Abstract
Schistosomiasis is of public health importance to an estimated one billion people in 79 countries. A vaccine is urgently needed. Here, we report the results of four independent, double-blind studies of an Sm-p80–based vaccine in baboons. The vaccine exhibited potent prophylactic efficacy against transmission of Schistosoma mansoni infection and was associated with significantly less egg–induced pathology, compared with unvaccinated control animals. Specifically, the vaccine resulted in a 93.45% reduction of pathology-producing female worms and significantly resolved the major clinical manifestations of hepatic/intestinal schistosomiasis by reducing the tissue egg–load by 89.95%. A 35-fold decrease in fecal egg excretion in vaccinated animals, combined with an 81.51% reduction in hatching of eggs into the snail-infective stage (miracidia), demonstrates the parasite transmission blocking potential of the vaccine. Substantially higher Sm-p80 expression in female worms and Sm-p80–specific antibodies in vaccinated baboons appear to play an important role in vaccine-mediated protection. Preliminary analyses of RNA sequencing revealed distinct molecular signatures of vaccine-induced effects in baboon immune effector cells. This study provides comprehensive evidence for the effectiveness of an Sm-p80–based vaccine for schistosomiasis.
Keywords: Sm-p80 vaccine, schistosomiasis, Schistosoma mansoni, baboons, systems biology, efficacy
Graphical abstract
Schistosomiasis is of public health importance to an estimated one billion people in 79 countries. A vaccine is urgently needed. Here, we report the results of four independent, double-blind studies of an Sm-p80–based vaccine in baboons. The vaccine exhibited potent prophylactic efficacy against transmission of Schistosoma mansoni infection and was associated with significantly less egg–induced pathology, compared with unvaccinated control animals.
Introduction
Schistosomiasis (Bilharzia) has afflicted humankind since at least the Pharaoh’s Middle Kingdom. Five species of Schistosoma are known to cause the disease in humans, with Schistosoma mansoni, S. haematobium and S. japonicum being the most clinically relevant. The disease is endemic in 79 countries and 200 million people are infected, with up to 800 million more being at risk to acquire the infection 1, 2, 3. These estimates are based on partially sensitive egg retrieval/detection techniques. Apparent “egg-negative” individuals may be carrying infections that are indiscernible using current schistosome egg detection procedures in feces/urine. Based on this assumption, the estimated number of people infected could be up to 600 million 4, 5, 6. Existing infection control measures have been suboptimal in reducing parasite transmission, morbidity and disease burden associated with schistosomiasis.
Dependence on mass drug administration (MDA) alone with praziquantel (PZQ) for the past several decades has not yielded satisfactory results, and infection rates continue to be high despite global PZQ coverage in 2016 of 54% 1, 2, 6, 7. Furthermore, large-scale PZQ use may lead to drug resistance in the parasite 4. An efficacious vaccine inducing prolonged protection would result in a substantial decrease in transmission of infection and morbidity, particularly if deployed concurrently with existing control measures 3.
A schistosomiasis vaccine is considered to be one of the ten vaccines urgently needed 5. Preferred product characteristics for a prophylactic schistosomiasis vaccine have been established to call for a substantial reduction in morbidity, rather than inducing sterilizing immunity8, 9. Mathematical modeling evaluating the impact of a vaccine on transmission dynamics has suggested that a partially protective vaccine with an efficacy as low as 60% could prevent transmission in low- and moderately-endemic areas 10. Compartmental model simulation of schistosomiasis transmission in endemic regions has predicted that compared to MDA-only programs, vaccination with a partially protective vaccine combined with MDA would be advantageous in reducing the acquisition of new worms and lowering egg release from residual worms in the environment 11. To date, there are three schistosomiasis vaccine candidates in various phases of human clinical trials: S. haematobium glutathione S-transferase (Sh28GST)12, S. mansoni tetraspanin, a 9-kDa surface antigen (Sm-TSP-2)13 and S. mansoni 14-kDa fatty acid-binding protein (Sm14) 14, 15.
To develop a viable schistosomiasis vaccine, the efficacy of an Sm-p80–based vaccine was systematically evaluated in baboons (Papio anubis), natural hosts of schistosomiasis and the most relevant nonhuman primate model of human clinical manifestations of both acute and chronic disease.16, 17, 18 Sm-p80 is the heavy chain of the S. mansoni calcium activated neutral protease (calpain), and is the only classical calpain among the non-human calpains.19, 20
Sm-p80 meets the requirements for a suitable schistosome vaccine candidate because it is present in the surface membranes and epithelial syncytium of the worm,21, 22 it is one of the immunodominant membrane antigens,22 and it displays no immunological cross-reactivity with human or other vertebrate calpains.22 The Sm-p80 protein plays an important role in the surface membrane biogenesis and turnover, a process utilized by hemo-helminths to escape host immune responses.22 Sm-p80 is also implicated in additional immune-evasion tactics utilized by the parasite, including cleavage of the host CD23,23 as well as degradation of fibronectin to inhibit blood clot formation around the worm in vivo.24
The present study reports the vaccine efficacy of Sm-p80 with a Toll-like receptor 4 (TLR4) agonist-based adjuvant, glucopyranosyl lipid A in an oil-in water emulsion (GLA-SE),25 in four independent double-blind experiments with 40-baboons. Reduction in worm burden (prophylactic efficacy), decrease in egg retention in tissues/organs (pathology-resolving efficacy), and egg expulsion in feces and lessening in egg hatching ability (parasite transmission blocking efficacy) were determined by quantifying each parameter separately followed by comparison between vaccinated and control groups of baboons.26, 27 We further present preliminary analyses of transcriptomic profiles and molecular mechanisms associated with Sm-p80 vaccination and post-infectious parasite challenge utilizing a systems biology approach, identifying gene–gene interactions and networks related to immunity and immunogenicity.28
Materials and methods
Animals and parasites
Specific pathogen-free (SPF) olive baboons (Papio anubis; 0.94 to 2.18 years old) were procured from the University of Oklahoma Health Sciences Center (OUHSC). Animals were kept in the Association for Assessment and Accreditation of Laboratory Animal Care International–accredited facilities at OUHSC. The baboon study was approved by the OUHSC Institutional Animal Care and Use Committee (IACUC; protocol number: 11–160-I). Baboons were prescreened for intestinal and blood parasites and for Sm-p80 cross-reactive antibodies. Only animals negative for both parasites and cross-reactive antibodies were used in the institutional animal care and use committee (IACUC) approved study. S. mansoni–infected snails (Biomphalaria glabrata) were obtained from the Schistosomiasis Resource Center, Biomedical Research Institute (Rockville, MD).
Sm-p80 vaccine preparation
Recombinant Sm-p80 antigen (Sm-p80) production using a prokaryotic expression system has been reported previously.29 The coding sequence of Sm-p80 was cloned into a pCold II vector (GenScript, Piscataway, NJ), and protein was expressed in Escherichia coli strain BL21 (DE3). The expressed Sm-p80 was purified by affinity purification followed by size exclusion chromatography. Sm-p80 expression and purity was confirmed via SDS-PAGE and western blot. Endotoxin levels in the Sm-p80 used for immunizations were lower than that approved by the Food and Drug Administration for human use (approximately 0.06 EU/mL). Prior to immunization, purified Sm-p80 was admixed with GLA-SE to make the final vaccine formulation.
Immunization strategies and parasite challenge
There were four independent double-blind trials with two groups of baboons (control and experimental) in each trial. A total of 10 baboons were randomly and equally divided into either control (n=5) or experimental (n=5) groups for each trial. Each baboon in the experimental group received 250 μg rSm-p80 formulated in 50 μg GLA-SE while baboons in the control group were immunized with 50 μg GLA-SE only. All animals received prime immunization followed by three boosts intramuscularly at 4-week intervals. Four weeks following the last immunization, each baboon was percutaneously challenged with 1000 S. mansoni cercariae. To allow for disease progression, all baboons remained under observation for 8 weeks.30 The protocol for the pre-clinical vaccine trials including animal randomization, immunizations, challenge infection and necropsy schedule for prophylactic efficacy determination is described in Supplementary Table 1.
Worm and egg burden determination
Necropsies and determination of percentage protection were performed as described previously. 26, 31 Eight weeks after challenge, the baboons were euthanized as previously described 26, 32 and adult schistosome parasites recovered by perfusion from the mesenteric vasculature and hepatic portal system.33 Protection (P) was calculated by comparing worm burdens from immunized (I) and control (C) baboons by a standard formula: % P = [(C–I)/C × 100]. After sacrifice, 3–5 g of liver and intestine samples were digested in KOH,34 the number of eggs per gram of tissue determined and the percent reduction in egg production calculated.
Weekly fecal egg output processing and monitoring
Fecal samples were collected from each baboon every week until animal euthanasia. A 3–5 g fecal specimen was emulsified in 10 mL of 10% formalin. After the emulsion was filtered and centrifuged, it was re-suspended in 12 mL of water with the addition of 3 mL of ethyl acetate. After vigorous shaking and centrifugation, the pellet was re-suspended in saline. Eggs present at each time point were determined for both control and experimental groups. The mean value corresponding to each group was then used to calculate the percent reduction in egg production35.
Liver egg hatching
For each baboon, 2.5–3 g sections of liver were collected at necropsy. Samples were kept in 1.2% NaCl and on ice during the procedure. Liver samples were blended on a low setting for 30 sec. Homogenized samples were then passed through sieves (106 μm and 45 μm) multiple times and eggs were collected from the top of 45 μm sieve and subsequently centrifuged. The supernatant was discarded and egg pellet was re-suspended in 5 mL of water. Resuspended eggs were then divided into 4 wells and placed under a light source for 15–30 min to stimulate hatching. Hatched eggs were counted using a light microscope for all samples.
Sm-p80 specific antibody titers
Samples of serum from each baboon in each trial were used to measure total immunoglobulin G (IgG), IgG1, IgA and IgM antibody titers 26, 36. Briefly, 96-well microtiter plates were coated with recombinant Sm-p80 (1.2 μg/well). Diluted baboon serum (1:500) from individual animals was used as the primary antibody and horseradish peroxidase (HRP)-labeled anti-monkey IgG (Thermo Scientific Rockford, IL) (1:50,000), HRP-labeled anti-human IgG1 (1:1,000), HRP-labeled anti-monkey IgA (1:5,000) and IgM (1:5,000) (Alpha Diagnostics, San Antonio, TX) were used as the secondary antibodies. All animal samples were measured in triplicate. Results were expressed as endpoint titers calculated from the curve of optical density versus serum dilution for the cutoff of 2 standard deviations above the control value 37.
Isolation of S. mansoni eggs from baboon livers for immunofluorescence
Livers washed with 1.2% NaCl solution were cut into small pieces and blended for 30 sec on low speed. The suspension was passed through a 425 μm and subsequently a 180, 106, and 45 μm sieves, and was well rinsed with 1.2% NaCl solution. The egg suspension was collected from the last sieve (45 μm), poured into a 50 mL tube and centrifuged for 5 min at 2,000 rpm at room temperature. The supernatant was discarded and 15 mL of 13% NaCl was added to the pellet and vortexed to get a homogeneous solution. The suspension was centrifuged at 3,600 × g for 5 min. The supernatant was discarded and 15 mL of 1.2 % NaCl solution was added to the pellet, then passed through a mesh into a 50 mL tube and rinsed with 1.2% NaCl. Then, the egg suspension was centrifuged for 3 min at 3,600 × g. The resulting pellet was resuspended in PBS and stored at −80 °C until required.
Immunofluorescence and confocal microscopy
S. mansoni adult worms obtained from control and experimental baboons were processed for immunofluorescence and confocal microscopy as previously described 29 with slight modifications. The primary antibody used in this study was rabbit anti–Sm-p80 polyclonal antibody (FabGennix, Texas) diluted in 1% BSA in PBS (1:300) and the secondary antibody was anti-rabbit Alexa-Fluor 647 (Molecular Probes, Thermo Fisher Scientific, Waltham, MD). All samples were mounted with an anti-fading reagent ProLong™ Gold antifade with DAPI (Molecular Probes, Thermo Fisher Scientific, Waltham, MD) and then cured for 24 hours. The isolated eggs from infected baboon livers were processed as published elsewhere 29 but were incubated with the same primary and secondary antibodies described above.
Quantification of immunofluorescence
Total corrected fluorescence (TCF) measurements were performed with open source Image J version 1.51s (https://imagej.nih.gov/ij/) using the following formula: TCF = integrated density – (area of region of interest × mean fluorescence of background). For each S. mansoni worm pair, an outline was drawn to measure integrated intensity, area and mean fluorescence of the adjacent background.
Total RNA extraction
Total RNA was used for RNA-Seq library preparation from individual samples of peripheral blood mononuclear cells (PBMCs), splenocytes and lymph node cells from each animal. Total RNA was extracted using GenElute™ mammalian total RNA miniprep kit (Millipore Sigma, St. Louis, MO) following manufacturer’s instructions. Total RNA concentrations were quantified using the Qubit® 3.0 Fluorometer and RNA HS assay kit (Thermo Fisher, MA). Quality of RNA was checked using RNA ScreenTapes on Agilent 2200 TapStation (Agilent, Santa Clara, CA).
Library preparations and RNA-sequencing
RNA-sequencing libraries were made using total RNA from PBMCs, spleen and lymph node cells. 1 μg total RNA was used for the cDNA library construction using TruSeq® Stranded mRNA LT kit (Illumina, San Diego) and epMotion robot (Eppendorf, Hamburg, Germany). Briefly, total RNA was incubated with magnetic oligo (dT) beads and heat denatured. Fragmented mRNA was primed with random hexamers and transcribed into first strand cDNA using SuperScript® III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA) and random primers, followed by second strand cDNA synthesis using DNA polymerase I. The resulting blunt-ended dscDNAs were then 3′ adenylated. Libraries were then single-indexed using unique indexes for each sample 30. Indexed libraries were then enriched via PCR to amplify DNA fragments that contained adapter molecules on both ends. The libraries were then purified using AMPure XP purification beads (Beckman Coulter, Brea, CA). Library construction produced single-indexed libraries with a median insert size of ~300 bp, which was validated by gel electrophoresis on an Agilent 2200 TapeStation instrument using D1000 Screen Tapes (Santa Clara, CA). All libraries were quantified in triplicate using SynergyH1 fluorescent plate reader (BioTek, Vermont). Prior to cluster generation for sequencing, samples were normalized to an equimolar concentration. Stock libraries were diluted in 10 mM Tris-HCl to 10 nM concentrations, pooled and quantified. The resulting libraries were then further diluted to 4 nM before denaturation with NaOH. The pooled denatured cDNA libraries were loaded on a cBot for cluster generation followed by 2 × 125 bp read length paired-end sequencing using HiSeq SBS Kits with V4 chemistry on a HiSeq 2500 sequencer (Illumina, San Diego) 30.
Bioinformatics
Sequencing run data containing base call information were demultiplexed using bcl2fastq software. Demultiplexed data were concatenated and unzipped on a Linux server. Raw reads quality was assessed using FastQC software (Babraham Bioinformatics). Quality filtered reads (both read 1 and 2) for each animal from each tissue sample were mapped to the Homo sapiens genome (GRCh37) using QSeq® version 15.0 software (DNASTAR, Madison, WI) for differential gene expression analysis using RPKM normalization. RNA-Seq data were analyzed by creating different replicate sets. Samples were separated according to tissue origin (PBMCs, lymph nodes, or spleen), group assignment (control or experimental), and, in the case of PBMC samples, separated by allocated time points. Differential gene expression analysis was performed by comparing grouped experimental samples to their corresponding grouped control samples. Genes were categorized as differentially expressed and statistically significant if they met 95% confidence (Student’s t-test and the Benjamini-Hochberg false discovery rate method) and a cutoff of 1.5-fold change. These differentially expressed genes (DEGs) were also filtered by signal from each sample group and were required to meet a signal cutoff of 0.1 in at least 1 of the 2 groups analyzed. These DEGs were then exported and loaded into the PANTHER Classification System for gene ontology 38 as well as into the Ingenuity Pathway Analysis (IPA) tool core analysis using standard settings with duplicates resolved. Fisher’s exact test (right-tailored) was used to calculate p-values, and activation z-score statistics was used to predict activation or inhibition of a process or the directional effect of one molecule on another. Bar graphs and heat maps were generated using GraphPad Prism version 7 (GraphPad Software, La Jolla, CA USA). IPA analysis comprised of ascertaining canonical pathways and upstream regulators. Additionally, immune-related DEGs were determined by gene lists from the Immunology Database and Analysis Portal and the Immunogenetic Related Information Source and were connected into networks using IPA.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was carried out to validate the expression profile of selected genes identified by RNA-Seq in PBMCs, spleen and lymph node (LN) cells. Candidate genes were selected based on two approaches. Specific primers for qRT-PCR were designed from mRNA sequences obtained from the NCBI for Papio anubis genes CSF1, CSF2, IGLL1, STAT6, PRDM1, TNFRSF, hIGHV; genes defining a TH1 profile (IFNG, IL1, IL2, IL12); a TH2 profile (IL4); and a TH17 profile (IL17A). The list of primer sequences used for qRT-PCR is presented in Supplementary Table 2. Briefly, PBMCs, splenocytes, or LN cells at necropsy were seeded at 1 × 106 cells/well in a 24-well Costar® plate (Corning Inc. Corning NY) in complete medium (RPMI-1640 supplemented with 10% fetal bovine serum, 100 μg/mL penicillin G, 100 μg/ml streptomycin and 10 μg/mL gentamycin). Seeded cells were incubated at 37 °C and 5% CO2 for 24 h. Total RNA was extracted from cells using GenElute™ Mammalian Total RNA Miniprep kit (Sigma Aldrich, St. Louis, MO) according to manufacturer’s instructions. First strand cDNA synthesis was carried out using Maxima First Strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA) according to manufacturer’s protocol. PCR amplification of selected genes was carried out using SYBR Premix Ex Taq™ (TIi RNase H Plus; Takara, Japan) on a StepOne™ plus Real-time PCR system (Thermo Fisher Scientific, Waltham, MA) in a reaction volume of 20 μL and primer concentration of 0.2 μM. Reaction conditions were initial denaturation at 95 °C for 5 mins and then amplification for 40 cycles at 95 °C for 5 seconds/60 °C for 30 seconds. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was utilized as a housekeeping gene. All reactions were carried out in triplicates and results analyzed using DataAssist™ software v3.0 (Thermo Fisher Scientific, Waltham, MA).
Data submission
Data from each experiment were added to an existing NCBI-SRA Database (PRJMA335367) containing Sm-p80 vaccine/animal models data.
Statistical analysis
For each parasite count outcomes (worm reduction and miracidia reduction), a pooled relative risk across study cohorts was estimated using generalized linear mixed negative binomial models with a random intercept for study cohort. A negative binomial model was chosen to adjust for over-dispersion. For miracidia, total eggs with miracidia was used as an offset term. For the normalized egg data (egg numbers in organs and in feces) generalized linear mixed model were estimated using a gamma family and log link with a random intercept for study cohort. For egg data, there were three replicates that were averaged using the mean before model input. For eggs obtained from feces, an additional mean was taken across technically (up to 4). Additionally, to compare egg burden in feces, the sum over 8 weeks post challenge was used as the outcome. Models, relative risks (1 – VE), and p-values were estimated using the lme4 package 39 in the R programming language. Standard errors and mean estimates were estimated from the models using the lsmeans package 40.
Results
Prophylactic, egg-induced pathology and transmission-blocking efficacy
The cumulative 40-baboon study yielded a statistically significant reduction of 65.90% (p <0.0001) in total worm pairs in animals vaccinated with Sm-p80/GLA-SE (Fig. 1A). This protection was consistent across four independent experiments (protection: trial 1, 61.96%; trial 2, 64.16%; trial 3, 69.35%; trial 4, 68.17%). The Sm-p80/GLA-SE vaccine was able to eliminate 93.45% (p<0.0001) of female parasites (Fig. 1A). The vaccine-mediated killing of female worms was similar among all four experiments (female worm killing: trial 1, 93.27%; trial 2, 91.16%; trial 3, 94.01%%; trial 4, 95.19%). The less than 7% of the egg-producing females that survived caused minimal pathology in host tissues/organs, as ascertained by statistically significant (p<0.0001) reduction in egg retention in liver (91.35 %) and small (86.50 %) and large intestine (91.1%) in vaccinated animals (Fig. 1B). Egg expulsion in feces was reduced 35-fold in vaccinated animals, with an average of 22.5 cumulative eggs per gram of feces (EPG) in vaccinated animals, compared with 799 cumulative EPG in control animals measured weekly over 8 weeks post-challenge (p <0.0001, Fig. 1C). In addition, the number of eggs hatching into miracidia (snail-infective stage) was reduced by 81.51% in vaccinated animals (p < 0.0001, Fig. 1D).
Fig. 1.
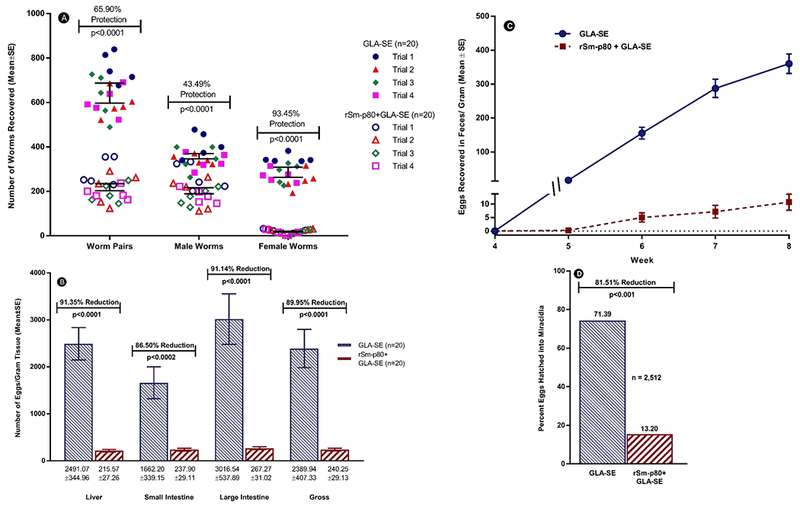
Prophylactic efficacy of Sm-p80–based vaccine. (A) Adult Schistosoma mansoni worm recovered per individual baboon in all four trials. (B) Egg burden per gram of tissue. (C) Weekly fecal egg per gram of feces. (D) Egg viability and hatching rate (n is the total number of eggs examined). Panels B, C and D depict cumulative data from all four trials.
Antibody profiles and kinetics in vaccinated baboons
We assessed the antibody response in baboons immunized with the Sm-p80 vaccine in the four independent trials. Sm-p80-specific serum total IgG was significantly higher in baboons immunized with Sm-p80/GLA-SE (maximum endpoint titers: 64,000–128,000) compared with GLA-SE control animals (maximum endpoint titers: 500–2,000) (Fig. 2A). We also observed a production of Sm-p80-specific IgG1 and IgA antibodies in experimental baboons, compared with the control animals (Fig. 2B and 2D). No significant difference was observed in serum IgM between experimental and control groups (Fig. 2C). In the four trials, we observed similar kinetics of Sm-p80–specific total IgG production following vaccination (Fig. 3); specifically, a significant increase in Sm-p80–specific total IgG was observed after the first immunization at week 4, which continued to increase following each booster at weeks 8 and 12 until challenge at week 16 (mean endpoint titers: trial 1: 107,200; trial 2: 20,400; trial 3: 32,000; trial 4: 48,000) (Fig. 3A–D).
Fig. 2.
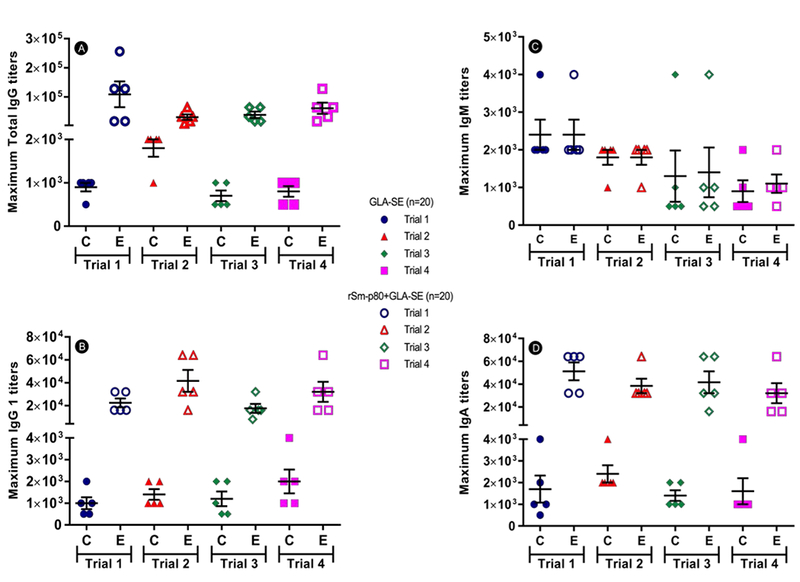
Maximum anti-Sm-p80 antibody titers in baboons following vaccination. (A) Maximum total IgG titers achieved by each baboon in trials 1–4, (B) Maximum IgM titers, (C) Maximum IgG1 titers and (D) maximum IgA titers for each baboon in Trials 1, 2, 3, and 4. “C” represents the control group (GLA-SE), while “E” represents the experimental group (rSm-p80 + GLA-SE). Serum samples were obtained from each baboon prior to immunizations, parasite challenge and at necropsy.
Fig. 3.

Kinetics of total Sm-p80–specific antibody in immunized baboons. Panels A–D show the production of Sm-p80–specific total IgG antibodies in control animals (panel C, GLA-SE) and experimental animals (panel E, rSm-p80 + GLA-SE) in trials 1, 2, 3 and 4, respectively. Endpoint titers were determined for week 0, week 4, week 8, week 12, week 16, and week 24.
Localization of Sm-p80 protein on adult worms
To investigate the expression and distribution of Sm-p80 protein on S. mansoni adult worms recovered from infected baboons (control and experimental groups), immunohistochemistry and confocal microscopy were performed on freshly perfused adult worms. Captured images of adult male and female worms are shown in Fig. 4; Sm-p80 is localized to the surface syncytium of the adult male and female worms, with higher expression observed in the female worms.
Fig. 4.
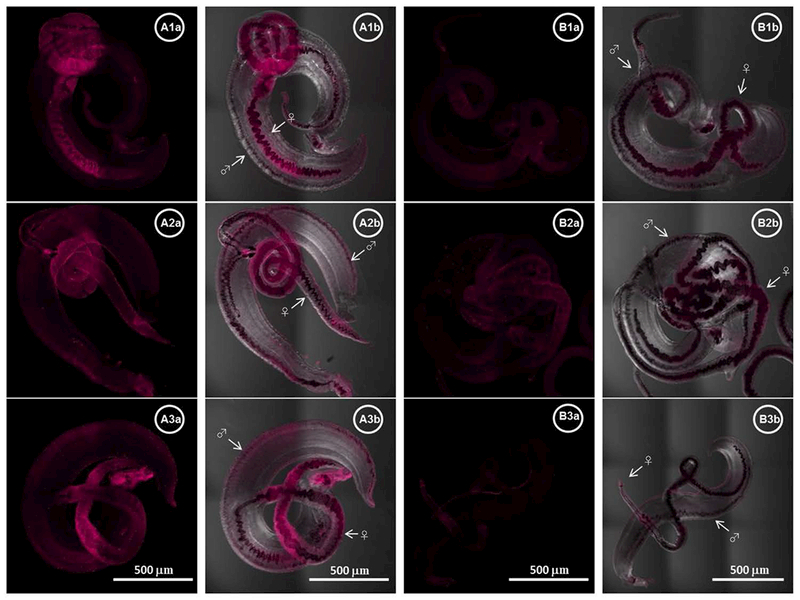
Localization of Sm-p80 in S. mansoni adult worms. Representative stitched images of adult worms are shown in panels A (control) and B (vaccinated). The distribution of Sm-p80 in adult worm recovered from three different baboons from control group is shown in A1a–3a respectively while images B1a–3a represent worms from three different vaccinated baboons (magenta). A1b–3b and B1b–3b show a merged of transmitted detector (TD) light and fluorescent Sm-p80. The images were taken using a Nikon T1-E confocal microscopy with a 10× objective and analyzed with NIS software. The stitched images represent a maximum projection intensity derived from a Z-stack. All images acquired with the same laser power and same gain. The arrows indicate the male (♂) and the female (♀) worms respectively.
Transcriptional profiles following vaccination and parasite challenge
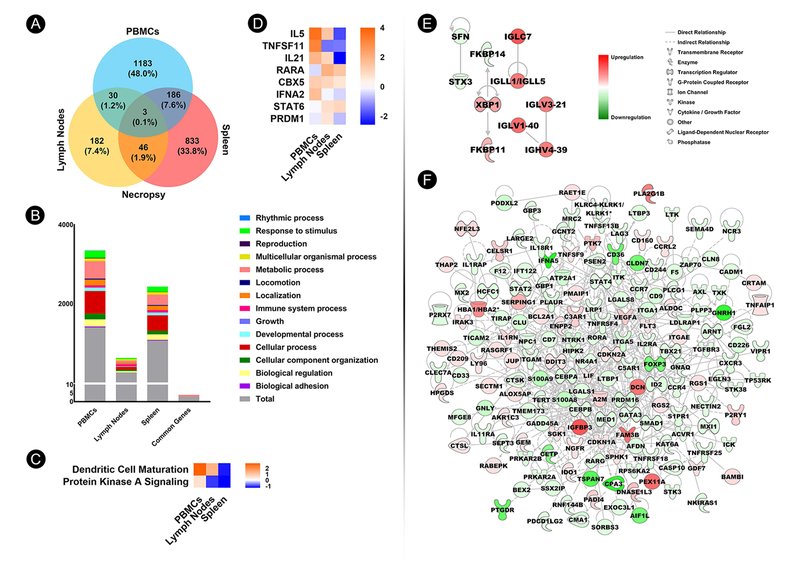
To identify unique transcriptional gene profiles associated with Sm-p80 vaccination that may be predictive of vaccine efficacy, we analyzed differentially expressed genes (DEGs) in PBMCs isolated from baboons at both vaccination and necropsy time points, compared with control animals as a baseline. Comparison of DEGs at vaccination and necropsy showed a total of 149 common to both time points, with 849 genes (37.7%) unique to Sm-p80 vaccination and 1,253 (55.7%) associated with S. mansoni infection in the vaccinated animals at necropsy (p<0.05 and 1.5-fold change cutoff) (Fig. 5A). The distribution of DEGs according to gene ontology classification is illustrated in Figure 5B. Ingenuity Pathway Analysis® (IPA) of common canonical pathways between vaccination and necropsy revealed increasing upregulation of several immune system–related pathways (Fig. 5C). For PBMCs, common upstream regulators were different between vaccination and necropsy (Fig. 5D). Network interaction mapping of immune-related DEGs in baboon PBMCs following Sm-p80 vaccination identified IFNG as a major node of interaction (Fig. 5E) and CD36 gene as a major node of interaction at necropsy (Fig. 5F). IPA also identified upstream regulators with functions associated with innate and TH1-biased immune responses, which were mostly up-regulated (Supplementary Table 3).
Fig. 5.

Identification of molecular signatures of Sm-p80–based vaccine-mediated immunogenicity and immunity in baboon PBMCs following vaccination and challenge infection. (A) Venn diagram depicting numbers and percentages of significant differentially expressed genes (DEGs) (p<0.05; 1.5-fold change cutoff) in Sm-p80–immunized baboons at vaccination and necropsy. (B) Distribution of DEGs (y-axis) in Sm-p80–immunized baboon PBMCs at vaccination and necropsy, as well as DEGs common to both time points according to ontology classification. (C) Heat map depicting IPA-identified common canonical pathways with activation z-scores for which z-score was available and not zero. (D) Heat map depicting IPA-identified common upstream regulators with activation z-scores for which scores were available and not zero. (E) Gene network derived from immune-related DEGs in baboon PBMCs following Sm-p80 vaccination. (F) Gene network derived from immune-related DEGs in baboon PBMCs at necropsy.
We observed variations in DEGs between PBMCs, lymph node, and spleen at necropsy. PBMCs exhibited the highest number of unique DEGs at 1,183 (48.0%), followed by spleen at 833 (33.8%), and lymph nodes at 182 (7.4%) (p<0.0.5 and 1.5-fold change cutoff) (Fig. 6A). The distribution of DEGs in Sm-p80–immunized baboon PBMCs and secondary lymphoid tissues according to ontology classification is depicted in Figure 6B. Gene expression profile across PBMCs and secondary lymphoid organs showed that only 3 DEGs (GDPD3, GYLTL1B, and ZNF331) were common across all tissues (Fig. 6A). The two common IPA-identified canonical pathways were up-regulated in the PBMCs and down-regulated in the spleen at necropsy (Fig. 6C), while the activation profiles of the common upstream regulators differed across all tissues (Fig. 6D). At necropsy, top canonical pathways, upstream regulators, and diseases and functions identified by IPA were mostly tissue specific (Supplementary Table 4). Molecular gene networks derived from immune system–related DEGs from immunized baboon lymph nodes showed activation of the transcription factor gene XBP1 as a major node of interaction (Fig. 6E), while the networks in spleen revealed IGFBP3 as an important node of interaction (Fig. 6F).
Fig. 6.
Systems biology approach to identify molecular signatures in PBMCs, lymph nodes, and spleen. (A) Venn diagram depicting numbers and percentages of significant differentially expressed genes (DEGs) (p<0.05; 1.5-fold change cutoff) in Sm-p80–immunized baboon PBMCs, lymph nodes, and spleen at necropsy. (B) Distribution of DEGs (y-axis) in Sm-p80–immunized baboon PBMCs and secondary lymphoid tissues, as well as DEGs common to all tissues at necropsy according to ontology classification. (C) Heat map depicting IPA-identified common canonical pathways with activation z-score for which scores were available and not zero. (D) Heat map depicting IPA-identified common upstream regulators with activation z-scores for which scores were available and not zero. (E) Gene network derived from immune-related DEGs in baboon lymph nodes at necropsy. (F) Gene network derived from immune-related DEGs in baboon spleen at necropsy.
DEGs validation by qRT-PCR
Twenty genes were selected from the RNA-Seq data and were validated by quantifying expression using qRT-PCR. The expression profile of all genes analyzed by qRT-PCR showed significant similarity to that obtained via RNA-Seq analysis and/or predicted by IPA (Supplementary Table 5). Specifically, the qRT-PCR data demonstrated high-degree matching in activation state (upregulation/downregulation) of all genes in all three tissues (PBMCs, spleen, and LN) to that obtained with RNA sequencing and/or predicted by IPA.
Discussion
The cumulative data obtained from the 40-baboon study described herein yielded a statistically significant reduction in worm pairs in animals vaccinated with Sm-p80/GLA-SE; this protection was consistent across four independent experiments. The uniqueness of these data is that the Sm-p80/GLA-SE vaccine was able to eliminate great majority of egg-producing female parasites, and this preferential killing was similar in all experiments. The ~7% of female worms that survived caused only minimal pathology, as ascertained by substantial reductions in eggs in the livers, small intestines and large intestines of vaccinated animals. Additionally, egg expulsion in feces and the ability of eggs to hatch into miracidia (snail-infective stage) were both significantly reduced in vaccinated animals.
Using these data and incorporating inhibitory effects on parasite mortality, fecundity and infection transmission, mathematical modeling was used to predict the overall potential efficacy of our vaccine on disease transmission dynamics. The macro-parasite transmission dynamics model for the transmission of the parasite to measure outcomes in human populations has predicted that an Sm-p80–based vaccine has the potential to contribute greatly to reducing the burden of schistosomiasis and parasite transmission in endemic regions 10.
Immunolocalization analyses of confocal images demonstrated that Sm-p80 protein is distributed in and on the surface epithelial syncytium of adult worms, with a 2.5-fold increased intensity (p<0.001) in Sm-p80–specific fluorescence in female worms compared with male counterparts. Given that Sm-p80 is the vaccine antigen, we postulate that the up-regulation and increased accessibility of Sm-p80 renders female worms more susceptible than male worms to immune-mediated killing 29. Prophylactic efficacy (Sm-p80/GLA-SE) trials have revealed that IgG and its subclasses appear to play an important role in Sm-p80–based vaccine-mediated protection consistent with previous studies41.
We have attempted to decipher responses following vaccination and challenge at the level of the transcriptomics by identifying molecular signatures and pathways involved in innate and adaptive immunity 28. Predicted activation of immune-related gene networks in PBMCs and secondary lymphoid tissues following vaccination revealed a complex and large repertoire of gene-gene interactions that provide an excellent starting point in identifying the correlates of protection. Important to note in these studies is the involvement of IFNG, which is implicated in killing of invading parasite larvae in lungs 42 and thus could be an important component in conferring immunity 26, 43. However, additional studies are necessary to support the association among the immune gene profiles identified and Sm-p80–mediated immune protection. Ultimately, systems biology approaches may provide more time-efficient and potentially noninvasive methods to predict, confirm, interpret and further investigate results of conventional methods of evaluating vaccine efficacy.
The Sm-p80–based vaccine provides a comprehensive protection against different stages of the parasitic life cycle, including egg excretion and hatching, schistosomula maturation and killing of adult worms, especially against female worms. Based on previously published data, the beneficial outcomes of the Sm-p80–based vaccine include prophylactic efficacy against Schistosoma mansoni (intestinal/hepatic schistosomiasis)26, relief from egg-induced tissue/organ pathology27, 44, post-exposure therapeutic efficacy by elimination of established adult worms in chronic disease45 and cross species-protection against both S. haematobium (urinary schistosomiasis)36 and S. japonicum (Asiatic/oriental disease).29 The vaccine induces long-term immune responses, with Sm-p80–specific IgG titers present in mice for up to 60 weeks in baboons for up to 5–8 years and maternal transfer of protective antibodies 46. Importantly, Sm-p80–specific IgE has not been detected in high-risk or infected populations from Africa 26 and South America 47, likely reducing the possibility of hypersensitivity following vaccination in humans. Furthermore, the GLA-SE adjuvant has been scaled up to millions of doses, is in multiple clinical trials for NTD vaccines and is used at a low 10 μg dose, thereby representing an economically feasible solution for a significant global health problem 14.
In summary, the Sm-p80–based vaccine results in preferential killing of female worms, considerably diminished egg-induced pathology and enhanced transmission blocking. We postulate that the Sm-p80–based vaccine, in addition to inducing adaptive immunity, will facilitate the continuation of natural immunity/resistance against schistosomes by retaining some non-pathogenic male worms. In the absence of female worms, growth of male parasites is stunted, with considerably shorter life spans, compared with that of paired worms (5 to 30 years) 48, 49, 50. Based on promising characteristics and analyses described above, the Sm-p80/GLA-SE vaccine is poised to enter phase 1 human clinical testing.
Supplementary Material
Supplementary Table 1. Protocol for pre-clinical vaccine trial in baboons: randomization, immunization, challenge and necropsy schedule for prophylactic efficacy determination.
Supplementary Table 2. List of primer nucleotide sequences used for qRT-PCR.
Supplementary Table 3. Top canonical pathways, upstream regulators, and diseases, and functions in baboon PBMCs at vaccination and necropsy.
Supplementary Table 4. Top canonical pathways, upstream regulators, and diseases and functions in baboon PBMCs, lymph node, and spleen at necropsy.
Supplementary Table 5. RT-qPCR validation of differentially expressed genes (DEGs) selected from Figs 5 and 6.
Acknowledgements
This work was supported by grants from the Bill and Melinda Gates Foundation Grant (OPP1097535) and from the NIAID/NIH SBIR (R43/R44 AI103983). The snails were supplied through a NIH-NIAID contract (HHSN2722010000051) to the Biomedical Research Institute. The Sm-p80/GLA-SE vaccine has been trademarked (D.C.) as SchistoShield®
Footnotes
Competing interests
All authors declare no competing interests.
References
- 1.Bergquist R, Utzinger J Keiser J. 2017. Controlling schistosomiasis with praziquantel: How much longer without a viable alternative? Infect Dis Poverty 6: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colley DG, Bustinduy AL, Secor WE et al. 2014. Human schistosomiasis. Lancet 383: 2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui AA & Siddiqui SZ. 2017. Sm-p80-Based Schistosomiasis Vaccine: Preparation for Human Clinical Trials. Trends Parasitol 33: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cioli D, Pica-Mattoccia L, Basso A et al. 2014. Schistosomiasis control: praziquantel forever? Mol Biochem Parasitol 195: 23–29. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J 2016. Unfilled Vials. Science 351: 16–19. [DOI] [PubMed] [Google Scholar]
- 6.King CH 2017. The evolving schistosomiasis agenda 2007–2017-Why we are moving beyond morbidity control toward elimination of transmission. PLoS Negl Trop Dis 11: e0005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergquist R, Zhou XN, Rollinson D et al. 2017. Elimination of schistosomiasis: the tools required. Infect Dis Poverty 6: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mo AX, Agosti JM, Walson JL et al. 2014. Schistosomiasis elimination strategies and potential role of a vaccine in achieving global health goals. Am J Trop Med Hyg 90: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mo AX & Colley DG. 2016. Workshop report: Schistosomiasis vaccine clinical development and product characteristics. Vaccine 34: 995–1001. [DOI] [PubMed] [Google Scholar]
- 10.Stylianou A, Hadjichrysanthou C, Truscott JE et al. 2017. Developing a mathematical model for the evaluation of the potential impact of a partially efficacious vaccine on the transmission dynamics of Schistosoma mansoni in human communities. Parasit Vectors 10: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alsallaq RA, Gurarie D, Ndeffo Mbah M et al. 2017. Quantitative assessment of the impact of partially protective anti-schistosomiasis vaccines. PLoS Negl Trop Dis 11: e0005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riveau G, Deplanque D, Remoue F et al. 2012. Safety and immunogenicity of rSh28GST antigen in humans: phase 1 randomized clinical study of a vaccine candidate against urinary schistosomiasis. PLoS Negl Trop Dis 6: e1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran MH, Pearson MS, Bethony JM et al. 2006. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat Med 12: 835–840. [DOI] [PubMed] [Google Scholar]
- 14.Santini-Oliveira M, Coler RN, Parra J et al. 2016. Schistosomiasis vaccine candidate Sm14/GLA-SE: Phase 1 safety and immunogenicity clinical trial in healthy, male adults. Vaccine 34: 586–594. [DOI] [PubMed] [Google Scholar]
- 15.Tendler M, Almeida M & Simpson A. 2015. Development of the Brazilian Anti Schistosomiasis Vaccine Based on the Recombinant Fatty Acid Binding Protein Sm14 Plus GLA-SE Adjuvant. Front Immunol 6: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqui AA, Ahmad G, Damian RT et al. 2008. Experimental vaccines in animal models for schistosomiasis. Parasitol Res 102: 825–833. [DOI] [PubMed] [Google Scholar]
- 17.Wilson RA, Li XH & Castro-Borges W. 2016. Do schistosome vaccine trials in mice have an intrinsic flaw that generates spurious protection data? Parasit Vectors 9: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddiqui AJ, Molehin AJ, Zhang W et al. 2018. Sm-p80-based vaccine trial in baboons: efficacy when mimicking natural conditions of chronic disease, praziquantel therapy, immunization, and Schistosoma mansoni re-encounter. Ann N Y Acad Sci. 10.1111/nyas.13866 [DOI] [PubMed] [Google Scholar]
- 19.Ono Y, Saido TC & Sorimachi H. 2016. Calpain research for drug discovery: challenges and potential. Nat Rev Drug Discov 15: 854–876. [DOI] [PubMed] [Google Scholar]
- 20.Sorimachi H, Ishiura S & Suzuki K. 1993. A novel tissue-specific calpain species expressed predominantly in the stomach comprises two alternative splicing products with and without Ca(2+)-binding domain. J Biol Chem 268: 19476–19482. [PubMed] [Google Scholar]
- 21.Braschi S & Wilson RA. 2006. Proteins exposed at the adult schistosome surface revealed by biotinylation. Mol Cell Proteomics 5: 347–356. [DOI] [PubMed] [Google Scholar]
- 22.Siddiqui AA, Zhou Y, Podesta RB et al. 1993. Characterization of Ca(2+)-dependent neutral protease (calpain) from human blood flukes, Schistosoma mansoni. Biochim Biophys Acta 1181: 37–44. [DOI] [PubMed] [Google Scholar]
- 23.Griffith Q, Liang Y, Whitworth P et al. 2015. Immuno-evasive tactics by schistosomes identify an effective allergy preventative. Exp Parasitol 153: 139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Da’dara AA & Skelly PJ. 2017. The human blood parasite Schistosoma mansoni expresses extracellular tegumental calpains that cleave the blood clotting protein fibronectin. Sci Rep 7: 12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois Cauwelaert N, Desbien AL, Hudson TE et al. 2016. The TLR4 Agonist Vaccine Adjuvant, GLA-SE, Requires Canonical and Atypical Mechanisms of Action for TH1 Induction. PLoS One 11: e0146372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad G, Zhang W, Torben W et al. 2011. Preclinical prophylactic efficacy testing of Sm-p80-based vaccine in a nonhuman primate model of Schistosoma mansoni infection and immunoglobulin G and E responses to Sm-p80 in human serum samples from an area where schistosomiasis is endemic. J Infect Dis 204: 1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Ahmad G, Torben W et al. 2010. Sm-p80-based DNA vaccine made in a human use approved vector VR1020 protects against challenge infection with Schistosoma mansoni in mouse. Parasite Immunol 32: 252–258. [DOI] [PubMed] [Google Scholar]
- 28.Haks MC, Bottazzi B, Cecchinato V et al. 2017. Molecular Signatures of Immunity and Immunogenicity in Infection and Vaccination. Front Immunol 8: 1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molehin AJ, Sennoune SR, Zhang W et al. 2017. Cross-species prophylactic efficacy of Sm-p80-based vaccine and intracellular localization of Sm-p80/Sm-p80 ortholog proteins during development in Schistosoma mansoni, Schistosoma japonicum, and Schistosoma haematobium. Parasitol Res 116: 3175–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rojo JU, Melkus MW, Kottapalli KR et al. 2017. Sm-p80-based schistosomiasis vaccine mediated epistatic interactions identified potential immune signatures for vaccine efficacy in mice and baboons. PLoS One 12: e0171677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmad G, Zhang W, Torben W et al. 2009. Protective and antifecundity effects of Sm-p80-based DNA vaccine formulation against Schistosoma mansoni in a nonhuman primate model. Vaccine 27: 2830–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmad G, Zhang W, Torben W et al. 2010. Protective effects of Sm-p80 in the presence of resiquimod as an adjuvant against challenge infection with Schistosoma mansoni in mice. Int J Infect Dis 14: e781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damian RT, Greene ND & Fitzgerald K. 1972. Schistosomiasis mansoni in baboons. The effect of surgical transfer of adult Schistosoma mansoni upon subsequent challenge infection. Am J Trop Med Hyg 21: 951–958. [PubMed] [Google Scholar]
- 34.Tucker MS, Karunaratne LB, Lewis FA et al. 2013. Schistosomiasis. Curr Protoc Immunol 103: Unit 19 11. [DOI] [PubMed] [Google Scholar]
- 35.Long EG, Tsin AT & Robinson BA. 1985. Comparison of the FeKal CON-Trate system with the formalin-ethyl acetate technique for detection of intestinal parasites. J Clin Microbiol 22: 210–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karmakar S, Zhang W, Ahmad G et al. 2014. Cross-species protection: Schistosoma mansoni Sm-p80 vaccine confers protection against Schistosoma haematobium in hamsters and baboons. Vaccine 32: 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frey A, Di Canzio J & Zurakowski D. 1998. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods 221: 35–41. [DOI] [PubMed] [Google Scholar]
- 38.Mi H, Muruganujan A, Casagrande JT et al. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bates D, Machler M, Bolker B et al. 2015. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- 40.Lenth RV 2016. Least-Squares Means: The R Package lsmeans. Journal of Statistical Software 69: 1–33. [Google Scholar]
- 41.Torben W, Ahmad G, Zhang W et al. 2011. Role of antibodies in Sm-p80-mediated protection against Schistosoma mansoni challenge infection in murine and nonhuman primate models. Vaccine 29: 2262–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mountford AP, Coulson PS, Pemberton RM et al. 1992. The generation of interferon-gamma-producing T lymphocytes in skin-draining lymph nodes, and their recruitment to the lungs, is associated with protective immunity to Schistosoma mansoni. Immunology 75: 250–256. [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson RA, Coulson PS & Mountford AP. 1999. Immune responses to the radiation-attenuated schistosome vaccine: what can we learn from knock-out mice? Immunol Lett 65: 117–123. [DOI] [PubMed] [Google Scholar]
- 44.Ahmad G, Zhang W, Torben W et al. 2009. Prime-boost and recombinant protein vaccination strategies using Sm-p80 protects against Schistosoma mansoni infection in the mouse model to levels previously attainable only by the irradiated cercarial vaccine. Parasitol Res 105: 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karmakar S, Zhang W, Ahmad G et al. 2014. Use of an Sm-p80-based therapeutic vaccine to kill established adult schistosome parasites in chronically infected baboons. J Infect Dis 209: 1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Ahmad G, Le L et al. 2014. Longevity of Sm-p80-specific antibody responses following vaccination with Sm-p80 vaccine in mice and baboons and transplacental transfer of Sm-p80-specific antibodies in a baboon. Parasitol Res 113: 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaze S, Driguez P, Pearson MS et al. 2014. An immunomics approach to schistosome antigen discovery: antibody signatures of naturally resistant and chronically infected individuals from endemic areas. PLoS Pathog 10: e1004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnon R 1990. Life span of parasite in schistosomiasis patients. Isr J Med Sci 26: 404–405. [PubMed] [Google Scholar]
- 49.Fulford AJ, Butterworth AE, Ouma JH et al. 1995. A statistical approach to schistosome population dynamics and estimation of the life-span of Schistosoma mansoni in man. Parasitology 110 ( Pt 3): 307–316. [DOI] [PubMed] [Google Scholar]
- 50.Kunz W 2001. Schistosome male-female interaction: induction of germ-cell differentiation. Trends Parasitol 17: 227–231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Protocol for pre-clinical vaccine trial in baboons: randomization, immunization, challenge and necropsy schedule for prophylactic efficacy determination.
Supplementary Table 2. List of primer nucleotide sequences used for qRT-PCR.
Supplementary Table 3. Top canonical pathways, upstream regulators, and diseases, and functions in baboon PBMCs at vaccination and necropsy.
Supplementary Table 4. Top canonical pathways, upstream regulators, and diseases and functions in baboon PBMCs, lymph node, and spleen at necropsy.
Supplementary Table 5. RT-qPCR validation of differentially expressed genes (DEGs) selected from Figs 5 and 6.