Abstract
Cognitive flexibility is a critical component of executive function and is strongly influenced by genetic factors. We conducted a genome-wide association study of cognitive flexibility (as measured by perseverative errors on the Wisconsin Card Sorting Test) in two sets of African American (AA) and European American (EA) subjects (Yale-Penn-1: 1,411 AAs/949 EAs; Yale-Penn-2: 1,178 AAs/1,335 EAs). We examined the association of cognitive flexibility with genotyped or imputed SNPs across the genome. In AAs, two correlated common SNPs (rs7165213/rs35633795) in the downstream region of the noncoding gene LOC101927286 on chromosome 15 showed genome-wide significant (GWS) associations with cognitive flexibility (Yale-Penn-1: P = 6.0×10−9/1.3×10−8; Yale-Penn-2: P = 0.029/0.010; meta-analysis: P = 4.2×10−7/1.0×10−7) in the same effect direction. In EAs, no GWS associations were observed. Enriched gene sets identified by DEPICT analysis of the top SNPs (Pmeta-analysis < 10−5) included the signalosome and ubiquitin specific peptidase 9, X-linked (USP9X) subnetwork in AAs, and abnormal frontal and occipital bone morphology in EAs. We also performed polygenic risk score (PRS) analysis to examine the genetic correlation of cognition-proxy phenotypes (general cognitive function, education attainment, childhood intelligence, and infant head circumference) and cognitive flexibility in EAs. The PRS derived from general cognitive function-associated SNPs was significantly associated with cognitive flexibility. Non-genetic factors (age, education, sex, and tobacco recency) also exerted significant effects on cognitive flexibility. Our study demonstrates that both genetic and non-genetic factors impact cognitive flexibility, and variants in genes involved in protein degradation and brain development may contribute to population variation in cognitive function.
Keywords: cognitive flexibility, Wisconsin Card Sorting Test, genome-wide association study, enrichment analysis, polygenic risk score
1 | INTRODUCTION
Cognitive flexibility is a vital component of executive function facilitating critical thinking and self-regulation. Inter-individual variability in executive function is largely influenced by genetic factors. Twin studies have demonstrated that genetic factors contributed to 59%, 63%, 29%, and 31% of the variance in working memory, verbal fluency, response inhibition, and cognitive flexibility (four specific measures of executive function), respectively (Lee et al., 2012). The correlation of these cognitive processes suggests the presence of shared genetic factors among them. Additionally, specific genetic factors may influence a particular executive function such as cognitive flexibility.
Cognitive flexibility is the ability to modify thinking and behavior in response to changing environmental conditions (Leber et al., 2008). Functional magnetic resonance imaging (fMRI) studies have shown that certain brain regions (such as the prefrontal cortex) are activated during cognitive flexibility tasks (Leber et al., 2008), i.e., specific brain regions are the essential determinants of cognitive flexibility or set-shifting capacity. An impairment in cognitive flexibility has been noted in a number of neuropsychiatric disorders, such as Alzheimer’s disease (Tatsuoka et al., 2013), obsessive-compulsive disorder (Gruner and Pittenger, 2017), schizophrenia (Minassian et al., 2005), autism (Romero-Munguia, 2008), attention/deficit hyperactivity disorder (Mary et al., 2016), and anorexia nervosa (Zastrow et al., 2009). Unraveling the genetic basis of cognitive flexibility can improve our understanding of the genetic mechanisms of these disorders and facilitate their early diagnosis and treatment.
Cognition is a complex genetic trait. Genome-wide association studies (GWAS) of cognitive functions or cognition-proxy phenotypes have been conducted. Two GWAS demonstrated genome-wide significant (GWS) associations between general cognitive function (often referred to as “general intelligence”) and variants in four Alzheimer’s disease-associated genes [the translocase of outer mitochondrial membrane 40 gene (TOMM40), the apolipoprotein E gene (APOE), the ATP binding cassette subfamily G member 1 gene (ABCG1), and the myocyte enhancer factor 2C gene (MEF2C)] (Davies et al., 2015) as well as the centromere protein O gene (CENPO) and the noncoding RNA gene LOC105378853 (Trampush et al., 2017). Two other GWAS that examined adult (Davies et al., 2011) and childhood (Benyamin et al., 2014) intelligence failed to detect any GWS SNPs. A GWAS on information processing speed identified a significant variant in the cell adhesion molecule 2 gene (CADM2) (Ibrahim-Verbaas et al., 2016), which is involved in glutamate signaling, gamma-aminobutyric acid (GABA) transport, and neuron cell-cell adhesion. Several GWAS on educational attainment (considered as a partial proxy phenotype for cognitive ability) have also been conducted. Rietveld et al. performed a GWAS on education attainment in a large sample and identified three genome-wide significant SNPs [rs9320913 near LOC100129158, rs11584700 near the leucine rich repeat neuronal 2 gene (LRRN2), and rs4851266 near LOC150577] (Rietveld et al., 2013). They also performed a two-stage study using the same proxy-phenotype approach and identified three SNPs [rs1487441 near LOC100129158, rs7923609 near the Jumonji domain containing 1C gene (JMJD1C), and rs2721173 near the leucine rich repeat containing 14 gene (LRRC14)] that were significantly associated with cognitive performance (assessed by total word recall and total mental status) (Rietveld et al., 2014). Davies et al. investigated the genetic contribution to variation in cognitive functions (verbal-numerical reasoning, memory, and reaction time) and educational attainment. They reported GWS associations in 20 genomic regions, including the ataxin 2 gene (ATXN2), the cytochrome P450 family 2 subfamily D member 6 gene (CYP2D6), the amyloid beta precursor protein binding family A member 1 gene (APBA1), and CADM2 (Davies et al., 2016). Additionally, a large GWAS conducted by Okbay et al. identified 74 loci associated with educational attainment (Okbay et al., 2016).
In the present study, we performed both a SNP- and a gene-based GWAS on cognitive flexibility (evaluated by the WCST) in African Americans (AAs; n = 2,589) and European Americans (EAs; n = 2,284). We also used a polygenic risk score (PRS) analysis to investigate the genetic correlation of cognitive flexibility with cognition-related phenotypes, including general cognitive function, educational attainment, childhood intelligence, and infant head circumference.
2 | METHODS
2.1 | Subjects
This study included 4,873 subjects, a subset of a larger sample recruited for studies of the genetics of substance dependence (Gelernter et al., 2015; Sherva et al., 2016). They were recruited through advertisements at the Yale University School of Medicine (New Haven, Connecticut, USA) (n = 2,789), the University of Connecticut Health Center (Farmington, Connecticut, USA) (n = 1,673), or the University of Pennsylvania Perelman School of Medicine (Philadelphia, Pennsylvania, USA) (n = 411). The subjects were divided into two groups [Yale-Penn-1: 1,411 African Americans (AAs)/949 European Americans (EAs); Yale-Penn-2: 1,178 AAs/1,335 EAs] genotyped using two different microarrays (Table 1). Subjects were interviewed using the Semi-Structured Assessment for Drug Dependence and Alcoholism (Pierucci-Lagha et al., 2005). Subjects gave written informed consent as approved by the institutional review board at each site. Information on sex, age, years of education, and tobacco recency was collected at the baseline interview. Characteristics of the participants are presented in Table 1.
TABLE 1.
Demographic characteristics of the sample
| Yale-Penn-1 (n=2,589)a
|
Yale-Penn-2 (n=2,284)b
|
|||
|---|---|---|---|---|
| AAs | EAs | AAs | EAs | |
| Number of subjects (n=4,873) | 1,411 | 949 | 1,178 | 1,335 |
| Males (%) | 791 (56.0%) | 559 (58.9%) | 739 (62.7%) | 818 (61.3%) |
| Age (years ± SD) | 42 (± 9) | 38 (± 11) | 41 (±11) | 39 (± 13) |
| Education (years ± SD) | 12 (±3) | 12 (±2) | 12 (±2) | 13 (±3) |
| Recency of tobacco use | ||||
| ≤ 2 weeks (current user) | 1,130 (80.1%) | 803 (84.6%) | 799 (67.8%) | 848 (63.5%) |
| 2 – 4 weeks | 5 (0.4%) | 8 (0.8%) | 10 (0.8%) | 26 (1.9%) |
| 1 – 6 months | 25 (1.8%) | 13 (1.4%) | 31 (2.6%) | 49 (3.7%) |
| 6 months – 1 year | 18 (1.2%) | 5 (0.5%) | 13 (1.1%) | 27 (2.0%) |
| > 1 Year | 233 (16.5%) | 120 (12.6%) | 325 (27.6%) | 385 (28.8%) |
Subjects in Yale-Penn-1 were genotyped on the Illumina HumanOmni1-Quad v1.0 microarray.
Subjects in Yale-Penn-2 were genotyped on the Illumina HumanCore Exome BeadChip.
AAs: African Americans; EAs: European Americans.
2.2 | Cognitive flexibility assessment and correlation with non-genetic factors
We used the 128-card computerized version of the Wisconsin Card Sorting Test (WCST) (Heaton, 1999) to evaluate subjects’ cognitive flexibility. Subjects were required to match response cards to four stimulus cards on three dimensions (color, form, or number) by pressing one of four number keys (1–4) on the computer keyboard. They needed to determine the correct sorting principle and change that principle when the test shifts it. The WCST index perseverative error has been demonstrated to be the most useful outcome measure in assessing executive function (Greve et al., 2005). We examined the influence of continuous variables (age and years of education) on perseverative errors using Pearson’s correlation analysis and the impact of dichotomous variables [sex and tobacco recency (≤ 2 vs. > 2 weeks)] on perseverative errors using Student’s t-test.
2.3 | DNA preparation and SNP genotyping
DNA was extracted from lymphoblastoid cell lines or directly from the participants’ blood or saliva. The Yale-Penn-1 sample (1,411 AAs/949 EAs) was genotyped using the Illumina HumanOmni1-Quad v1.0 microarray (for 988,306 autosomal SNPs) (Illumina, San Diego, California, USA) at the Center for Inherited Disease Research (Baltimore, Maryland, USA) or the Yale Center for Genomic Analysis (West Haven, Connecticut, USA). The Yale-Penn-2 sample (1,178 AAs/1,335 EAs) was genotyped at the West Haven VA Medical Center using the Illumina HumanCoreExome-12 v1.0 BeadChip (Illumina, San Diego, California, USA) for genotyping 538,448 SNPs (including 263,929 exome-focused SNPs and 274,519 tagging SNPs which allow genome-wide imputation). Genotypes obtained from both genotyping platforms were called using GenomeStudio software V2011.1 and genotype module V1.8.4 (Illumina, San Diego, CA, USA). SNPs were excluded if their genotyping or subject call rates were <98%.
2.4 | Population stratification and genotype imputation
To confirm the self-reported races of subjects, we compared the GWAS data from all subjects with genotypes from the 1000 Genomes reference panel (http://www.1000genomes.org/), which contains phased haplotypes for 1,092 individuals of various ancestries: 379 of European descent, 286 of Asian descent, 181 admixed Americans, and 246 of African descent. We conducted principal component (PC) analysis to explore population structure using Eigensoft (Price et al., 2006). After pruning GWAS SNPs for linkage disequilibrium (LD) with r2 > 0.8, 145,472 SNPs common to our GWAS and the 1000 Genomes reference panel datasets were used to characterize the underlying genetic architecture of our samples. The first three PCs were used to distinguish AAs and EAs, and outliers were excluded. A second set of PC analyses within AAs and EAs was conducted, from which the first five PCs were used in all subsequent analyses to correct for residual population stratification. To account for genetic relationships between subjects, we calculated the pair-wise identity by descent using PLINK (Purcell et al., 2007).
Genotype imputation of our GWAS samples was performed using the program IMPUTE2 (Howie et al., 2009) with the 1000 Genomes phase 1 data set as the reference. Genotypes of AAs and EAs were imputed separately. The imputed genotype was used in subsequent genetic association analyses. SNPs with Hardy-Weinberg equilibrium P-value < 10−5, minor allele frequency < 5%, or imputation accuracy < 0.8 were excluded. In the Yale-Penn-1 sample, 8,933,065 SNPs in AAs and 6,489,050 SNPs in EAs were included in association analyses; in the Yale-Penn-2 sample, 6,774,960 SNPs in AAs and 5,244,066 SNPs in EAs were considered.
2.5 | SNP-based association analysis and meta-analysis
We performed SNP-based association analyses using a linear random effects model embedded in a generalized estimating equation (GEE) (Ziegler et al., 1998) to correct for correlations among related individuals. The R package GWAF (Chen and Yang, 2010) was used to test the association of both genotyped and imputed SNPs with WCST index %PE under an additive model with age, sex, years of education, tobacco recency (≤ 2 weeks vs. > 2 weeks), and the first five PCs as covariates. Because the Shapiro-Wilks tests (Shapiro and Wilk, 1965) demonstrated that the distribution of %PE departed from normality, we used the Box-Cox Transformation (Box and Cox, 1964) to normalize %PE before data analysis (Supplementary Figure S1). Regional genetic associations were plotted using LocusZoom (Pruim et al., 2010).
The association results for 9,039,803 SNPs in each of the two AA samples (Yale-Penn-1 and Yale-Penn-2) and 6,550,705 SNPs in each of the two EA samples (Yale-Penn-1 and Yale-Penn-2) were meta-analyzed within each population using the inverse variance method implemented in the program METAL (Willer et al., 2010). To explore the association and the heterogeneity across the two ancestral groups, we performed meta-analysis of 9,722,417 SNPs in AAs and EAs. Two heterogeneity measurements, Cochran’s Q statistic and I2, were estimated across the groups. Top SNP signals (P < 10−5) obtained from meta-AA, meta-EA, or meta-AA+EA analyses were clumped by LD based on the 1000 Genomes dataset using PLINK (with r2 < 0.2 and SNPs within a 200-kb window). The 1000 Genomes dataset from the African populations were used as the reference for meta-AA SNP clumping, while the 1000 Genomes dataset from the European populations were used as the reference for both meta-EA and meta-AA+EA SNP clumping.
2.6 | SNP functional prediction
The program PROMO (Messeguer et al., 2002) was applied to predict whether GWS SNPs were located in transcription factor binding sites (TFBS) as defined in the TRANSFAC database (Matys et al., 2006). The Brain eQTL Almanac (http://www.braineac.org) was queried to see whether GWS SNPs were eQTLs for genes expressed in 10 different brain regions. Considering the important role of the frontal cortex (Neubert et al., 2014) and the hippocampus (Burghardt et al., 2012) in cognitive processes, we focused on eQTLs influencing gene expression in these two brain regions.
2.7 | Interpretation of GWAS findings using predicted gene functions
We used DEPICT (Data-driven Expression-Prioritized Integration for Complex Traits) (Pers et al., 2015) to predict whether genes located in associated regions were enriched for reconstituted gene sets (i.e., known pathways and protein-protein interaction subnetworks). We also used DEPICT to identify the type of tissues/cells in which genes located in associated regions are highly expressed. We ran DEPICT for gene set and tissue/cell expression enrichment analysis using all independent SNPs (clumped in 500-kb windows and with LD r2 < 0.1) with Pmeta ≤ 10−5 in AA and EA meta-analyses. Nominal P-values and false discovery rates (FDR) were calculated for each gene set and tissue/cell type.
2.8 | Gene-based association analysis
We performed gene-based tests using the program VEGAS2 (Mishra and Macgregor, 2015), which combines P-values of a set of SNPs within a gene region (from 10 kb upstream of the 5′UTR to 10 kb downstream of the 3′UTR) to obtain an overall P value for the association of the entire gene with Box-Cox normalized %PE. Both gene length and SNP LD were taken into consideration. It was conducted in AAs and EAs separately, with P-values obtained from meta-AA and meta-EA analyses. The GWS levels for gene-based tests were set to 2.18×10−6 (0.05/22,888 autosomal genes) for AAs and 2.21×10−6 (0.05/22,659 autosomal genes) for EAs.
2.9 | Polygenic risk score analysis
We investigated whether polygenic risk scores (PRS) for cognition-related traits correlated with cognitive flexibility. Summary statistics of GWAS on general cognitive function (Trampush et al., 2017), educational attainment (Okbay et al., 2016), childhood intelligence (Benyamin et al., 2014), and infant head circumference (Taal et al., 2012) of subjects of European descent were downloaded from the websites of the Social Science Genetic Association Consortium (http://www.thessgac.org/data) and the Cognitive Genomics Consortium (Trampush et al., 2017). SNPs associated with cognition-related phenotypes were clumped by LD with r2 < 0.2 in a 200-kb window. The PRS was calculated as previously described (Reeves et al., 2010). Five P-value thresholds (PT = 0.00001, 0.0001, 0.001, 0.01, and 0.05) were considered. We used the linear regression model embedded in GEE (Ziegler et al., 1998) to examine the association between Box-Cox-transformed %PE and the PRS. The WCST index %PE was the dependent variable, the PRS (rescaled to range from 0 to 2) was the independent variable, and age, sex, education year, tobacco recency, and the first five PCs were covariates. The PRS analysis was performed in the two sets of EA samples, respectively, followed by meta-analysis. An association with P < 0.0025 (0.05/4*5 to correct for the testing of four PRS analyses at five PT values) was considered to be significant.
3 | RESULTS
3.1 | Effects of non-genetic factors on cognitive flexibility
The influence of non-genetic factors on cognitive flexibility as measured by percentages of perseverative errors (Box-Cox transformed %PE) on the WCST is presented in Table 2. Age was significantly correlated with perseverative errors in EAs (r = 0.16, P < 0.001) (i.e., older age predicted worse cognitive flexibility) but not in AAs. Years of education was negatively correlated with perseverative errors in both AAs (r = −0.09, P < 0.001) and EAs (r = −0.21, P < 0.001) (i.e., the more education, the greater the cognitive flexibility). Male EAs made significantly fewer perseverative errors (t = −2.32, P = 0.021) than female EAs. There were no significant differences in WCST performance between male and female AAs. Recent tobacco use was associated with greater %PE in both AAs (t = −5.70, P < 0.001) and EAs (%PE: t = −9.97, P < 0.001).
TABLE 2.
Effects of non-genetic factors on cognitive Inflexibility
| %PE in AAsa | %PE in EAsb | |
|---|---|---|
| Pearson correlation analysis: | ||
| Age | r = −0.03, P = 0.144 | r = 0.16, P < 0.001 |
| Years of education | r = −0.09, P < 0.001 | r = −0.21, P < 0.001 |
|
| ||
| Student’s t-test: | ||
| Sex | ||
| Male (Mean ± SD) | 2.25 ± 0.43 | 1.51 ± 0.23 |
| Female (Mean ± SD) | 2.26 ± 0.44 | 1.53 ± 0.24 |
| t = −0.80, P = 0.422 | t = −2.32, P = 0.021 | |
| Tobacco recency | ||
| ≤ 2 weeks | 2.28 ± 0.44 | 1.55 ± 0.24 |
| > 2 weeks | 2.17 ± 0.41 | 1.44 ± 0.21 |
| t = −5.70, P < 0.001 | t = −9.97, P < 0.001 | |
%PE: The percentage of perseverative errors (as measured by the WCST) was normalized using the Box-Cox transformation.
2,589 African Americans (AAs; 1,411 AAs from Yale-Penn-1 and 1,178 AAs from Yale-Penn-2).
2,284 European Americans (EAs; 949 EAs from Yale-Penn-1 and 1,335 EAs from Yale-Penn-2).
3.2 | SNPs associated with cognitive flexibility
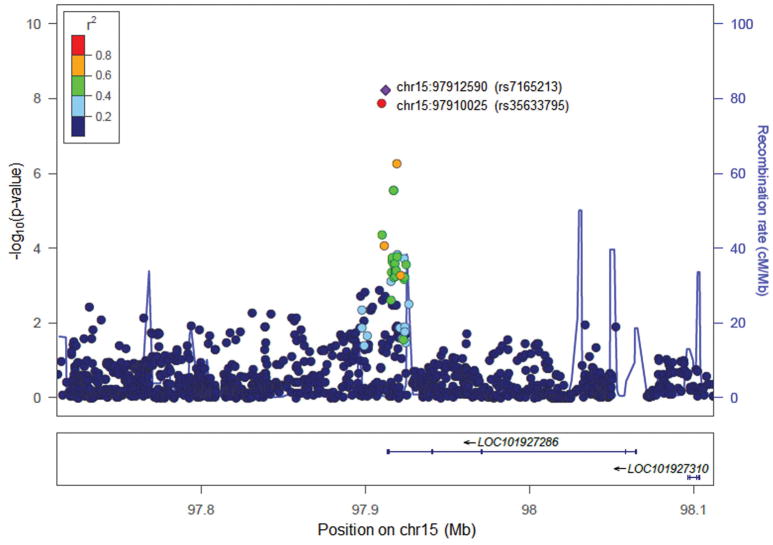
The SNP-based GWAS of cognitive flexibility (denoted as Box-Cox-transformed %PE) was performed in AAs and EAs separately, followed by meta-analysis. GWAS results for AAs are shown in Figure 1 and Supplementary Figure S2. Rs7165213, which is 1,011 bp downstream of the noncoding RNA gene LOC101927286 on chromosome 15, showed a GWS association with %PE in the Yale-Penn-1 AA sample (β = 0.08, P = 6.0×10−9) and a nominally significant association with %PE in the Yale-Penn-2 AA sample (β = 0.02, P = 0.029). This finding was supported by a GWS association with %PE at rs35633795 [2,565 bp from rs7165213 and in high LD with rs7165213 (r2 = 0.907)] in the Yale-Penn-1 AA sample (β = 0.08, P = 1.3×10−8) and a nominally significant association with %PE in the Yale-Penn-2 AA sample (β = 0.02, P = 0.010). Three additional common SNPs that are 4,843–6,856 bp from rs7165213 at the 3′ end of LOC101927286 showed non-GWS associations with %PE (rs12592141: β = −0.07, P = 5.6×10−7; rs201950623: β = 0.07, P = 2.8×10−6; rs75006937: β = 0.07, P = 2.8×10−6). There were no GWS findings obtained in the two sets of EA samples (Supplementary Figure S3). Meta-analyses of AA, EA, or all AA+EA samples did not reveal GWS associations with %PE. Nevertheless, a meta-analysis in the two sets of AA samples showed that the associations of the two top SNPs identified in the Yale-Penn-1 AA sample with cognitive flexibility were close to significance at the GWS level (rs7165213: β = 0.08, Pmet = 4.2×10−7; rs35633795: β = 0.08, Pmet = 1.0×10−7) (Supplementary Figure S4). However, when P-values and the direction of effects were applied in the meta-analysis, the results reached GWS (rs7165213: Pmet = 7.9×10−9; rs35633795: Pmet = 2.9×10−9), although this is not the preferred method for GWAS meta-analysis. There was little evidence for inflation (λ = 1.01–1.03) of P-values from either SNP-based GWAS or meta-analyses in the AA, EA, or AA+EA samples (Supplementary Figure S5).
FIGURE 1.
Regional Manhattan plot of SNPs rs7165213 and rs35633795 in association with perseverative errors.
Regional association (Manhattan) plot shows SNP-based GWAS results for association of perseverative errors and single nucleotide polymorphisms (SNPs) mapped to the LOC101927286 region on chromosome 15 in African Americans (AAs). The SNPs are color coded according to r2 with the most significant SNP rs7165213 shown in purple. The light blue line and right Y-axis display the observed recombination rate in the HapMap YRI sample.
3.3 | Annotated function of GWS SNPs
Functional prediction of the above two GWS SNPs (rs7165213 and rs35633795) by PROMO indicated that they were potentially located in transcription factor binding sites (TFBS) (rs7165213: HNF1B and HNF1C; rs35633795: GR-beta and XBP1). The BRAINEAC eQTL analysis showed that rs7165213 and rs9301456 were putative eQTLs for gene expression in the frontal cortex and/or the hippocampus. Rs7165213 was shown to be a cis-eQTL for the expression of a proximal gene “family with sequence similarity 169 member B” (FAM169B) in the frontal cortex (Pnominal = 4.3×10−3), while SNP rs35633795 was a likely cis-eQTL for the expression of FAM169B in the frontal cortex (Pnominal = 4.2×10−3) and the non-coding gene LOC145945 in the hippocampus (Pnominal = 9.4×10−3). However, the results did not withstand correction for multiple testing.
3.4 | DEPICT enrichment analysis results
We used DEPICT to interpret GWAS findings based on predicted gene functions. In AAs, 197 independent SNPs (Pmeta < 10−5) identified from the meta-AA analysis yielded 38 unique and non-overlapping loci mapped to 41 unique genes. The enriched gene sets included “signalosome” and “ubiquitin specific peptidase 9, X-linked (USP9X) subnetwork” and the enriched tissue/cell types included osteoblasts and skin (Supplementary Table S1). In EAs, 27 independent SNPs (Pmeta < 10−5) identified from the meta-EA analysis yielded 12 unique and non-overlapping loci mapped to 15 unique genes. The enriched gene sets included “abnormal frontal bone morphology” and “abnormal occipital bone morphology” and the enriched tissue/cell types included exocrine glands and prostate. The enrichment of gene set “abnormal frontal bone morphology” survived multiple testing correction (Supplementary Table S2).
3.5 | Gene-based association analysis results
Following the per-SNP GWAS, we performed gene-based association tests to identify genes that harbor more cognitive flexibility-associated SNPs (with small P-values) than by chance. Genes with gene-level P-values less than 10−4 are listed in Supplementary Table S3. In AAs, we identified four perseverative error-associated genes (P < 10−4): the CPEB2 antisense RNA 1 (head To head) gene (CPEB2-AS1), the cytoplasmic polyadenylation element binding protein 2 gene (CPEB2), the cysteine rich secretory protein LCCL domain containing 1 gene (CRISPLD1), and the transforming growth factor beta 1 induced transcript 1 gene (TGFB1I1). In EAs, we identified five perseverative errors-associated genes (P < 10−4): the gamma-aminobutyric acid type A receptor pi subunit gene (GABRP), the noncoding RNA gene LOC102031319, the enhancer of polycomb homolog 1 gene (EPC1), the microRNA 548c gene (MIR548C), and the microRNA 548z gene (MIR548Z). However, none of these genes survived correction for multiple testing.
3.6 | Polygenic risk scores (PRS)
We performed PRS analysis to examine genetic effects shared by cognition-proxy phenotypes and cognitive flexibility. The PRS analysis results for five levels of PT (0.00001, 0.0001, 0.001, 0.01, and 0.05) from the EA samples are summarized in Supplementary Table S4. The results with P ≤ 0.050/4*5 = 0.002 (correcting for four PRS analyses with five levels of PT) were considered significant. The PRS of general cognitive function was significantly associated with perseverative errors (PT = 0.05: β = −0.03, Pmeta = 0.002; PT = 0.01: β = −0.03, P meta= 0.001). The PRS of years of education (PT = 0.05: β = −0.02, Pmeta = 0.025; PT = 0.01: β = −0.02, Pmeta = 0.027; PT = 0.001: β = −0.02, Pmeta = 0.044) and the PRS of infant head circumference (PT = 0.05: β = −0.03, Pmeta = 0.005) showed nominally significant associations with perseverative errors. Childhood intelligence did not show a significant genetic correlation with perseverative errors.
We performed an additional PRS analysis using the summary statistics from a GWAS on educational attainment (Okbay et al., 2016) to predict the years of education in our two EA samples. As shown in Supplementary Table S5, the PRS of educational attainment from Okbay et al. (2016) was significantly associated with years of education in our two sets of EA subjects (PT = 0.05: β = 1.83, Pmeta = 5.2×10−23; PT = 0.01: β = 1.69, Pmeta = 1.3×10−20; PT = 0.001: β = 1.49, Pmeta = 7.4×10−19; PT = 0.0001: β = 1.37, Pmeta = 3.3×10−15; PT = 0.00001: β = 1.37, Pmeta = 7.8×10−13).
4 | DISCUSSION
In the present study, we identified cognitive flexibility-associated genetic loci using SNP- and gene-based GWAS as well as PRS analysis. We also explored the correlation of non-genetic factors with cognitive flexibility. Our study identified both genetic and non-genetic factors that influence cognitive flexibility, measured using perseverative errors on the WCST.
Through SNP-based GWAS, we identified cognitive flexibility-associated SNPs in AAs. Two nearby and tightly linked SNPs (rs7165213 and rs35633796), located downstream of a non-coding RNA (ncRNA) gene LOC101927286, were significantly associated with perseverative errors in AAs (Figure 1). Although ncRNAs are often non-functional, some of them can regulate gene expression at the level of transcription or post-transcription (Huttenhofer et al., 2005). Because multiple SNPs in the 3′ end of LOC101927286 were associated with cognitive flexibility at GWS or sub-GWS levels (Figure 1), LOC101927286 or other genes in this region may regulate the expression of cognition-relevant genes or participate in biological pathways underlying variation in cognitive functioning.
It is a challenge to unravel the function of GWAS-identified disease-associated genetic variants because their effect size may be small and they are often located in noncoding regions, such as the two cognitive flexibility-associated SNPs identified here (rs7165213 and rs35633796). We used DEPICT to show Gene Ontology, KEGG, and Reactome pathways that are enriched for genes tagged by cognitive flexibility-associated SNPs. Enriched gene sets or pathways in AAs included “signalosome”, which is a protein complex with isopeptidase activity, and “USP9X subnetwork”, which consists of proteins that are similar to ubiquitin-specific proteases. Enriched gene sets or pathways in EAs included “abnormal frontal bone morphology” and “abnormal occipital bone morphology”. These findings suggest that genes carrying cognitive flexibility-associated variants may be involved in protein degradation or brain development. In addition, we examined whether cognition-related genes reported in previous GWAS (see Introduction) harbored SNPs that were associated with cognitive flexibility in our samples. As shown in Supplementary Table S6, three previously GWAS-identified genes (CADM2, LOC105378853, and ABCG1) had more than 20 SNPs with meta-analysis P-values less than 0.05 in our AA, EA, or AA+EA samples. However, none of these genes harbored SNPs that were associated with cognitive flexibility at the GWS level.
We then performed gene-based GWAS to evaluate the effect of significant SNPs in a gene region on cognitive flexibility. We identified novel potential cognitive flexibility-associated genes, but none reached GWS. The top genes (with gene-level P-values < 10−4) associated with cognitive flexibility were CPEB2-AS1, CPEB2, CRISPLD1, and TGFB1I1 in AAs as well as GABRP, LOC102031319, EPC1, MIR548C, and MIR548Z in EAs (Supplementary Table S3). The majority of these potential cognitive flexibility-associated genes are involved in the regulation of gene expression or neurotransmission, but further research to validate this hypothesis is warranted.
Cognitive flexibility may be pleiotropic with other components of cognition. We used the PRS method to evaluate genetic variants that could underlie the phenotypic link between cognitive flexibility and other cognitive phenotypes. General cognitive function encompasses a number of cognitive abilities. Educational attainment is strongly associated with levels of cognitive function (Wilson et al., 2009). Childhood intelligence is a significant predictor of various health outcomes such as educational attainment, income, and lifespan (Benyamin et al., 2014). Infant head circumference has been correlated with developmental cognition (Gale et al., 2006). Because GWAS of general cognitive function (Trampush et al., 2017), educational attainment (Okbay et al., 2016), childhood intelligence (Benyamin et al., 2014), and infant head circumference (Taal et al., 2012) were performed in European populations, we performed the PRS analysis only in our EA sample (as the replication sample). The PRS derived from the above proxy phenotype-associated SNPs was associated with cognitive flexibility (represented by perseverative errors), but only the results from the general cognitive function PRS survived correction for multiple testing. This finding is consistent with a pleiotropic effect of genetic variants on both cognitive flexibility and general cognitive function. We did not perform the PRS analysis in our AA sample because the GWAS data for cognition-proxy phenotypes was not available for AAs.
Apart from heritable genetic factors, brain development and function are also susceptible to perturbation by environmental factors. We observed significant effects of non-genetic factors (age, education, sex, and tobacco recency) on cognitive flexibility in AAs and EAs. The consistent findings in both populations were that education attainment was negatively but recent tobacco use was positively correlated with perseverative errors.
This study is limited in several respects. First, our findings should be verified in a larger independent sample. If WCST data from other research groups becomes available, we could also meta-analyze results from multiple samples. Second, subjects for the present study were mainly recruited for genetic studies of substance dependence rather than from a population. The comparability of our sample on cognitive measures with other samples is nevertheless supported by the PRS analysis regarding years of education. Third, the identified SNPs with GWS associations with cognitive flexibility are located in noncoding region. The function of these noncoding SNPs in regulating gene expression needs to be validated. In addition, we need to explore the interactive effects of genetic and environmental factors on cognitive flexibility. Even though two groups of subjects both carry genetic risk factors for cognitive flexibility, significant associations may only be shown in one group of subjects but not in another if relevant environmental factors differ. The different percentages of recent AA smokers in Yale-Penn-1 (80.1%) and Yale-Penn-2 (67.8%) samples (refer to Table 1) may partially explain the different findings in these two sets of AA samples, i.e., GWS signals were observed in Yale-Penn-1 AAs but not in Yale-Penn-2 AAs (although the result was nominally significant); we know smoking recency is a substantive environmental factor. Similarly, the reason that the meta-analysis of the two AA datasets (Yale-Penn-1 and -2 AAs) did not generate a more significant result may be partially due to the interaction of genetic variants and environmental factors including tobacco recency. Thus, further gene-environment interaction analysis is warranted.
To summarize, the present study demonstrated an influence of both genetic and non-genetic factors on cognitive flexibility, and identified specific risk alleles important in modulating cognitive flexibility in AAs. It suggests that genes with cognitive flexibility-associated variants could be potential targets for the treatment of cognitive dysfunction. To ameliorate cognitive deficits in patients with psychiatric disorders, pharmacogenetic, psychological, and behavioral approaches could potentially be guided by both genetic and environmental predictors of cognitive flexibility.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health grants R21 AA023068 (HZ), R01 AA025080 (HZ), R01 AA017535 (JG), R01 DA012690 (JG), and P50 AA12870 (JG). Dr. Kranzler has been an advisory board member, consultant, or CME speaker for Alkermes, Indivior and Lundbeck. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative (ACTIVE), which was supported in the past three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor and Amygdala Neurosciences.
We appreciate the work in recruitment and assessment provided at the Yale University School of Medicine and the APT Foundation, the University of Connecticut Health Center, and the University of Pennsylvania Perelman School of Medicine. Genotyping services were provided by the Center for Inherited Disease Research (CIDR) and the Yale University Center for Genome Analysis (YCGA). The CIDR is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle and Christa Robinson at Yale University School of Medicine for their excellent technical assistance, to the SSADDA interviewers led by Yari Nunez at the APT Foundation, and to John Farrell at Boston University School of Medicine for database management assistance.
Footnotes
DISCLOSURE: The other authors report no biomedical financial interests or potential conflicts of interest.
References
- Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion MJ, … Visscher PM. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Molecular psychiatry. 2014;19:253–258. doi: 10.1038/mp.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box GEP, Cox DR. An analysis of transformations. J R Stat Soc Series B. 1964;26:211–252. [Google Scholar]
- Burghardt NS, Park EH, Hen R, Fenton AA. Adult-born hippocampal neurons promote cognitive flexibility in mice. Hippocampus. 2012;22:1795–1808. doi: 10.1002/hipo.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, … Deary IJ. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949) Molecular psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, … Deary IJ. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151) Molecular psychiatry. 2016;21:758–767. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, … Deary IJ. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale CR, O’Callaghan FJ, Bredow M, Martyn CN Avon Longitudinal Study of P, Children Study T. The influence of head growth in fetal life, infancy, and childhood on intelligence at the ages of 4 and 8 years. Pediatrics. 2006;118:1486–1492. doi: 10.1542/peds.2005-2629. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, … Farrer LA. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77:493–503. doi: 10.1016/j.biopsych.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve KW, Stickle TR, Love JM, Bianchini KJ, Stanford MS. Latent structure of the Wisconsin Card Sorting Test: a confirmatory factor analytic study. Arch Clin Neuropsychol. 2005;20:355–364. doi: 10.1016/j.acn.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Gruner P, Pittenger C. Cognitive inflexibility in Obsessive-Compulsive Disorder. Neuroscience. 2017;345:243–255. doi: 10.1016/j.neuroscience.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK. Wisconsin Card Sorting Test (Computer Version 3 for Windows Research Edition) Psychological Assessment Resources, Inc; 1999. [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Ibrahim-Verbaas CA, Bressler J, Debette S, Schuur M, Smith AV, Bis JC, … Mosley TH. GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Molecular psychiatry. 2016;21:189–197. doi: 10.1038/mp.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber AB, Turk-Browne NB, Chun MM. Neural predictors of moment-to-moment fluctuations in cognitive flexibility. Proc Natl Acad Sci U S A. 2008;105:13592–13597. doi: 10.1073/pnas.0805423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Mosing MA, Henry JD, Trollor JN, Ames D, Martin NG, … Team OR. Genetic influences on four measures of executive functions and their covariation with general cognitive ability: the Older Australian Twins Study. Behav Genet. 2012;42:528–538. doi: 10.1007/s10519-012-9526-1. [DOI] [PubMed] [Google Scholar]
- Mary A, Slama H, Mousty P, Massat I, Capiau T, Drabs V, Peigneux P. Executive and attentional contributions to Theory of Mind deficit in attention deficit/hyperactivity disorder (ADHD) Child Neuropsychol. 2016;22:345–365. doi: 10.1080/09297049.2015.1012491. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, … Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Minassian A, Granholm E, Verney S, Perry W. Visual scanning deficits in schizophrenia and their relationship to executive functioning impairment. Schizophr Res. 2005;74:69–79. doi: 10.1016/j.schres.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Mishra A, Macgregor S. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- Neubert FX, Mars RB, Thomas AG, Sallet J, Rushworth MF. Comparison of human ventral frontal cortex areas for cognitive control and language with areas in monkey frontal cortex. Neuron. 2014;81:700–713. doi: 10.1016/j.neuron.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, … Benjamin DJ. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, … Franke L. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, … Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug and alcohol dependence. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, … Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, … Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GK, Travis RC, Green J, Bull D, Tipper S, Baker K … Million Women Study C. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010;304:426–434. doi: 10.1001/jama.2010.1042. [DOI] [PubMed] [Google Scholar]
- Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, … Koellinger PD. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111:13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, … Koellinger PD. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Munguia MA. Mnesic imbalance: a cognitive theory about autism spectrum disorders. Ann Gen Psychiatry. 2008;7:20. doi: 10.1186/1744-859X-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, … Gelernter J. Genome-wide Association Study of Cannabis Dependence Severity, Novel Risk Variants, and Shared Genetic Risks. JAMA Psychiatry. 2016;73:472–480. doi: 10.1001/jamapsychiatry.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taal HR, St Pourcain B, Thiering E, Das S, Mook-Kanamori DO, Warrington NM … Early Growth Genetics C. Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat Genet. 2012;44:532–538. doi: 10.1038/ng.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuoka C, Tseng H, Jaeger J, Varadi F, Smith MA, Yamada T … Alzheimer’s Disease Neuroimaging I. Modeling the heterogeneity in risk of progression to Alzheimer’s disease across cognitive profiles in mild cognitive impairment. Alzheimers Res Ther. 2013;5:14. doi: 10.1186/alzrt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trampush JW, Yang ML, Yu J, Knowles E, Davies G, Liewald DC, … Lencz T. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Molecular psychiatry. 2017;22:336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RS, Hebert LE, Scherr PA, Barnes LL, Mendes de Leon CF, Evans DA. Educational attainment and cognitive decline in old age. Neurology. 2009;72:460–465. doi: 10.1212/01.wnl.0000341782.71418.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastrow A, Kaiser S, Stippich C, Walther S, Herzog W, Tchanturia K, … Friederich HC. Neural correlates of impaired cognitive-behavioral flexibility in anorexia nervosa. Am J Psychiatry. 2009;166:608–616. doi: 10.1176/appi.ajp.2008.08050775. [DOI] [PubMed] [Google Scholar]
- Ziegler A, Blettner M, Kastner C, Chang-Claude J. Identifying influential families using regression diagnostics for generalized estimating equations. Genet Epidemiol. 1998;15:341–353. doi: 10.1002/(SICI)1098-2272(1998)15:4<341::AID-GEPI2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.