to be an allosteric inhibitor of GLS1 and has been widely studied [122-126]. It has been reported to have anti-tumor activity in lymphoma, breast cancer, glioblastoma, and lung cancer. Furthermore, it has been shown to inhibit cell proliferation and sensitize paclitaxel in ovarian cancer [127].
6. Inhibition of Isocitrate Dehydrogenase (IDH)
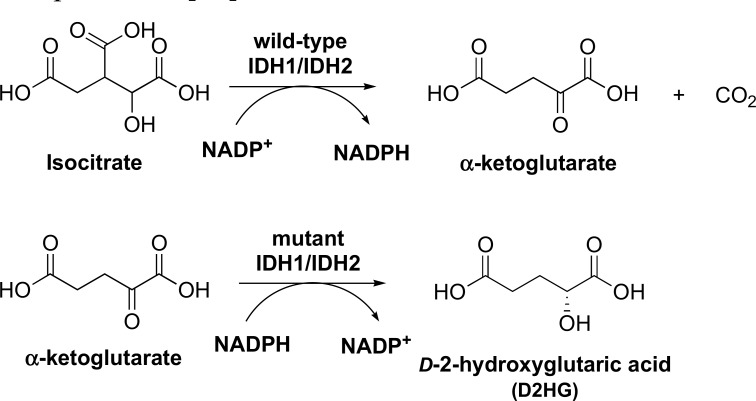
Isocitrate dehydrogenase catalyzes the conversion of isocitrate to α-ketoglutarate, a key reaction in the citric acid cycle, which contributes to the overall enhanced production of α-ketoglutarate in cancer cells [128]. There are three isoforms of IDH: cytosolic NADP-dependent IDH1, mitochondrial NADP-dependent IDH2, and mitochondrial NAD-dependent IDH3 [129]. Of the three isoforms, mutations in IDH1 and IDH2 are not present in most common tumors, but have been identified in significant percentages of several rare tumor types, including acute lymphoblastic leukemia and gliomas [128], and have been suggested to be responsible for the initiation and/or maintenance of these cancers [130]. Mutant IDH1/IDH2 are found to almost lose the wild-type enzyme activity; instead, these mutant enzymes catalyze a new reaction, the reduction of α-ketoglutarate to D-2-hydroxyglutaric acid (D2HG) (Fig. 5), which then inhibits many α-ketoglutarate-dependent dioxygenases, including histone and DNA demethylases [130].
Fig. (5).
Enzyme reactions catalyzed by wild-type IDH1/IDH2 and mutant IDH1/IDH2.
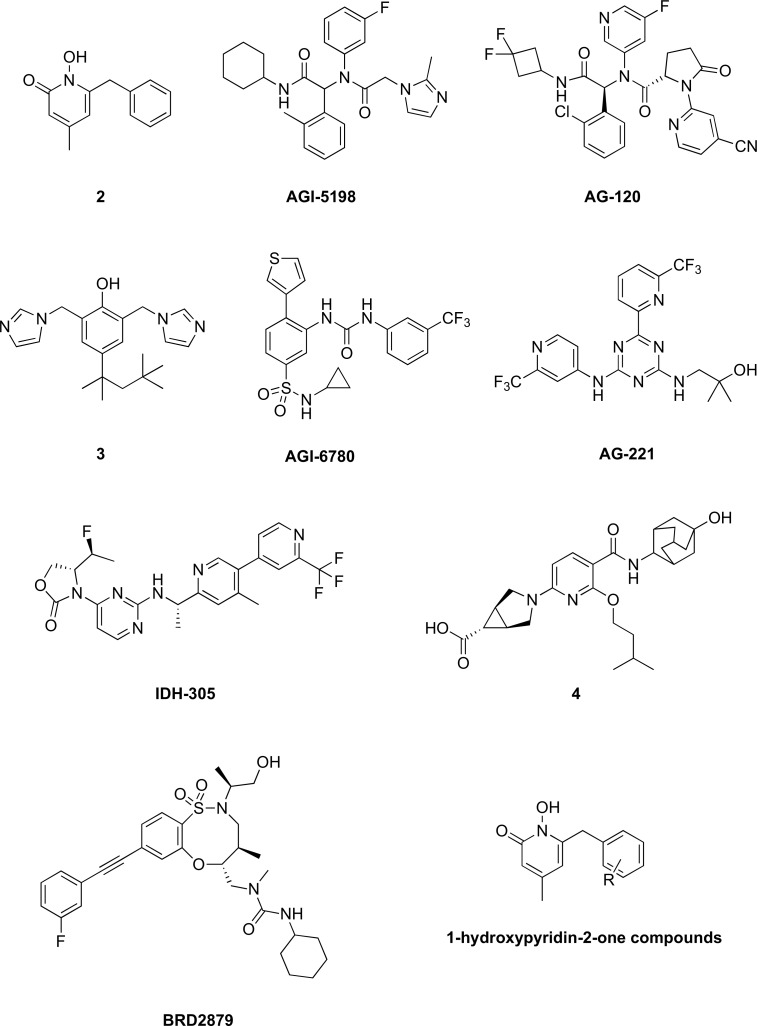
While no potent or specific inhibitors of wild-type IDH have been reported, several potent inhibitors have been identified for mutant IDH1 (2, AGI-5198, AG-120, and 3, Fig. 6) [5,131,132] and mutant IDH2 (AGI-6780 and AG-221, Fig. 6) [130,131]. AGI-5198 (Fig. 6) was reported to inhibit D2HG accumulation in IDH1-mutated glioma cells in vivo [133], reverse histone methylation, and delay growth and promote differentiation of glioma cells [134]. Compounds 2 and AGI-6780 (Fig. 6) were reported to inhibit the proliferation of IDH1- and IDH2-mutated cancer cells, respectively [130,135]. AG-120 and AG-221 (Fig. 6) are the latest development of mutant IDHs inhibitors [131]. AG-120 and AG-221 are selective for mutant IDH1 and IDH2, respectively. AG-120 is currently in Phase 1 clinical trials for patients with IDH1 mutant-positive advanced solid tumors, including glioma, chondrosarcoma and cholangiocarcinoma [14]. AG-221 is currently in Phase 2 clinical trials for patients with IDH2 mutant-positive acute myeloid leukemia, who are in second or later relapse, refractory to second-line induction or reinduction treatment, or have relapsed after allogeneic transplantation [14].
Fig. (6).
Structures of known isocitrate dehydrogenase inhibitors.
Over the last several years, more inhibitors of mutant IDH1/IDH2 have been discovered [15,135-137]. The clinical candidate, IDH-305, was shown to be a brain penetrable mutant IDH1 inhibitor. Mutations at Arg132 in IDH1 have been linked to various types of cancer, including glioma, glioblastoma, AML, chondrasarcoma, and cholangiocarcinoma [138-141]. Tested against wild-type IDH, IDH-305 was shown to have a 200-fold higher selectivity towards the mutant enzyme. IDH-305 was tested in various xenograft models and was shown to inhibit the growth of IDH1-mutated PDX melanoma model. It was also shown to have an improved brain penetration, suggesting potential utility against brain cancers. IDH-305 is currently in clinical trials to treat IDH1 mutant-positive tumors [15]. Compound 4 (Fig. 6) was recently discovered from high-throughput screening and structure-activity relationship to be an allosteric inhibitor of mutant IDH1 and display potent activity against proliferation of human acute myeloid leukemia cells [142]. In addition, 4 did not show any activity against wild-type IDH. In another study, 8-membered ring sulfonamides were discovered to be inhibitors of mutant IDH1, with BRD2879 (Fig. 6) being the most potent compound from the study [143]. However, the low solubility and poor metabolic stability of BRD2879 prevented its use in vivo. BRD2879 was also tested against wild-type IDH and was shown to have no activity. Other 1-hydroxypyridin-2-one compounds like 2 (Fig. 6) were also found to inhibit mutant IDH1 [135]. Of the 61 derivatives synthesized, several potent inhibitors were identified. These compounds exhibited potent and selective activity in inhibiting the proliferation of IDH1-mutated BT-142 glioma cells. When tested against wild-type IDH, these compounds showed little or no activity.
CONCLUSION
Cancer cells have a very different metabolism from that of normal cells from which they are derived. They depend on aerobic glycolysis, fatty acid synthesis, and glutaminolysis. The difference in their metabolism promotes cell proliferation and decreases drug-induced apoptosis, leading to therapeutic resistance. The enzymes that catalyze deregulated transformations in cancer metabolism become very attractive targets for cancer therapy. The more the studies on these important metabolic targets, the better the understanding and the clearer the picture on the differences in cancer cell metabolism and normal cell metabolism to assess selectivity and toxicity. The amount of work being done in this field is growing and making great progress in the development of small molecule anticancer agents.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
We are grateful to the American Association of Colleges of Pharmacy (a 2018 New Investigator Award to H.V.L.) and the Department of BioMolecular Sciences at the University of Mississippi School of Pharmacy for the support.
conflict of interest
The authors declare no conflict of interest, financial or otherwise.
REFEReNCES
- 1.Stewart B.W., Wild C.P. World Cancer Rep., 2014. IARC; 2014. [Google Scholar]
- 2.Tennant D.A., Durán R.V., Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Li C., Zhang G., Zhao L., Ma Z., Chen H. Metabolic reprogramming in cancer cells: Glycolysis, glutaminolysis, and bcl-2 proteins as novel therapeutic targets for cancer. World J. Surg. Oncol. 2015;14:15. doi: 10.1186/s12957-016-0769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng G., Shen J., Yin M., McManus J., Mathieu M., Gee P., He T., Shi C., Bedel O., McLean L.R., Le-Strat F., Zhang Y., Marquette J-P., Gao Q., Zhang B., Rak A., Hoffmann D., Rooney E., Vassort A., Englaro W., Li Y., Patel V., Adrian F., Gross S., Wiederschain D., Cheng H., Licht S. Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by an allosteric small molecule. J. Biol. Chem. 2015;290:762–774. doi: 10.1074/jbc.M114.608497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson J.W., Cerione R.A. Glutaminase: a hot spot for regulation of cancer cell metabolism? Oncotarget. 2010;1:734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R., Fleming M.D., Schreiber S.L., Cantley L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 8.Emadi A., Jun S.A., Tsukamoto T., Fathi A.T., Minden M.D., Dang C.V. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp. Hematol. 2014;42:247–251. doi: 10.1016/j.exphem.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Xiang Y., Stine Z.E., Xia J., Lu Y., O’Connor R.S., Altman B.J., Hsieh A.L., Gouw A.M., Thomas A.G., Gao P., Sun L., Song L., Yan B., Slusher B.S., Zhuo J., Ooi L.L., Lee C.G.L., Mancuso A., McCallion A.S., Le A., Milone M.C., Rayport S., Felsher D.W., Dang C.V. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 2015;125:2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W., Le A., Hancock C., Lane A.N., Dang C.V., Fan T.W-M., Phang J.M. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. USA. 2012;109:8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le A., Lane A.N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C.J., Slusher B.S., Zhang H., Zimmerman L.J., Liebler D.C., Slebos R.J.C., Lorkiewicz P.K., Higashi R.M., Fan T.W.M., Dang C.V. Glucose-independent glutamine metabolism via tca cycling for proliferation and survival in b cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdel-Magid A.F. Glutaminase GLS1 inhibitors as potential cancer treatment. ACS Med. Chem. Lett. 2016;7:207–208. doi: 10.1021/acsmedchemlett.6b00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Botton S., Mondesir J., Willekens C., Touat M. IDH1 and IDH2 mutations as novel therapeutic targets: Current perspectives. J. Blood Med. 2016;7:171–180. doi: 10.2147/JBM.S70716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho Y.S., Levell J.R., Liu G., Caferro T., Sutton J., Shafer C.M., Costales A., Manning J.R., Zhao Q., Sendzik M., Shultz M., Chenail G., Dooley J., Villalba B., Farsidjani A., Chen J., Kulathila R., Xie X., Dodd S., Gould T., Liang G., Heimbach T., Slocum K., Firestone B., Pu M., Pagliarini R., Growney J.D. Discovery and evaluation of clinical candidate IDH305, a Brain penetrant mutant IDH1 Inhibitor. ACS Med. Chem. Lett. 2017;8:1116–1121. doi: 10.1021/acsmedchemlett.7b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. https://clinicaltrials.gov/ct2/show/
- 17.Shapiro R.A., Clark V.M., Curthoys N.P. Inactivation of Rat Renal Phosphate-Dependent Glutaminase with 6-Diazo-5-Oxo-L-Norleucine. Evidence for interaction at the glutamine binding site. J. Biol. Chem. 1979;254:2835–2838. [PubMed] [Google Scholar]
- 18.Gatzemeier U., Cavalli F., Häussinger K., Kaukel E., Koschel G., Martinelli G., Neuhauss R., von Pawel J. Phase III trial with and without lonidamine in non-small cell lung cancer. Semin. Oncol. 1991;18:42–48. [PubMed] [Google Scholar]
- 19.Mohanti B.K., Rath G.K., Anantha N., Kannan V., Das B.S., Chandramouli B.A.R., Banerjee A.K., Das S., Jena A., Ravichandran R., Sahi U.P., Kumar R., Kapoor N., Kalia V.K., Dwarakanath B.S., Jain V. Improving cancer radiotherapy with 2-Deoxy-D-Glucose: Phase I/II clinical trials on human cerebral gliomas. Int. J. Radiat. Oncol. Biol. Phys. 1996;35:103–111. doi: 10.1016/s0360-3016(96)85017-6. [DOI] [PubMed] [Google Scholar]
- 20.Di Cosimo S., Ferretti G., Papaldo P., Carlini P., Fabi A., Cognetti F. Lonidamine: Efficacy and safety in clinical trials for the treatment of solid tumors. Drugs Today (Barc) 2003;39:157–174. doi: 10.1358/dot.2003.39.3.799451. [DOI] [PubMed] [Google Scholar]
- 21. https://clinicaltrials.gov/ct2/show/NCT00633087
- 22.Mueller C., Al-Batran S., Jaeger E., Schmidt B., Bausch M., Unger C., Sethuraman N. 2008. [Google Scholar]
- 23.Prabhakara S., Kalia V.K. Optimizing radiotherapy of brain tumours by a combination of temozolomide & lonidamine. Indian J. Med. Res. 2008;128:140–148. [PubMed] [Google Scholar]
- 24. https://clinicaltrials.gov/ct2/show/NCT02381886
- 25.Macheda M.L., Rogers S., Best J.D. Molecular and Cellular Regulation of Glucose Transporter (GLUT) Proteins in Cancer. J. Cell. Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 26.Wood T.E., Dalili S., Simpson C.D., Hurren R., Mao X., Saiz F.S., Gronda M., Eberhard Y., Minden M.D., Bilan P.J., Klip A., Batey R.A., Schimmer A.D. A novel inhibitor of glucose uptake sensitizes cells to fas-induced cell death. Mol. Cancer Ther. 2008;7:3546–3555. doi: 10.1158/1535-7163.MCT-08-0569. [DOI] [PubMed] [Google Scholar]
- 27.Chan D.A., Sutphin P.D., Nguyen P., Turcotte S., Lai E.W., Banh A., Reynolds G.E., Chi J-T., Wu J., Solow-Cordero D.E., Bonnet M., Flanagan J.U., Bouley D.M., Graves E.E., Denny W.A., Hay M.P., Giaccia A.J. Targeting GLUT1 and the warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci. Transl. Med. 2011;3:94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBrayer S.K., Cheng J.C., Singhal S., Krett N.L., Rosen S.T., Shanmugam M. Multiple myeloma exhibits novel dependence on GLUT4, GLUT8, and GLUT11: Implications for glucose transporter-directed therapy. Blood. 2012;119:4686–4697. doi: 10.1182/blood-2011-09-377846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalva-Aydemir S., Bajpai R., Martinez M., Adekola K.U.A., Kandela I., Wei C., Singhal S., Koblinski J.E., Raje N.S., Rosen S.T., Shanmugam M. Targeting the metabolic plasticity of multiple myeloma with fda-approved ritonavir and metformin. Clin. Cancer Res. 2015;21:1161–1171. doi: 10.1158/1078-0432.CCR-14-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra R.K., Wei C., Hresko R.C., Bajpai R., Heitmeier M., Matulis S.M., Nooka A.K., Rosen S.T., Hruz P.W., Schiltz G.E., Shanmugam M. In silico modeling-based identification of glucose transporter 4 (GLUT4)-Selective inhibitors for cancer therapy. J. Biol. Chem. 2015;290:14441–14453. doi: 10.1074/jbc.M114.628826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srirangam A., Milani M., Mitra R., Guo Z., Rodriguez M., Kathuria H., Fukuda S., Rizzardi A., Schmechel S., Skalnik D.G., Pelus L.M., Potter D.A. The human immunodeficiency virus protease inhibitor ritonavir inhibits lung cancer cells, in part, by inhibition of survivin. J. Thorac. Oncol. 2011;6:661–670. doi: 10.1097/JTO.0b013e31820c9e3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu K-H., Ho C-T., Chen Z-F., Chen L-C., Whang-Peng J., Lin T-N., Ho Y-S. The apple polyphenol phloretin inhibits breast cancer cell migration and proliferation via inhibition of signals by type 2 glucose transporter. Yao Wu Shi Pin Fen Xi. 2018;26:221–231. doi: 10.1016/j.jfda.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M-S., Kwon J.Y., Kang N.J., Lee K.W., Lee H.J. Phloretin induces apoptosis in H-Ras MCF10A human breast tumor cells through the activation of p53 via JNK and p38 mitogen-activated protein kinase signaling. Ann. N. Y. Acad. Sci. 2009;1171:479–483. doi: 10.1111/j.1749-6632.2009.04692.x. [DOI] [PubMed] [Google Scholar]
- 34.Martin H.J., Kornmann F., Fuhrmann G.F. The inhibitory effects of flavonoids and antiestrogens on the glut1 glucose transporter in human erythrocytes. Chem. Biol. Interact. 2003;146:225–235. doi: 10.1016/j.cbi.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Wang S., El-Deiry W.S. TRAIL and apoptosis induction by tnf-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 36.Mawji I.A., Simpson C.D., Hurren R., Gronda M., Williams M.A., Filmus J., Jonkman J., Da Costa R.S., Wilson B.C., Thomas M.P., Reed J.C., Glinsky G.V., Schimmer A.D. Critical role for fas-associated death domain-like interleukin-1-converting enzyme-like inhibitory protein in anoikis resistance and distant tumor formation. J. Natl. Cancer Inst. 2007;99:811–822. doi: 10.1093/jnci/djk182. [DOI] [PubMed] [Google Scholar]
- 37.Wu C-H., Ho Y-S., Tsai C-Y., Wang Y-J., Tseng H., Wei P-L., Lee C-H., Liu R-S., Lin S-Y. In vitro and in vivo study of phloretin-induced apoptosis in human liver cancer cells involving inhibition of type ii glucose transporter. Int. J. Cancer. 2009;124:2210–2219. doi: 10.1002/ijc.24189. [DOI] [PubMed] [Google Scholar]
- 38.Nelson J.A.S., Falk R.E. The efficacy of phloridzin and phloretin on tumor cell growth. Anticancer Res. 1993;13:2287–2292. [PubMed] [Google Scholar]
- 39.Yang K.C., Tsai C.Y., Wang Y.J., Wei P.L., Lee C.H., Chen J.H., Wu C.H., Ho Y.S. Apple polyphenol phloretin potentiates the anticancer actions of paclitaxel through induction of apoptosis in human hep g2 cells. Mol. Carcinog. 2009;48:420–431. doi: 10.1002/mc.20480. [DOI] [PubMed] [Google Scholar]
- 40.Cao X., Fang L., Gibbs S., Huang Y., Dai Z., Wen P., Zheng X., Sadee W., Sun D. Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia. Cancer Chemother. Pharmacol. 2007;59:495–505. doi: 10.1007/s00280-006-0291-9. [DOI] [PubMed] [Google Scholar]
- 41.Lin S-T., Tu S-H., Yang P-S., Hsu S-P., Lee W-H., Ho C-T., Wu C-H., Lai Y-H., Chen M-Y., Chen L-C. Apple polyphenol phloretin inhibits colorectal cancer cell growth via inhibition of the type 2 glucose transporter and activation of p53-mediated signaling. J. Agric. Food Chem. 2016;64:6826–6837. doi: 10.1021/acs.jafc.6b02861. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y., Cao Y., Zhang W., Bergmeier S., Qian Y., Akbar H., Colvin R., Ding J., Tong L., Wu S., Hines J., Chen X. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol. Cancer Ther. 2012;11:1672–1682. doi: 10.1158/1535-7163.MCT-12-0131. [DOI] [PubMed] [Google Scholar]
- 43.Le Calvé B., Rynkowski M., Le Mercier M., Bruyère C., Lonez C., Gras T., Haibe-Kains B., Bontempi G., Decaestecker C., Ruysschaert J-M., Kiss R., Lefranc F. Long-term in vitro treatment of human glioblastoma cells with temozolomide increases resistance in vivo through up-regulation of glut transporter and aldo-keto reductase enzyme akr1c expression. Neoplasia. 2010;12:727–739. doi: 10.1593/neo.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bork P., Sander C., Valencia A. Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson J.E. Hexokinases. Rev. Physiol. Biochem. Pharmacol. 1995;126:65–198. doi: 10.1007/BFb0049776. [DOI] [PubMed] [Google Scholar]
- 46.Cárdenas M.L., Cornish-Bowden A., Ureta T. Evolution and regulatory role of the hexokinases. Biochim. Biophys. Acta - Mol. Cell Res. 1998;1401:242–264. doi: 10.1016/s0167-4889(97)00150-x. [DOI] [PubMed] [Google Scholar]
- 47.Bustamante E., Pedersen P.L. High aerobic glycolysis of rat hepatoma cells in culture: Role of mitochondrial hexokinase. Proc. Natl. Acad. Sci. USA. 1977;74:3735–3739. doi: 10.1073/pnas.74.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rempel A., Mathupala S.P., Griffin C.A., Hawkins A.L., Pedersen P.L. Glucose catabolism in cancer cells: Amplification of the gene encoding type II Hexokinase. Cancer Res. 1996;56:2468–2471. [PubMed] [Google Scholar]
- 49.Ko Y.H., Pedersen P.L., Geschwind J.F. Glucose catabolism in the rabbit vx2 tumor model for liver cancer: Characterization and targeting hexokinase. Cancer Lett. 2001;173:83–91. doi: 10.1016/s0304-3835(01)00667-x. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q., Zhang Y., Zhang P., Chao Z., Xia F., Jiang C., Zhang X., Jiang Z., Liu H., Hexokinase I.I. Inhibitor, 3-BrPA induced autophagy by Stimulating ROS formation in human breast cancer cells. Genes Cancer. 2014;5:100–112. doi: 10.18632/genesandcancer.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jae H.J., Chung J.W., Park H.S., Lee M.J., Lee K.C., Kim H.C., Yoon J.H., Chung H., Park J.H. The antitumor effect and hepatotoxicity of a hexokinase II Inhibitor 3-Bromopyruvate: In vivo investigation of intraarterial administration in a rabbit VX2 hepatoma model. Korean J. Radiol. 2009;10:596–603. doi: 10.3348/kjr.2009.10.6.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W., Zheng M., Wu S., Gao S., Yang M., Li Z., Min Q., Sun W., Chen L., Xiang G., Li H. Benserazide, a dopadecarboxylase inhibitor, suppresses tumor growth by targeting hexokinase 2. J. Exp. Clin. Cancer Res. 2017;36:58. doi: 10.1186/s13046-017-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao L., Wei L., Liu Y., Ding Y., Liu X., Zhang X., Wang X., Yao Y., Lu J., Wang Q., Hu R. Gen-27, a newly synthesized flavonoid, inhibits glycolysis and induces cell apoptosis via suppression of hexokinase II in human breast cancer cells. Biochem. Pharmacol. 2017;125:12–25. doi: 10.1016/j.bcp.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Du Q., Wang Y., Liu C., Wang H., Fan H., Li Y., Wang J., Zhang X., Lu J., Ji H., Hu R., Du Q., Wang Y., Liu C., Wang H., Fan H., Li Y., Wang J., Zhang X., Lu J., Ji H., Hu R. Chemopreventive Activity of GEN-27, a Genistein Derivative, in Colitis-Associated Cancer Is Mediated by p65-CDX2-β-Catenin Axis. Oncotarget. 2016;7:17870–17884. doi: 10.18632/oncotarget.7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai W., Wang F., Lu J., Xia Y., He L., Chen K., Li J., Li S., Liu T., Zheng Y., Wang J., Lu W., Zhou Y., Yin Q., Abudumijiti H., Chen R., Zhang R., Zhou L., Zhou Z., Zhu R., Yang J., Wang C., Zhang H., Zhou Y., Xu L., Guo C. By reducing hexokinase 2, resveratrol induces apoptosis in HCC cells addicted to aerobic glycolysis and inhibits tumor growth in mice. Oncotarget. 2015;6:13703–13717. doi: 10.18632/oncotarget.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W., Hao J., Zhang L., Cheng Z., Deng X., Shu G. Astragalin reduces hexokinase 2 through increasing miR-125b to inhibit the proliferation of hepatocellular carcinoma cells in Vitro and in Vivo. J. Agric. Food Chem. 2017;65:5961–5972. doi: 10.1021/acs.jafc.7b02120. [DOI] [PubMed] [Google Scholar]
- 57.Xu D., Jin J., Yu H., Zhao Z., Ma D., Zhang C., Jiang H. Chrysin inhibited tumor glycolysis and induced apoptosis in hepatocellular carcinoma by targeting hexokinase-2. J. Exp. Clin. Cancer Res. 2017;36:44. doi: 10.1186/s13046-017-0514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu R.H., Pelicano H., Zhou Y., Carew J.S., Feng L., Bhalla K.N., Keating M.J., Huang P. Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005;65:613–621. [PubMed] [Google Scholar]
- 59.Kim W., Yoon J-H., Jeong J-M., Cheon G-J., Lee T-S., Yang J-I., Park S-C., Lee H-S. Apoptosis-inducing antitumor efficacy of hexokinase II inhibitor in hepatocellular carcinoma. Mol. Cancer Ther. 2007;6:2554–2562. doi: 10.1158/1535-7163.MCT-07-0115. [DOI] [PubMed] [Google Scholar]
- 60.Ganapathy-Kanniappan S., Vali M., Kunjithapatham R., Buijs M., Syed L.H., Rao P.P., Ota S., Kwak B.K., Loffroy R., Geschwind J.F. 3-bromopyruvate: A new targeted antiglycolytic agent and a promise for cancer therapy. Curr. Pharm. Biotechnol. 2010;11:510–517. doi: 10.2174/138920110791591427. [DOI] [PubMed] [Google Scholar]
- 61.Kwiatkowska E., Wojtala M., Gajewska A., Soszyński M., Bartosz G., Sadowska-Bartosz I. Effect of 3-bromopyruvate acid on the redox equilibrium in non-invasive MCF-7 and Invasive MDA-MB-231 breast cancer cells. J. Bioenerg. Biomembr. 2016;48:23–32. doi: 10.1007/s10863-015-9637-5. [DOI] [PubMed] [Google Scholar]
- 62.Sun Y., Liu Z., Zou X., Lan Y., Sun X., Wang X., Zhao S., Jiang C., Liu H. Mechanisms underlying 3-bromopyruvate-induced cell death in colon cancer. J. Bioenerg. Biomembr. 2015;47:319–329. doi: 10.1007/s10863-015-9612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ihrlund L.S., Hernlund E., Khan O., Shoshan M.C. 3-bromopyruvate as inhibitor of tumour cell energy metabolism and chemopotentiator of platinum drugs. Mol. Oncol. 2008;2:94–101. doi: 10.1016/j.molonc.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chong D., Ma L., Liu F., Zhang Z., Zhao S., Huo Q., Zhang P., Zheng H., Liu H. Synergistic antitumor effect of 3-Bromopyruvate and 5-fluorouracil against human colorectal cancer through cell cycle arrest and induction of apoptosis. Anticancer Drugs. 2017;28:831–840. doi: 10.1097/CAD.0000000000000517. [DOI] [PubMed] [Google Scholar]
- 65.Nakano A., Tsuji D., Miki H., Cui Q., El Sayed S.M., Ikegame A., Oda A., Amou H., Nakamura S., Harada T., Fujii S., Kagawa K., Takeuchi K., Sakai A., Ozaki S., Okano K., Nakamura T., Itoh K., Matsumoto T., Abe M. Glycolysis Inhibition Inactivates ABC Transporters to Restore Drug Sensitivity in Malignant Cells. PLoS One. 2011;6:e27222. doi: 10.1371/journal.pone.0027222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feldwisch-Drentrup H. Candidate cancer drug suspected after death of three patients at an alternative medicine clinic. http://www.sciencemag.org/news/2016/08/candidate-cancer-drug-suspected-after-death-three-patients-alternative-medicine-clinic
- 67.Maher J.C., Krishan A., Lampidis T.J. Greater Cell Cycle Inhibition and Cytotoxicity Induced by 2-Deoxy-D-Glucose in tumor cells treated under hypoxic vs aerobic conditions. Cancer Chemother. Pharmacol. 2004;53:116–122. doi: 10.1007/s00280-003-0724-7. [DOI] [PubMed] [Google Scholar]
- 68.Kurtoglu M., Maher J.C., Lampidis T.J. Differential Toxic Mechanisms of 2-Deoxy-D-Glucose versus 2-Fluorodeoxy-D -glucose in hypoxic and normoxic tumor cells. Antioxid. Redox Signal. 2007;9:1383–1390. doi: 10.1089/ars.2007.1714. [DOI] [PubMed] [Google Scholar]
- 69.Jain V.K., Kalia V.K., Sharma R., Maharajan V., Menon M. Effects of 2-Deoxy-D-glucose on glycolysis, proliferation kinetics and radiation response of human cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 1985;11:943–950. doi: 10.1016/0360-3016(85)90117-8. [DOI] [PubMed] [Google Scholar]
- 70.Dwarakanath B.S., Jain V.K. Modification of the radiation induced damage by 2-Deoxy-d-glucose in organ cultures of human cerebral gliomas. Int. J. Radiat. Oncol. Biol. Phys. 1987;13:741–746. doi: 10.1016/0360-3016(87)90293-8. [DOI] [PubMed] [Google Scholar]
- 71.Yamaguchi R., Janssen E., Perkins G., Ellisman M., Kitada S., Reed J.C. Efficient elimination of cancer cells by deoxyglucose-ABT-263/737 combination therapy. PLoS One. 2011;6:e24102. doi: 10.1371/journal.pone.0024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nancolas B., Guo L., Zhou R., Nath K., Nelson D.S., Leeper D.B., Blair I.A., Glickson J.D., Halestrap A.P. The anti-tumour agent lonidamine is a potent inhibitor of the mitochondrial pyruvate carrier and plasma membrane monocarboxylate transporters. Biochem. J. 2016;473:929–936. doi: 10.1042/BJ20151120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhutia Y.D., Babu E., Ganapathy V. Re-programming tumour cell metabolism to treat cancer: no lone target for lonidamine. Biochem. J. 2016;473:1503–1506. doi: 10.1042/BCJ20160068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gallo-Curcio C., Verturo I., Rinaldi M., Ambesi Impiombato F., Del Vecchio M.R., Tonachella R., Tropea F., Antilli A., Ciottoli G.B., De Gregorio M. Chemotherapy/radiation therapy plus/minus lonidamine in the treatment of non-small cell lung cancer (limited disease): preliminary results. Semin. Oncol. 1988;15:26–31. [PubMed] [Google Scholar]
- 75.Raaphorst G.P., Feeley M.M., Martin L., Danjoux C.E., Maroun J., Desanctis A.J., Ko D. Enhancement of sensitivity to hyperthermia by lonidamine in human cancer cells. Int. J. Hyperthermia. 1991;7:763–772. doi: 10.3109/02656739109056445. [DOI] [PubMed] [Google Scholar]
- 76.Magno L., Terraneo F., Bertoni F., Tordiglione M., Bardelli D., Rosignoli M.T., Ciottoli G.B. Double-blind randomized study of lonidamine and radiotherapy in head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 1994;29:45–55. doi: 10.1016/0360-3016(94)90225-9. [DOI] [PubMed] [Google Scholar]
- 77.Pui C-H., Evans W.E. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 78.Hulleman E., Kazemier K.M., Holleman A., VanderWeele D.J., Rudin C.M., Broekhuis M.J.C., Evans W.E., Pieters R., Den Boer M.L. Inhibition of glycolysis modulates prednisolone resistance in acute lymphoblastic leukemia cells. Blood. 2009;113:2014–2021. doi: 10.1182/blood-2008-05-157842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alli E., Solow-Cordero D., Casey S.C., Ford J.M. Therapeutic targeting of BRCA1-mutated breast cancers with agents that activate DNA repair. Cancer Res. 2014;74:6205–6215. doi: 10.1158/0008-5472.CAN-14-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao L., Wang W. miR-125b suppresses the proliferation of hepatocellular carcinoma cells by targeting sirtuin7. Int. J. Clin. Exp. Med. 2015;8:18469–18475. [PMC free article] [PubMed] [Google Scholar]
- 81.Muñoz-Pinedo C., El Mjiyad N., Ricci J-E. Cancer metabolism: Current perspectives and future directions. Cell Death Dis. 2012;3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi H. shan; Li, D.; Zhang, J.; Wang, Y. sheng; Yang, L.; Zhang, H. long; Wang, X. huo; Mu, B.; Wang, W.; Ma, Y.; Guo, F. chun; Wei, Y. quan. Silencing of pkm2 Increases the Efficacy of docetaxel in human lung cancer xenografts in mice. Cancer Sci. 2010;101:1447–1453. doi: 10.1111/j.1349-7006.2010.01562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu S., Le H. Dual Roles of PKM2 in cancer metabolism. acta Biochim. Biophys. Sin. (Shanghai) 2013;45:27–35. doi: 10.1093/abbs/gms106. [DOI] [PubMed] [Google Scholar]
- 84.Christofk H.R., Vander Heiden M.G., Wu N., Asara J.M., Cantley L.C. Pyruvate Kinase M2 Is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 85.Yoo B.C., Ku J-L., Hong S-H., Shin Y-K., Park S.Y., Kim H.K., Park J-G. Decreased pyruvate kinase M2 activity linked to cisplatin resistance in human gastric carcinoma cell lines. Int. J. Cancer. 2004;108:532–539. doi: 10.1002/ijc.11604. [DOI] [PubMed] [Google Scholar]
- 86.Arango D., Wilson A.J., Shi Q., Corner G.A., Arañes M.J., Nicholas C., Lesser M., Mariadason J.M., Augenlicht L.H. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br. J. Cancer. 2004;91:1931–1946. doi: 10.1038/sj.bjc.6602215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shin Y-K., Yoo B.C., Hong Y.S., Chang H.J., Jung K.H., Jeong S-Y., Park J-G. Upregulation of glycolytic enzymes in proteins secreted from human colon cancer cells with 5-fluorouracil resistance. Electrophoresis. 2009;30:2182–2192. doi: 10.1002/elps.200800806. [DOI] [PubMed] [Google Scholar]
- 88.Martinez-Balibrea E., Plasencia C., Ginés A., Martinez-Cardús A., Musulén E., Aguilera R., Manzano J.L., Neamati N., Abad A. A proteomic approach links decreased pyruvate kinase m2 expression to oxaliplatin resistance in patients with colorectal cancer and in human cell lines. Mol. Cancer Ther. 2009;8:771–778. doi: 10.1158/1535-7163.MCT-08-0882. [DOI] [PubMed] [Google Scholar]
- 89.Duffy M.J., van Dalen A., Haglund C., Hansson L., Holinski-Feder E., Klapdor R., Lamerz R., Peltomaki P., Sturgeon C., Topolcan O. Tumour markers in colorectal cancer: European group on tumour markers (EGTM) guidelines for clinical use. Eur. J. Cancer. 2007;43:1348–1360. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 90.Ryan K.M., Phillips A.C., Vousden K.H. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 2001;13:332–337. doi: 10.1016/s0955-0674(00)00216-7. [DOI] [PubMed] [Google Scholar]
- 91.Vogelstein B., Lane D., Levine A.J. Surfing the p53 Network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 92.Boxer M.B., Jiang J.K., Vander Heiden M.G., Shen M., Skoumbourdis A.P., Southall N., Veith H., Leister W., Austin C.P., Park H.W., Inglese J., Cantley L.C., Auld D.S., Thomas C.J. Evaluation of substituted N,N′-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate Kinase. J. Med. Chem. 2010;53:1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu H., Wang Y., Wu C., Yang P., Li H., Li Z. Resveratrol induces cancer cell apoptosis through MiR-326/PKM2-Mediated ER stress and mitochondrial fission. J. Agric. Food Chem. 2016;64:9356–9367. doi: 10.1021/acs.jafc.6b04549. [DOI] [PubMed] [Google Scholar]
- 94.Shan S., Shi J., Yang P., Jia B., Wu H., Zhang X., Li Z. Apigenin Restrains Colon Cancer Cell Proliferation via Targeted Blocking of Pyruvate Kinase M2-Dependent Glycolysis. J. Agric. Food Chem. 2017;65:8136–8144. doi: 10.1021/acs.jafc.7b02757. [DOI] [PubMed] [Google Scholar]
- 95.Hitosugi T., Kang S., Vander Heiden M.G., Chung T-W., Elf S., Lythgoe K., Dong S., Lonial S., Wang X., Chen G.Z., Xie J., Gu T-L., Polakiewicz R.D., Roesel J.L., Boggon T.J., Khuri F.R., Gilliland D.G., Cantley L.C., Kaufman J., Chen J. Tyrosine phosphorylation inhibits pkm2 to promote the warburg effect and tumor growth. Sci. Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thangavelu K., Chong Q.Y., Low B.C., Sivaraman J. Structural basis for the active site inhibition mechanism of human kidney-type glutaminase (KGA). Sci. Rep. 2014;4:3827. doi: 10.1038/srep03827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shukla K., Ferraris D.V., Thomas A.G., Stathis M., Duvall B., Delahanty G., Alt J., Rais R., Rojas C., Gao P., Xiang Y., Dang C.V., Slusher B.S., Tsukamoto T. Design, synthesis, and pharmacological evaluation of bis-2-(5-Phenylacetamido-1,2,4-Thiadiazol-2-Yl)ethyl Sulfide 3 (BPTES) analogs as glutaminase inhibitors. J. Med. Chem. 2012;55:10551–10563. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu W., Zhang C., Wu R., Sun Y., Levine A., Feng Z. Glutaminase 2, a Novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. USA. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xiang L., Xie G., Liu C., Zhou J., Chen J., Yu S., Li J., Pang X., Shi H., Liang H. Knock-down of Glutaminase 2 expression decreases glutathione, NADH, and sensitizes cervical cancer to ionizing radiation. Biochim. Biophys. Acta - Mol. Cell Res. 2013;1833:2996–3005. doi: 10.1016/j.bbamcr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 100.Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T., Zeller K.I., De Marzo A.M., Van Eyk J.E., Mendell J.T., Dang C.V. C-Myc Suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qie S., Chu C., Li W., Wang C., Sang N. ErbB2 activation upregulates glutaminase 1 expression which promotes breast cancer cell proliferation. J. Cell. Biochem. 2014;115:498–509. doi: 10.1002/jcb.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van den Heuvel A.P.J., Jing J., Wooster R.F., Bachman K.E. Analysis of glutamine dependency in non-small cell lung cancer. Cancer Biol. Ther. 2012;13:1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seltzer M.J., Bennett B.D., Joshi A.D., Gao P., Thomas A.G., Ferraris D.V., Tsukamoto T., Rojas C.J., Slusher B.S., Rabinowitz J.D., Dang C.V., Riggins G.J. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Newcomb R.W. 2002.
- 105.Wang J. Bin; Erickson, J.W.; Fuji, R.; Ramachandran, S.; Gao, P.; Dinavahi, R.; Wilson, K.F.; Ambrosio, A.L.B.; Dias, S.M.G.; Dang, C. V.; Cerione, R.A. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ferreira A.P.S., Cassago A., De Almeida Goncalves K., Dias M.M., Adamoski D., Ascenção C.F.R., Honorato R.V., De Oliveira J.F., Ferreira I.M., Fornezari C., Bettini J., Oliveira P.S.L., Leme A.F.P., Portugal R.V., Ambrosio A.L.B., Dias S.M.G. Active glutaminase c self-assembles into a supratetrameric oligomer that can be disrupted by an allosteric inhibitor. J. Biol. Chem. 2013;288:28009–28020. doi: 10.1074/jbc.M113.501346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Delabarre B., Gross S., Fang C., Gao Y., Jha A., Jiang F., Song J. J.; Wei, W.; Hurov, J.B. Full-Length Human Glutaminase in Complex with an Allosteric Inhibitor. Biochemistry. 2011;50:10764–10770. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- 108.Thangavelu K., Pan C.Q., Karlberg T., Balaji G., Uttamchandani M., Suresh V., Schuler H., Low B.C., Sivaraman J. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (kga) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc. Natl. Acad. Sci. USA. 2012;109:7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J., Chen L., Goyal B., Laidig G., Stanton T.F., Sjogren E.B. 2013.
- 111.Gross M.I., Demo S.D., Dennison J.B., Chen L., Chernov-Rogan T., Goyal B., Janes J.R., Laidig G.J., Lewis E.R., Li J., MacKinnon A.L., Parlati F., Rodriguez M.L.M., Shwonek P.J., Sjogren E.B., Stanton T.F., Wang T., Yang J., Zhao F., Bennett M.K. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 112.Study of the Glutaminase Inhibitor CB-839 in Solid Tumors https://clinicaltrials.gov/ct2/show/NCT02071862
- 113.Prajda N. Enzyme targets of antiglutamine agents in cancer chemotherapy. Adv. Enzyme Regul. 1985;24:207–223. doi: 10.1016/0065-2571(85)90077-9. [DOI] [PubMed] [Google Scholar]
- 114.Rosenfeld H., Roberts J. Enhancement of antitumor activity of glutamine antagonists 6-Diazo-5-Oxo-L-norleucine and acivicin in cell culture by glutaminase-asparaginase. Cancer Res. 1981;41:1324–1328. [PubMed] [Google Scholar]
- 115.Hidalgo M., Rodriguez G., Kuhn J.G., Brown T., Weiss G., MacGovren J.P., Von Hoff D.D., Rowinsky E.K. A Phase I and pharmacological study of the glutamine antagonist acivicin with the amino acid solution aminosyn in patients with advanced solid malignancies. Clin. Cancer Res. 1998;4:2763–2770. [PubMed] [Google Scholar]
- 116.Allen L., Meck R., Yunis A. The Inhibition of Gamma-Glutamyl Transpeptidase from Human Pancreatic Carcinoma Cells by (Alpha S,5S)-Alpha-Amino-3-Chloro-4,5-Dihydro-5-Isoxazoleacetic Acid (AT-125; NSC-163501). Res. Commun. Chem. Pathol. Pharmacol. 1980;27:175–182. [PubMed] [Google Scholar]
- 117.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rahman A., Smith F., Luc P-V., Woolley P. Phase I study and clinical pharmacology of 6-Diazo-5-Oxo-L-Norleucine (DON). Invest. New Drugs. 1985;3:369–374. doi: 10.1007/BF00170760. [DOI] [PubMed] [Google Scholar]
- 119.Earhart R.H., Amato D.J., Chang A.Y., Borden E.C., Shiraki M., Dowd M.E., Comis R.L., Davis T.E., Smith T.J., Phase I.I. Trial of 6-Diazo-5-Oxo-L-norleucine versus aclacinomycin-a in advanced sarcomas and mesotheliomas. Invest. New Drugs. 1990;8:113–119. doi: 10.1007/BF00216936. [DOI] [PubMed] [Google Scholar]
- 120.Yeh T.K., Kuo C.C., Lee Y.Z., Ke Y.Y., Chu K.F., Hsu H.Y., Chang H.Y., Liu Y.W., Song J.S., Yang C.W., Lin L.M., Sun M., Wu S.H., Kuo P.C., Shih C., Chen C.T., Tsou L.K., Lee S.J. Design, synthesis, and evaluation of thiazolidine-2,4-dione derivatives as a novel class of glutaminase inhibitors. J. Med. Chem. 2017;60:5599–5612. doi: 10.1021/acs.jmedchem.7b00282. [DOI] [PubMed] [Google Scholar]
- 121.Zimmermann S.C., Wolf E.F., Luu A., Thomas A.G., Stathis M., Poore B., Nguyen C., Le A., Rojas C., Slusher B.S., Tsukamoto T. Allosteric glutaminase inhibitors based on a 1,4-Di(5-Amino-1,3,4-Thiadiazol-2-Yl)butane Scaffold. ACS Med. Chem. Lett. 2016;7:520–524. doi: 10.1021/acsmedchemlett.6b00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Simpson N.E., Tryndyak V.P., Beland F.A., Pogribny I.P. An in Vitro investigation of metabolically sensitive biomarkers in breast cancer progression. Breast Cancer Res. Treat. 2012;133:959–968. doi: 10.1007/s10549-011-1871-x. [DOI] [PubMed] [Google Scholar]
- 123.Katt W.P., Ramachandran S., Erickson J.W., Cerione R.A. Dibenzophenanthridines as Inhibitors of Glutaminase C and Cancer Cell Proliferation. Mol. Cancer Ther. 2012;11:1269–1278. doi: 10.1158/1535-7163.MCT-11-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Simpson N.E., Tryndyak V.P., Pogribna M., Beland F.A., Pogribny I.P. Modifying metabolically sensitive histone marks by inhibiting glutamine metabolism affects gene expression and alters cancer cell phenotype. Epigenetics. 2012;7:1413–1420. doi: 10.4161/epi.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kahlert U.D., Cheng M., Koch K., Marchionni L., Fan X., Raabe E.H., Maciaczyk J., Glunde K., Eberhart C.G. Alterations in cellular metabolome after pharmacological inhibition of notch in glioblastoma cells. Int. J. Cancer. 2016;138:1246–1255. doi: 10.1002/ijc.29873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xie C., Jin J., Bao X., Zhan W-H., Han T-Y., Gan M., Zhang C., Wang J. Inhibition of mitochondrial glutaminase activity reverses acquired erlotinib resistance in non-small cell lung Cancer. Oncotarget. 2016;7:610–621. doi: 10.18632/oncotarget.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yuan L., Sheng X., Clark L.H., Zhang L., Guo H., Jones H.M., Willson A.K., Gehrig P.A., Zhou C., Bae-Jump V.L. Glutaminase inhibitor compound 968 inhibits cell proliferation and sensitizes paclitaxel in ovarian cancer. Am. J. Transl. Res. 2016;8:4265–4277. [PMC free article] [PubMed] [Google Scholar]
- 128.Zheng B., Yao Y., Liu Z., Deng L., Anglin J.L., Jiang H., Prasad B.V.V., Song Y. Crystallographic investigation and selective inhibition of mutant isocitrate dehydrogenase. ACS Med. Chem. Lett. 2013;4:542–546. doi: 10.1021/ml400036z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang B., Zhong C., Peng Y., Lai Z., Ding J. Molecular mechanisms of “off-on Switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 2010;20:1188–1200. doi: 10.1038/cr.2010.145. [DOI] [PubMed] [Google Scholar]
- 130.Wang F., Travins J., DeLaBarre B., Penard-Lacronique V., Schalm S., Hansen E., Straley K., Kernytsky A., Liu W., Gliser C., Yang H., Gross S., Artin E., Saada V., Mylonas E., Quivoron C., Popovici-Muller J., Saunders J.O., Salituro F.G., Yan S., Murray S., Wei W., Gao Y., Dang L., Dorsch M., Agresta S., Schenkein D.P., Biller S.A., Su S.M., de Botton S., Yen K.E. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 131.Dimitrov L., Hong C.S., Yang C., Zhuang Z., Heiss J.D. New Developments in the Pathogenesis and Therapeutic Targeting of the IDH1 Mutation in Glioma. Int. J. Med. Sci. 2015;12:201–213. doi: 10.7150/ijms.11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wu F., Jiang H., Zheng B., Kogiso M., Yao Y., Zhou C., Li X.N., Song Y. Inhibition of cancer-associated mutant isocitrate dehydrogenases by 2-thiohydantoin compounds. J. Med. Chem. 2015;58:6899–6908. doi: 10.1021/acs.jmedchem.5b00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Popovici-Muller J., Saunders J.O., Salituro F.G., Travins J.M., Yan S., Zhao F., Gross S., Dang L., Yen K.E., Yang H., Straley K.S., Jin S., Kunii K., Fantin V.R., Zhang S., Pan Q., Shi D., Biller S.A., Su S.M. Discovery of the first potent inhibitors of mutant idh1 that lower tumor 2-HG in Vivo. ACS Med. Chem. Lett. 2012;3:850–855. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rohle D., Popovici-Muller J., Palaskas N., Turcan S., Grommes C., Campos C., Tsoi J., Clark O., Oldrini B., Komisopoulou E., Kunii K., Pedraza A., Schalm S., Silverman L., Miller A., Wang F., Yang H., Chen Y., Kernytsky A., Rosenblum M.K., Liu W., Biller S.A., Su S.M., Brennan C.W., Chan T.A., Graeber T.G., Yen K.E., Mellinghoff I.K. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu Z., Yao Y., Kogiso M., Zheng B., Deng L., Qiu J.J., Dong S., Lv H., Gallo J.M., Li X-N., Song Y. Inhibition of cancer-associated mutant isocitrate dehydrogenases: Synthesis, structure-activity relationship, and selective antitumor activity. J. Med. Chem. 2014;57:8307–8318. doi: 10.1021/jm500660f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Law J.M., Stark S.C., Liu K., Liang N.E., Hussain M.M., Leiendecker M., Ito D., Verho O., Stern A.M., Johnston S.E., Zhang Y-L., Dunn G.P., Shamji A.F., Schreiber S.L. Discovery of 8-membered ring sulfonamides as inhibitors of oncogenic mutant isocitrate dehydrogenase 1. ACS Med. Chem. Lett. 2016;7:944–949. doi: 10.1021/acsmedchemlett.6b00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jones S., Ahmet J., Ayton K., Ball M., Cockerill M., Fairweather E., Hamilton N., Harper P., Hitchin J., Jordan A., Levy C., Lopez R., McKenzie E., Packer M., Plant D., Simpson I., Simpson P., Sinclair I., Somervaille T.C.P., Small H., Spencer G.J., Thomson G., Tonge M., Waddell I., Walsh J., Waszkowycz B., Wigglesworth M., Wiseman D.H., Ogilvie D. Discovery and optimization of allosteric inhibitors of mutant isocitrate dehydrogenase 1 (R132H IDH1) displaying activity in human acute myeloid leukemia cells. J. Med. Chem. 2016;59:11120–11137. doi: 10.1021/acs.jmedchem.6b01320. [DOI] [PubMed] [Google Scholar]
- 138.Paschka P., Schlenk R.F., Gaidzik V.I., Habdank M., Krönke J., Bullinger L., Späth D., Kayser S., Zucknick M., Götze K., Horst H-A., Germing U., Döhner H., Döhner K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with npm1 mutation without flt3 internal tandem duplication. J. Clin. Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 139.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K.W., Velculescu V.E., Vogelstein B., Bigner D.D. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I-M., Gallia G.L., Olivi A., McLendon R., Rasheed B.A., Keir S., Nikolskaya T., Nikolsky Y., Busam D.A., Tekleab H., Diaz L.A., Hartigan J., Smith D.R., Strausberg R.L., Marie S.K.N., Shinjo S.M.O., Yan H., Riggins G.J., Bigner D.D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 141.Cairns R.A., Mak T.W. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 142.Jones S., Ahmet J., Ayton K., Ball M., Cockerill M., Fairweather E., Hamilton N., Harper P., Hitchin J., Jordan A., Levy C., Lopez R., McKenzie E., Packer M., Plant D., Simpson I., Simpson P., Sinclair I., Somervaille T.C.P., Small H., Spencer G.J., Thomson G., Tonge M., Waddell I., Walsh J., Waszkowycz B., Wigglesworth M., Wiseman D.H., Ogilvie D. Discovery and optimization of allosteric inhibitors of mutant isocitrate dehydrogenase 1 (R132H IDH1) displaying activity in human acute myeloid leukemia cells. J. Med. Chem. 2016;59:11120–11137. doi: 10.1021/acs.jmedchem.6b01320. [DOI] [PubMed] [Google Scholar]
- 143.Law J.M., Stark S.C., Liu K., Liang N.E., Hussain M.M., Leiendecker M., Ito D., Verho O., Stern A.M., Johnston S.E., Zhang Y-L., Dunn G.P., Shamji A.F., Schreiber S.L. Discovery of 8-membered ring sulfonamides as inhibitors of oncogenic mutant isocitrate dehydrogenase 1. ACS Med. Chem. Lett. 2016;7:944–949. doi: 10.1021/acsmedchemlett.6b00264. [DOI] [PMC free article] [PubMed] [Google Scholar]