Abstract
Jun N-terminal kinase (JNK) is a stress-activated protein kinase that can be induced by inflammatory cytokines, bacterial endotoxin, osmotic shock, UV radiation, and hypoxia. We report the identification of an anthrapyrazolone series with significant inhibition of JNK1, -2, and -3 (Ki = 0.19 μM). SP600125 is a reversible ATP-competitive inhibitor with >20-fold selectivity vs. a range of kinases and enzymes tested. In cells, SP600125 dose dependently inhibited the phosphorylation of c-Jun, the expression of inflammatory genes COX-2, IL-2, IFN-γ, TNF-α, and prevented the activation and differentiation of primary human CD4 cell cultures. In animal studies, SP600125 blocked (bacterial) lipopolysaccharide-induced expression of tumor necrosis factor-α and inhibited anti-CD3-induced apoptosis of CD4+ CD8+ thymocytes. Our study supports targeting JNK as an important strategy in inflammatory disease, apoptotic cell death, and cancer.
Inflammatory disease is characterized by greatly increased expression of multiple immune proteins that frequently result in irreparable pathology. A major focus of drug discovery efforts is the identification of small molecules that have disease-modifying activity in addition to relieving clinical symptoms. Because individual transcription factors are capable of regulating the expression of multiple genes, the signaling pathways that control the activity of these transcription factors are prime candidates for therapeutic intervention. c-Jun N-terminal kinase (JNK) is a serine threonine protein kinase that phosphorylates c-Jun (1, 2), a component of the transcription factor activator protein-1 (AP-1; refs. 3 and 4). In complex with other DNA binding proteins, AP-1 regulates the transcription of numerous genes including cytokines [e.g., IFN-γ, IL-2, and tumor necrosis factor (TNF)-α; refs. 5 and 6], growth factors [e.g., vascular endothelial growth factor (VEGF); ref. 7], immunoglobulins (e.g., κ light chain; ref. 8), inflammatory enzymes (e.g., COX-2; ref. 9), and matrix metalloproteinases (e.g., MMP-13; ref. 10). JNK is a member of the mitogen-activated protein kinase (MAPK) family that includes the extracellular regulated kinases (ERKs) and p38 kinases. Three JNK genes (JNK1, -2, and -3) have been identified in humans; however, splice variants result in a total of 10 isoforms (11). JNK1 and JNK2 have a broad tissue distribution, whereas JNK3 seems primarily localized to neuronal tissues and the cardiac myocyte (12). Mice lacking JNK1 or JNK2 exhibit deficits in T-helper (CD4+) cell function (13–15). Double knockout animals are embryonic lethal, although fibroblasts from these animals are viable in vitro and exhibit a remarkable resistance to radiation-induced apoptosis (16). The JNK3 knockout mouse exhibits resistance to kainic acid-induced apoptosis in the hippocampus and to subsequent seizures (17). Therefore, JNK activity seems critical for both the immune response and for programmed cell death. Therapeutic inhibition of JNK may provide clinical benefit in diseases as diverse as arthritis, inflammatory bowel disease, chronic obstructive pulmonary disease, graft vs. host disease, stroke, Parkinson's disease, ischemic injury, and myocardial infarction.
Materials and Methods
Biochemical Characterization of JNK Enzyme Activity.
We have described in detail methods for the expression and purification of recombinant proteins, glutathione S-transferase-c-Jun, and JNK, along with methods for a complete kinetic evaluation (18). Double-reciprocal analysis was used to assess the kinetic mechanism. To obtain kinetic constants, the data were fit to the equation for a sequential mechanism by the nonlinear least-squares method of Cleland (19). ERK1 and p38–2 kinetic assays were identical to the JNK assay except in the choice of phosphoacceptor. The ERK assay measured the phosphorylation of myelin basic protein, and the p38–2 assay measured the phosphorylation of glutathione S-transferase-activating transcription factor (ATF). The time-resolved fluorescence assay for JNK has been described (20).
Use of SP600125 in Cell Culture.
SP600125 (molecular weight = 220) is poorly soluble in water. Stock solutions of at least 20 mM can be made by using 100% dimethyl sulfoxide. As a general guide, for every 10 μM SP600125, it is recommended to include 0.1% DMSO in the culture media (e.g., 30 μM/0.3% DMSO). Prewarming of media and the use of serum protein may enhance solubility. SP600125 typically precipitates as fine needles that are visible at 50-fold magnification.
CD4+ Cell Culture.
Purified (>95%) naive CD4+ cells were isolated from human peripheral blood mononuclear cells (PBMCs) and differentiated into T helper (Th)-1 and Th2 subsets as described (18). Cells were stained for CD4, CD25, CD45RA, CD45RO, CCR5, and CXCR4 with specific mAbs labeled with FITC, phycoerythrin, CyChrome, or PerCP from BD PharMingen. Stained cells were measured on a Coulter XL flow cytometer, using SYSTEM II acquisition software (Beckman Coulter). Data were later analyzed by using flowjo software (TreeStar, San Marcos, CA).
Cytokine Analysis: ELISA and Simultaneous Analyte Reagent Technology (SMaRT).
Culture supernatants were collected and stored at −80°C then tested by an ELISA according to the manufacturer's guidelines (Endogen, Cambridge, MA). SMaRT is an Ab bead capture system for quantifying analytes that uses a flow cytometer. SMaRT was developed by Beckman Coulter and was beta-tested by several sites including the Signal Research Division of the Celgene Corporation. In brief, microspheres containing 7 different intensities of a fluorescent emitter are each coated with an Ab specific for 1 of 7 cytokines: IL-1β, -4, -6, -10, -12, IFN-γ, and TNF-α. A second set of 7 cytokine-specific biotin-conjugated Abs was mixed with the coated microspheres and culture supernatant, incubated, and then washed by using a 96-well filter plate. In the second step, streptavidin conjugated to a second fluorescent emitter (PC5) was mixed with the microspheres–Ab–analyte–Ab–biotin complex, incubated, and washed, resulting in a solution containing seven different microsphere populations each with Ab-captured cytokine that is equivalent to the amount of bound PC5. The flow cytometer is able to discriminate the seven bead populations and quantify the PC5 signal. In our hands the technology achieved all control parameters required of the beta-testing protocol. Data collected for IL-4 and IFN-γ was confirmed by using an ELISA (Endogen). The SD of triplicate samples never exceeded 20% and was typically less than 10% of the mean.
Preparation of Human CD4+ Cells.
Processed buffy coats (50 ml) were purchased from the San Diego Blood Bank. PBMCs were isolated by gradient centrifugation (900 × g), using LYMPHOPREP (Nycomed, Oslo), and washed with Miltenyi Biotec (Auburn, CA) buffer (2 mM EDTA/0.5% FCS in PBS). PBMCs were incubated with anti-CD4 microbeads (Miltenyi Biotec) for 15 min on ice and fractionated by the Miltenyi MidiMACS columns following manufacturer's protocol.
Multiplex Analysis of mRNA.
Multiarray plate screening of mRNA was performed by High Throughput Genomics (HTG, Tucson, AZ). In brief, cell lysates were prepared by using a single-step proprietary lysis buffer. Lysates were incubated with a 16-gene capture array manufactured into each well of a 96-well plate. Detection was by luminescence and was performed by HTG. SDs for triplicate samples were typically 3–8% for samples with high levels of gene expression and 15–25% for samples with very low (near-threshold) levels of cytokine gene expression.
mRNA Analysis.
Determination of mRNA half-life was performed essentially as described (21), except that CD14+ cells were stimulated with (bacterial) lipopolysaccharide (LPS; 50 ng/ml) for 2 h before addition of actinomycin D (5 μg/ml). SP600125 (25 μM) or vehicle (0.5% DMSO vol/vol) was added immediately following the actinomycin D. Analysis was performed by using real-time reverse transcription (RT)-PCR. Total RNA was extracted with an RNeasy Mini kit (Qiagen, Valencia, CA). TNF mRNA was measured by real time RT-PCR (7700 Sequence Detection; phycoerythrin), using a TNF Taqman probe. All data were normalized by using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. The TNF-α forward primer was 5′-CTGGCCCAGGCAGTCAGAT-3′ and the reverse primer was 5′-TATCTCTCAGCTCCACGCCATT-3′. The Taqman probe sequence was 5′-FAM-CCTGTAGCCCATGTTGTAGCAAACCCTCA-TAMRA-3′.
For glyceraldehyde-3-phosphate dehydrogenase, the forward primer was 5′-GAAGGTGAAGGTCGGAGTC-3′ and the reverse primer was 5′-GAAGATGGTGATGGGATTTC-3′, and the Taqman probe sequence was 5′-Joe-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′.
Animal Studies.
Mouse LPS/TNF assay was performed as follows: Female CD-1 mice (8–10 weeks of age from Charles River Laboratories, Hollister, CA) were dosed i.v. or per os with SP600125 in PPCES vehicle (30% PEG-400/20% polypropylene glycol/15% Cremophor EL/5% ethanol/30% saline), final volume of 5 ml/kg, 15 min before i.v. injection with LPS in saline (0.5 mg/kg; Escherichia coli 055:B5; Westphal method; Difco). At 90 min, a terminal bleed was obtained from the abdominal vena cava, and the serum was recovered. Samples were analyzed for mouse TNF-α by using an ELISA (BioSource International, Camarillo, CA).
The in-life phase of the thymocyte apoptosis assay was performed in female C57BL/6 mice (Harlan, San Diego). SP600125 was administered at 0, 12, 24, and 36 h, 15 mg/kg s.c. in PPCES vehicle. Anti-CD3 (50 μg) i.p. (clone 145-2C11, BD PharMingen) was administered as a single dose immediately after SP600125 at time 0. After 48 h, mice were killed, and the thymus was dissected for thymocyte isolation. Treated and untreated mice thymuses were excised and immediately placed in complete medium (RPMI medium 1640 with 10% FBS, penicillin/streptomycin, and l-glutamine) on ice. Each thymus was then pressed between the frosted ends of 2 microscope slides to form a single cell suspension and collected through a 30 μm nylon mesh (Partec GmbH, Germany). Cells were stained for cell surface CD4 and CD8 (22) and apoptosis (23) and measured by flow cytometry.
Results
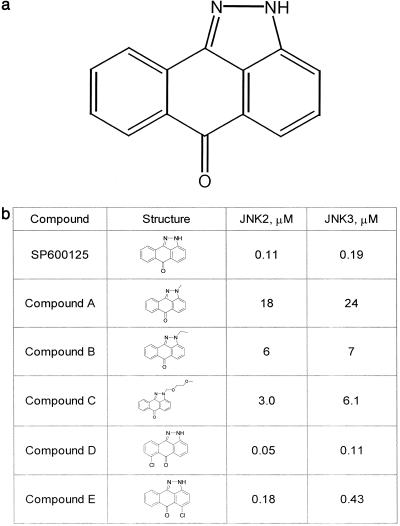
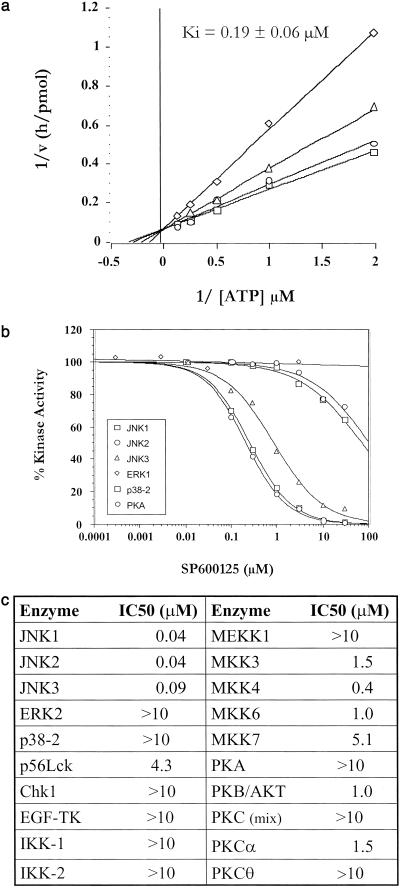
To identify novel small-molecule inhibitors of JNK, we established a high-throughput time-resolved fluorescence screening assay based on the in vitro phosphorylation of glutathione S-transferase (GST)-c-Jun (1–79) by recombinant human JNK2 (20). Screening of a proprietary diversity library identified a compound, SP600125, anthra[1,9-cd]pyrazol-6(2H)-one (Fig. 1a), with a Ki = 190 nM in this assay. Although anthrapyrazolones have been described in an anticancer screen (24), they had not been reported as kinase inhibitors. However, the highly planar structure of SP600125 and the presence of a nitrogen-containing ring system for interaction with key residues in the kinase active site were consistent with many kinase inhibitors (25). SP600125 was poorly soluble in aqueous solvents (0.0012 mg/ml in water) but was soluble in 100% DMSO up to at least 20 mM as a stock solution. Initial biochemical characterization of SP600125 showed inhibition of JNK2 to be competitive with respect to ATP (Fig. 2a) and fully reversible (not shown). Screening a limited selectivity panel showed that SP600125 inhibited JNK1, -2, and -3 isoforms with similar potency but exhibited greater than 300-fold selectivity against related MAP kinases ERK1 and p38–2, and the serine threonine kinase PKA (Fig. 2b). More extensive analysis of p38–2 inhibition revealed that SP600125 was ATP-competitive (Ki = 33 μM) but noncompetitive with substrate, GST-ATF2 (Kis = 38 μM, Kii = 35 μM; data not shown). An expanded selectivity panel (Fig. 2c), using freshly purified SP600125, indicated 10-fold selectivity (MKK4), 25-fold selectivity (MKK3, MKK6, PKB, and PKCα), and 100-fold selectivity for other kinases examined. No significant inhibitory activity was observed against 18 inflammatory enzymes with diverse catalytic activities at a concentration of 10 μM, a 100-fold higher concentration than the IC50 for JNK2. This data has been reported elsewhere (26).
Figure 1.
Characterization of SP600125. (a) Chemical structure. (b) Structure-activity relationship study of anthrapyrazolones (IC50 values).
Figure 2.
Inhibitory profile of SP600125. (a) SP600125 is an ATP-competitive inhibitor of JNK2 with a Ki value of 0.19 ± 0.06 μM. At fixed concentrations of JNK2 (9 nM) and c-Jun (2 μM), both ATP and SP600125 concentrations were varied; (□) 0 nM, (○) 50 nM, (▵) 100 nM, and (◊) 300 nM SP600125. The Ki value was derived from a nonlinear least-squares fit of the data to the kinetic equation for competitive inhibition. (b) SP600125 is a selective inhibitor of JNK. The inhibition profiles of SP600125 vs. related MAP kinases (ERK2 and p38–2) and an unrelated serine threonine kinase (PKA) were performed at Km levels of substrates. (c) Kinase selectivity data for SP600125.
Substructure searching of compound libraries revealed several compounds with related structures (Fig. 1b). Preliminary structure–activity relationship studies revealed the importance of the free NH group in the pyrazoloanthrone structure. All of the derivatives having N-alkyl substituents showed significant loss in inhibitory activity. Placement of a chlorine atom at the 8-position of SP600125 resulted in 2-fold improvement in potency whereas the corresponding 6-isomer was slightly less potent. It is highly plausible that, based on the ATP-competitive nature of inhibition, the pyrazole moiety of SPC600125 is involved in forming a critical hydrogen-bonding interaction at the ATP-binding site of JNK.
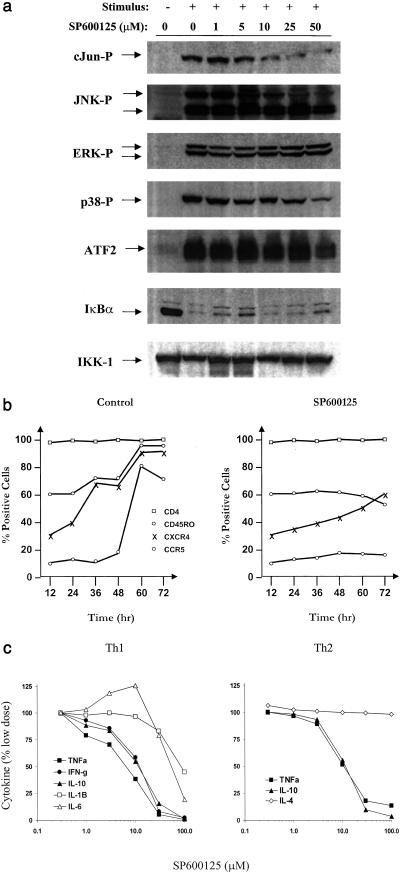
On the basis of the promising biochemical profile exhibited by SP600125, we proceeded to assess the activity of this compound in cell-based assays. Jurkat T cells were treated with increasing concentrations of SP600125 and stimulated with phorbol 12-myristate 13-acetate (PMA) plus anti-CD3 and anti-CD28 to maximally stimulate multiple pathways. Cell lysates were fractionated and probed with different Abs targeting the JNK, p38, ERK, and I kappa B kinase (IKK) signaling pathways (Fig. 3a). SP600125 blocked the phosphorylation of c-Jun with an IC50 of 5–10 μM. The observed increase in IC50 between the biochemical assay (JNK2) and cell-based assay is most likely a reflection of high ATP levels present in the cell. At a concentration of 50 μM, SP600125 did not block the phosphorylation of ERK1 or -2 and did not inhibit the degradation of IκBα. Partial inhibition of phospho-p38 and ATF2 was observed at 50 μM but not at 25 μM. This inhibitory activity is possibly due to the inhibition of MKK3 and MKK6 enzyme activity (Fig. 2c). Interestingly, we observed inhibition of phospho-JNK that was equivalent to phospho-c-Jun. Although this may be due in part to MKK4 inhibitory activity, it may also be due to inhibition of JNK autophosphorylating activity. These data show that SP600125 blocks JNK and not other inflammatory-signaling cascades, and seems to have a selectivity of at least 10-fold for the JNK pathway. In Jurkat T cells, JNK has been reported to regulate the transcription of the IL-2 gene (5, 27). In cells stimulated with PMA and phytohemagglutinin, SP600125 dose-dependently blocked both IL-2 and IFN-γ expression with IC50 values of 6 and 7 μM, respectively (data not shown). This concentration was consistent with the inhibition of c-Jun phosphorylation observed in Fig. 3. No cell toxicity, as monitored by MTS (Owen's reagent) conversion, was observed at the concentrations used in these experiments.
Figure 3.
Activity in T cells. (a) Jurkat T cells were resuspended in growth medium and stimulated with phorbol 12-myristate 13-acetate (PMA; 50 ng/ml), anti-CD3 (0.5 μg/ml), and anti-CD28 (2 μg/ml) for 30 min. SP600125 was added as a 10-min pretreatment at the concentrations indicated. The same cell lysates were examined by using different Abs targeting mitogen-activated protein kinase and NF-κB signaling pathways. Levels of nonphosphorylated IKK-1 were used as an indication of equal loading. (b) Inhibition of CD4+ cell activation and differentiation. Th0 cells isolated from either human cord or peripheral blood were cultured with anti-CD3/antiCD28, IL-12, anti-IL-4 in the absence or presence of SP600125 (30 μM). Culture aliquots were analyzed at different times for the expression of cell surface markers CD4, CD45RO, CXCR4, and CCR5. (c) CD4+ cells were differentiated into either Th1 or Th2 cultures over 5 days by using polarizing conditions before stimulating with anti-CD3/anti-CD28 in the presence of increasing concentrations of SP600125 for 15 h in triplicate. Culture supernatants were analyzed by simultaneous analyte reagent technology multiplex cytokine bead array. Data shown represent one of the duplicate experiments. Results are expressed as the mean of triplicate samples.
Targeted disruption of JNK1 or -2 has revealed a consistent immunologic phenotype featuring defective CD4+ cell function (13–15). Therefore, we examined the effect of SP600125 on CD4+ cell activation and differentiation. Naïve human T-cells (Th0) are characterized as CD45RAhi and CD45ROlo and do not express activation markers such as CD25, CD69, or the chemokine receptors CXCR4 and CCR5. Stimulation of Th0 cells with anti-CD3 and anti-CD28 causes cells to increase markedly in size, to express early activation markers such as CD69 and CD25, and become highly proliferative. In the presence of polarizing cytokines (IL-12 or -4) and quenching Abs (anti-IL4 or -IL12), CD4+ cells will terminally differentiate into either Th1 or Th2 subsets over a period of 4–5 days in vitro. To examine the effect of SP600125 on CD4+ cell activation, we isolated primary human Th0 cells and cultured them under Th1 polarizing conditions in the absence or presence of SP600125 (25 μM). Fig. 3b shows that after 5 days, a majority of cells were CD45RO-positive and expressed high levels of CCR5 [receptor for macrophage inflammatory protein-1α and regulated on activation of normal T cell expressed and secreted (RANTES)] and CXCR4 (receptor for stromal cell-derived factor). In contrast, although cells exposed to SP600125 remained CD4-positive, they showed no increase in the activation markers shown or in CD69 or CD25 (data not shown). Total (absolute) cell number on day 5 was essentially the same as that observed on day 1 and no apoptosis was observed. Cell cycle analysis revealed that cells failed to proliferate because of a G2/M block. Interestingly, addition of exogenous IL-2 did not overcome the cell cycle block; however, withdrawal of compound restored cell cycling and proliferation, indicating that compound effects were fully reversible.
To further extend our studies in CD4+ cells, we first fully differentiated Th0 cells into Th1 and Th2 subsets. Then, in the presence of anti-CD3 and anti-CD28 stimulation, we incubated cells with increasing concentrations of SP600125 and measured cytokine levels in culture supernatants (Fig. 3c). Th1 cells are characterized as expressing high levels of IL-2, IFN-γ, and TNFα, whereas Th2 cultures express high levels of IL-4 and low levels of IL-2, TNF-α, and IL-10. As shown in Fig. 3c, SP600125 potently blocked IFN-γ, TNF-α, and IL-10 (IC50 7–12 μM) but only weakly blocked IL-1β and -6 (IC50 > 30 μM) in Th1 cells. In Th2 cells, SP600125 potently blocked TNF-α and IL-10 but had no effect on the Th2-specific cytokine IL-4. SP600125 also dose-dependently inhibited IL-2 expression in Th1 and Th2 cultures with an IC50 of 5 μM (data not shown). Therefore, SP600125 exhibits a differential cytokine inhibitory profile in CD4+ cells with cytokines strongly, weakly, or not inhibited at 30 μM.
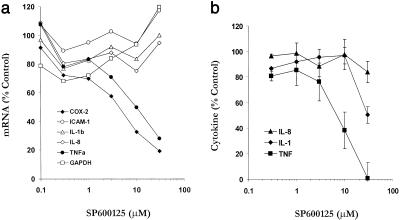
To further understand the role of SP600125 in inhibiting the expression of inflammatory genes and to compare its inhibitory profile in a different cell type, we performed experiments with PBMCs and primary human monocytes (CD14+) stimulated with LPS. We first examined the selectivity of SP600125 inhibition by monitoring the steady-state levels of mRNA in LPS-stimulated PBMCs (Fig. 4a). The levels of COX-2 (IC50 = 5 μM) and TNF-α (IC50 = 10 μM) mRNA were dose-dependently inhibited by SP600125. In contrast, mRNA levels of GAPDH, and LPS-induced, ICAM-1, IL-1β, and IL-8, were not inhibited by SP600125. In a separate experiment with purified CD14+ cells and real-time PCR, we confirmed the dose-dependent inhibition of TNF-α mRNA with SP600125 (data not shown). Therefore, SP600125 selectively blocks the mRNA expression of particular inflammatory genes in monocytes at concentrations that are consistent with those observed for inhibition of c-Jun phosphorylation and cytokine inhibition in T cells.
Figure 4.
Inhibitory activity in primary human monocytes. (a) Human PBMCs isolated from peripheral blood were stimulated with LPS (100 ng/ml) for 4 h in the presence of increasing concentrations of SP600125. Cell lysates were prepared and analyzed by using multiplex mRNA detection (Systems Integration Drug Discovery Company). Results are expressed as the mean of triplicate determinations. (b) Primary human monocytes were isolated by positive selection (CD14) and stimulated with LPS (100 ng/ml) in the presence of increasing concentrations of SP600125. After 15 h, culture supernatants were analyzed for expression of IL-1β, -8, and TNF-α with an ELISA. Values are mean and SD for quadruplicate samples.
By using purified CD14+ monocytes isolated from PBMCs, we measured the level of LPS-induced IL-1β, -8, and TNF-α in culture supernatants (Fig. 4b). Similar to what was observed with the mRNA analysis and to what was observed in Th1 cells, SP600125 potently blocked TNF-α, weakly inhibited IL-1β, and failed to inhibit the expression of IL-8, even at 30 μM. Previous reports have suggested that inhibition of the JNK pathway inhibits IL-2 expression by means of a mechanism that may involve destabilization of IL-2 mRNA (21). We performed TNF-α mRNA half-life experiments in CD14+ cells in the absence or presence of SP600125 (data not shown). TNF-α mRNA exhibited a basal half-life decay of 75 min. However, in the presence of SP600125, TNF-α mRNA levels were only 35% of control within the first 30 min, giving an estimated initial phase half-life of 20 min. These data suggest that JNK may play a role in mRNA stabilization of several induced inflammatory genes including IL-2 and TNF-α.
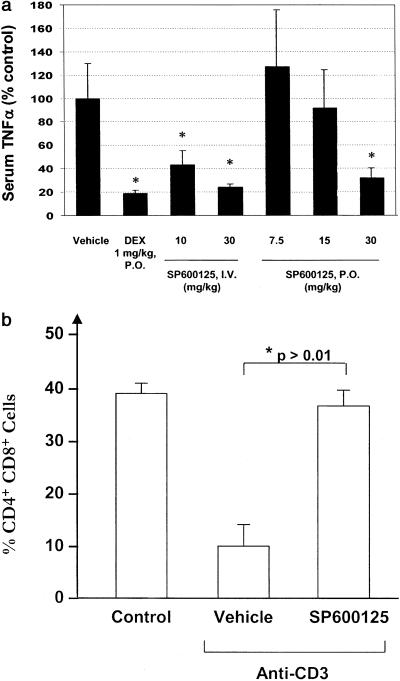
Because multiple cell-based assays confirmed the potent inhibition of TNF-α with SP600125, we proceeded to test SP600125 inhibitory activity in a mouse model of LPS-induced TNF-α expression. Mice were pretreated with SP600125 per os or i.v. administration and then challenged with bacterial endotoxin (LPS). Dexamethasone 21-acetate was used as a positive control. Administration of SP600125 at 15 or 30 mg/kg i.v. significantly inhibited TNF-α serum levels, whereas oral administration dose-dependently blocked TNF-α expression with significant inhibition observed at 30 mg/kg per os (Fig. 5a). Therefore, SP600125 demonstrates efficacy in an in vivo model of endotoxin-induced inflammation.
Figure 5.
In vivo activity of SP600125. (a) CD-1 mice were dosed with SP600125 i.v. 15 min before injection with LPS or per os 30 min before injection with LPS. At 90 min, a blood sample was recovered, and the serum was obtained. Samples were analyzed for mouse TNF-α by using an ELISA (BioSource). Results are expressed as the mean and standard error with 4 animals per compound treatment group and 6 animals in vehicle control group. Asterisk (*) indicates P ≤ 0.05. DEX, dexamethasone 21-acetate. (b) Apoptosis in murine thymocytes. Thymocytes were isolated from C57BL/6 mice 48 h after injection with anti-CD3 (50 μg) i.p. and analyzed for CD4 and CD8 by flow cytometry. Control animals received no anti-CD3 or vehicle. SP600125 was administered at 0, 12, 24, and 36 h, 15 mg/kg s.c. Anti-CD3 administration was at a single time immediately after the first dosing of SP600125. Data for double-positive (CD4+ CD8+ ) cells is shown expressed as the mean percentage of total thymocytes (n = 4 per group).
Genetic mutants have also revealed a role for JNK in the apoptotic cell death of immature T cells in the thymus (15). Compared with wild-type animals, JNK2 knock-out mice exhibited almost complete resistance to apoptosis of double-positive thymocytes 48 h after injection with CD3 Ab. We repeated this study to observe the effect of the JNK inhibitor, SP600125 (Fig. 5b). In control animals, CD4+ CD8+ thymocytes represented just less than 40% of total thymocytes. Forty-eight hours after exposure to CD3 Ab in vivo, the percentage of CD4+ CD8 + cells had declined to 10%. Remarkably, mice receiving SP600125 showed almost complete resistance to CD3 Ab-mediated apoptosis with CD4+ CD8+ numbers the same as control animals. This study further demonstrated the in vivo efficacy of SP600125 and showed consistent data to that observed in JNK knock-out animals.
Discussion
We report the identification and characterization of a small molecule that acts as a novel inhibitor of JNK catalytic activity. SP600125, an anthrapyrazole was identified in a high-throughput biochemical screen by using purified recombinant JNK2 and c-Jun. In characterizing the activity of SP600125 we have based our experiments on published observations made with JNK knock-out mice and cells deficient in JNK1, -2, or -3. In this regard, the inhibitory activity of SP600125 was remarkably consistent with the role JNK has been reported to play in CD4+ cell activation and differentiation, CD14+ cell gene expression, and in thymocyte death. SP600125 demonstrated notable selectivity in enzyme panel and in cell assays presented here. Concentrations of SP600125 that inhibited c-Jun phosphorylation in cells also inhibited COX-2, IL-2, IFN-γ, TNF-α, IL-10, and also MMP gene expression (26). Genes weakly inhibited included ICAM-1, IL-1β, -4, -6, and -8. Although studies using biochemical and promoter analysis have indicated a role for JNK and/or Jun and/or AP-1 in the expression of each of these genes (28–30), pharmacologic intervention with SP600125 suggests that JNK is not a critical mediator of expression, at least in the stimuli and cell types we have used. The weak inhibition of IL-1β and -6 that we observed may be due to the partial p38 pathway inhibition we observed with SP600125, because the high-affinity p38 inhibitor, SB203580, potently inhibits the expression of these genes (31).
In in vivo studies in mice, SP600125 inhibited LPS-induced TNF-α expression. The regulation of TNF-α expression is controlled by both transcriptional and posttranscriptional mechanisms including mRNA stability and protein processing (32). The p38 inhibitor SB203580 has been reported to inhibit TNF-α by targeting mRNA stabilization (33), and we observed that treatment of CD14+ cells with SP600125 caused a significant decrease in TNF-α mRNA half-life, suggesting that JNK may also be involved in promoting stability of mRNA transcripts. Indeed, JNK has been implicated in stabilizing IL-2 mRNA (21). Sequence and deletion analysis of the TNF-α promoter has identified DNA binding elements for the transcription factors AP-1, AP-2, ATF-2/c-Jun, Egr-1, Ets-1, C/EBPβ, CRE, and NF-κB (34). Activation of monocytic cells with inflammatory stimuli such as LPS leads to changes in the composition and arrangement of transcriptional proteins associated with the TNF-α promoter. Major protranscriptional changes include the binding of NF-κB and the binding and activation of c-Jun, which is a component of c-Jun/c-Fos (AP-1) and ATF-2/c-Jun transcription factors. As a major cytokine of the vertebrate immune response, TNF-α has been implicated in a large number of diseases. Therapies that interfere with TNF-α function, such as soluble TNF-α receptor (Etanercept) and TNF-α Ab (Infliximab), have demonstrated proven clinical efficacy in inflammatory diseases such as rheumatoid arthritis and inflammatory bowel disease. Of note, SP600125 has shown efficacy in models of adjuvant-induced arthritis and antigen-induced lung inflammation in rats (ref. 26 and I. M. Adcock, unpublished data). Although protein therapeutics are currently administered by injection, development of small-molecule inhibitors should provide advantages in both delivery and cost.
In a second animal model, we observed that SP600125 prevented anti-CD3-mediated thymocyte apoptosis. This result mirrored the finding in JNK2−/− mice, which were resistant to anti-CD3-mediated apoptosis of double-positive thymocytes (15). This result supports a role for JNK in regulating apoptosis, perhaps by regulating the activity of bcl-2-related proteins that promote cytochrome-c release from mitochondria (16). Published work describing CEP-1347 (34), an indolocarbazole that blocks the activation of the JNK pathway by inhibiting the mitogen-activated protein kinase (MAPKKK), MLK3. CEP-1347 has mostly been characterized in neuronal systems modeling apoptosis and has shown efficacy in a primate model of Parkinson's disease. These findings support the hypothesis that JNK inhibitors may have clinical benefits in diseases involving cell death, including infarction, stroke, ischemia reperfusion injury, and chronic neuronal cell death.
An additional experimental finding was the observation that SP600125 blocked cell proliferation in CD4+ cells but did not kill these cells. Therefore, SP600125 was described as having a “cytostatic” effect on T cell proliferation. Several anthrapyrazoles, including doxorubicin, have been identified as chelators of DNA. However, we performed ethidium bromide displacement assays (35) but observed no competitive binding (data not shown), suggesting SP600125 is not a strong interchelator of DNA. Consistent with this finding, we observed no apoptotic cell death with SP600125, and anthrapyrazoles with unsubstituted side chains (e.g., SP600125) performed poorly in murine leukemia cell death assays compared with highly substituted structures such as mitoxantrone (24). This observation clearly supports further studies examining the effect of JNK inhibitors in proliferation and tumor progression. c-Jun has been strongly implicated in the regulation of cell growth and proliferation. c-Jun was originally described as a protooncogene because of its homology to avian sarcoma virus 17 and its transforming potential in cells (36, 37). Regenerating livers show increased c-Jun expression after partial hepatectomy (38). Studies with antisense oligonucleotides (39) and fibroblasts deficient in c-Jun (40) support the requirement of c-Jun for G1 progression. Interestingly, it has been reported that G1 progression does not require the phosphorylation of serines 63–73 on c-Jun, suggesting that c-Jun-mediated progression may be independent of JNK activity (40). However, antisense to JNK2, but not JNK1, was able to block growth factor-induced proliferation of lung carcinoma cells, suggesting that JNK is important for cell proliferation; however, this activity may be distinct from the regulation of the c-Jun transactivation domain (41).
In conclusion, we have characterized SP600125 as a small-molecule inhibitor of JNK. Our pharmacologic studies indicate JNK plays important roles in transcription factor activation, mRNA stabilization, apoptotic progression, and cell proliferation. Combined with published reports describing JNK knock-out mice and JNK-deficient cells, therapeutic inhibition of JNK has significant potential in inflammatory diseases, acute organ failure due to cell death, and cancer.
Acknowledgments
We acknowledge the enthusiasm and encouragement of Vincent Fert (SMaRT) and Bruce Seligmann (HTG). We thank Bernd Stein for critical review of this manuscript.
Abbreviations
- JNK
c-Jun N-terminal kinase
- LPS
lipopolysaccharide
- AP-1
activator protein-1
- ERK
extracellular regulated kinase
- PBMC
peripheral blood mononuclear cell
- Th0
naïve human T cell
- TNF
tumor necrosis factor
- ATF
activating transcription factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Derijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 2.Kallunki T, Su B, Tsigelny I, Sluss H, Derijard B, Moore G, Davis R, Karin M. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 3.Karin M. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 4.Ip Y T, Davis R J. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 5.Jain J, Valge-Archer V E, Rao A. J Immunol. 1992;148:1240–1250. [PubMed] [Google Scholar]
- 6.Foletta V C, Segal D H, Cohen D R. J Leukocyte Biol. 1998;63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- 7.Finkenzeller G, Technau A, Marme D. Biochem Biophys Res Commun. 1995;208:432–439. doi: 10.1006/bbrc.1995.1356. [DOI] [PubMed] [Google Scholar]
- 8.Schanke J T, Marcuzzi A, Podzorski R P, Van Ness B. Nucleic Acids Res. 1994;22:5425–5432. doi: 10.1093/nar/22.24.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Knethen A, Callsen D, Brune B. Mol Cell Biol. 1999;10:361–372. doi: 10.1091/mbc.10.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pendas A M, Balbin M, Llano E, Jimenez M G, Lopez-Otin C. Genomics. 1997;40:222–233. doi: 10.1006/geno.1996.4554. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H, Derijard B, Davis R J. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 12.Martin J H, Mohit A A, Miller C A. Brain Res Mol Brain Res. 1996;35:47–57. doi: 10.1016/0169-328x(95)00181-q. [DOI] [PubMed] [Google Scholar]
- 13.Dong C, Yang D, Wysk M, Whitmarsh A, Davis R, Flavell R. Science. 1998;282:2092–2095. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 14.Yang D, Whitmarsh A, Conze D, Barrett T, Davis R, Rincon M, Flavell R. Immunity. 1998;9:575–585. doi: 10.1016/s1074-7613(00)80640-8. [DOI] [PubMed] [Google Scholar]
- 15.Sabapathy K, Hu Y, Kallunki T, Schreiber M, David J, Jochum W, Wagner E, Karin M. Curr Biol. 1999;9:116–125. doi: 10.1016/s0960-9822(99)80065-7. [DOI] [PubMed] [Google Scholar]
- 16.Tournier C, Hess P, Yang D D, Xu J, Turner T K, Nimnual A, Bar-Sagi D, Jones S N, Flavell R A, Davis R J. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 17.Yang D, Kuan C, Whitmarsh A, Rincon M, Zheng T, Davis R, Rakic P, Flavell R. Nature (London) 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- 18.Murray B W, Bennett B L, Sasaki D. Methods Enzymol. 2001;332:432–452. doi: 10.1016/s0076-6879(01)32220-6. [DOI] [PubMed] [Google Scholar]
- 19.Cleland W W. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 20.Gaarde W, Hunter T, Brady H, Murray B W, Goldman M. J Biomol Screening. 1997;2:213–223. [Google Scholar]
- 21.Chen C-Y, Del Gatto-Konczak F, Wu Z, Karin M. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, Bissonnette R, Parfrey N, Szalay M, Kubo R, Green D. J Immunol. 1991;146:3340–3346. [PubMed] [Google Scholar]
- 23.Nicoletti I, Migliorati G, Pagliacci M, Grignani F, Riccardi C. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 24.Showalter H D H, Johnson J L, Hoftiezer J M, Turner W R, Werbel L M, Leopold W R, Shillis J L, Jackson R C, Elslager E F. J Med Chem. 1987;30:121–131. doi: 10.1021/jm00384a021. [DOI] [PubMed] [Google Scholar]
- 25.McMahon G, Sun L, Liang C, Tang C. Curr Opin Drug Discovery Dev. 1998;1:131–146. [PubMed] [Google Scholar]
- 26.Han H, Boyle D, Chang L, Bennett B, Karin M, Manning A, Firestein G. J Clin Invest. 2001;108:73–81. doi: 10.1172/JCI12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faris M, Kokot N, Lee L, Nel A. J Biol Chem. 1996;271:27366–27373. doi: 10.1074/jbc.271.44.27366. [DOI] [PubMed] [Google Scholar]
- 28.De Cesaris P, Starace D, Starace G, Filippini A, Stefanini M, Ziparo E. J Biol Chem. 1999;274:28978–28982. doi: 10.1074/jbc.274.41.28978. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Tournier C, Davis R J, Flavell R A. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holtmann H, Winzen R, Holland P, Eickemeier S, Hoffmann E, Wallach D, Malinin N, Cooper J, Resch K, Kracht M. Mol Cell Biol. 1999;19:6742–6753. doi: 10.1128/mcb.19.10.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Laydon J, McDonnell P, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M, Heys J, Landvatter S, et al. Nature (London) 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 32.Vilcek J, Lee T H. J Biol Chem. 1991;266:7313–7316. [PubMed] [Google Scholar]
- 33.Rutault K, Hazzalin C A, Mahadevan L C. J Biol Chem. 2000;276:6666–6674. doi: 10.1074/jbc.M005486200. [DOI] [PubMed] [Google Scholar]
- 34.Kaneko M, Saito Y, Saito H, Matsumoto T, Matsuda Y, Vaught J L, Dionne C A, Angeles T S, Glicksman M A, Neff N T, et al. J Med Chem. 1997;40:1863–1869. doi: 10.1021/jm970031d. [DOI] [PubMed] [Google Scholar]
- 35.Hartley J A, Reszka K, Zuo E T, Wilson W D, Morgan A R, Lown J W. Mol Pharmacol. 1988;33:265–271. [PubMed] [Google Scholar]
- 36.Maki Y, Bos T J, Davis C, Starbuck M, Vogt P K. Proc Natl Acad Sci USA. 1987;84:2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angel P, Allegretto E A, Okino S T, Hattori K, Boyle W J, Hunter T, Karin M. Nature (London) 1988;332:166–171. doi: 10.1038/332166a0. [DOI] [PubMed] [Google Scholar]
- 38.Morello D, Fitzgerald M J, Babinet C, Fausto N. Mol Cell Biol. 1990;10:3185–3193. doi: 10.1128/mcb.10.6.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soprano K J, Cosenza S C, Yumet G, Soprano D R. Ann NY A Sci. 1992;660:231–239. doi: 10.1111/j.1749-6632.1992.tb21075.x. [DOI] [PubMed] [Google Scholar]
- 40.Wisdom R, Johnson R S, Moore C. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bost F, McKay R, Bost M, Potapova O, Dean N M, Mercola D. Mol Cell Biol. 1999;19:1938–1949. doi: 10.1128/mcb.19.3.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]