Abstract
Mitochondria are key cell organelles with a prominent role in both energetic metabolism and the maintenance of cellular homeostasis. Since mitochondria harbor their own genome, which encodes a limited number of proteins critical for oxidative phosphorylation and protein translation, their function and biogenesis strictly depend upon nuclear control. The yeast Saccharomyces cerevisiae has been a unique model for understanding mitochondrial DNA organization and inheritance as well as for deciphering the process of assembly of mitochondrial components. In the last three decades, yeast also provided a powerful tool for unveiling the communication network that coordinates the functions of the nucleus, the cytosol and mitochondria. This crosstalk regulates how cells respond to extra- and intracellular changes either to maintain cellular homeostasis or to activate cell death. This review is focused on the key pathways that mediate nucleus–cytosol–mitochondria communications through both transcriptional regulation and proteostatic signaling. We aim to highlight yeast that likely continues to serve as a productive model organism for mitochondrial research in the years to come.
Keywords: yeast, mitochondrial biogenesis, mitochondria–nucleus communication, mPOS, proteostasis, retrograde regulation
An overview of our current knowledge on nucleus–cytosol–mitochondria cross-talk in yeast highlighting this unicellular organism as a premier model for mitochondrial research.
ABBREVIATIONS
- AMPK:
AMP-activated protein kinase
- ATFS-1:
activating transcription factor associated with stress
- ATP:
adenosine Triphosphate
- cAMP:
cyclic AMP
- CCR:
carbon catabolite repression
- ER:
endoplasmic reticulum
- ERα:
estrogen receptor alpha
- ETC:
electron transport chain
- FOXOs:
forkhead family of transcription factors
- HSPs:
heat shock proteins
- IMM:
inner mitochondrial membrane
- ISC:
iron-sulfur clusters
- MAD:
mitochondria-associated degradation
- MAGIC:
mitochondria as guardian in cytosol
- mPOS:
mitochondrial precursor overaccumulation stress
- mtDNA:
mitochondrial DNA
- OMM:
outer mitochondrial membrane
- OXPHOS:
oxidative phosphorylation
- PDR:
pleiotropic drug resistance
- PDS:
postdiauxic shift
- PKA:
protein Kinase A
- RCD:
regulated cell death
- rDNA:
extrachromosomal ribosomal-DNA
- RLS:
replicative lifespan
- ROS:
reactive oxygen species
- RTG:
retrograde genes or pathway
- SAPK:
stress-activated protein kinase
- SLIK:
histone acetyltransferase-coactivator SAGA-like complex
- TCA:
tricarboxylic acid cycle
- TOR:
target of Rapamycin
- UPRam:
unfolded protein response activated by mistargeting of proteins
- UPRmt:
mitochondrial unfolded protein response
INTRODUCTION
Since the discovery of cytochrome c release from mitochondria as a key step in the initiation of apoptotic cell death more than 20 years ago (Liu et al.1996), mitochondrial research has experienced a tremendous boost. Researchers have been gathering a growing wealth of knowledge recognizing the central role of these organelles in the maintenance of eukaryotic cell homeostasis. This role is not restricted to the generation of intermediary metabolites and the production of ATP1 through the tricarboxylic acid cycle (TCA) and oxidative phosphorylation (OXPHOS). Not only can mitochondria synthesize fundamental molecules, such as heme and iron-sulfur clusters (Sun, Cheng and Chen 2015; Braymer and Lill 2017), but they are also major sites of amino acid, nucleotide and fatty acid metabolism and can receive, integrate and relay intracellular signals (Goldenthal and Marin-Garcia 2004; Chandel 2014). Moreover, it is becoming increasingly clear that mitochondrial and cytosolic proteostasis are intimately related (Wang et al.2008; Wang and Chen 2015; Suhm et al.2018). These processes can also play a role in shaping cell fate. In view of this and with the discovery of pathogenic mitochondrial DNA defects in the 1980’s, mitochondrial dysfunction is now recognized as a common factor underlying many pathological conditions (Picard, Wallace and Burelle 2016). Many of these advancements would not have been possible without the model organism Saccharomyces cerevisiae.
Mitochondria are semiautonomous organelles. They carry their own small DNA genome (mtDNA), a legacy of the endosymbiosis, the process which plausibly originated mitochondria and the eukaryotic cell (Lane and Martin 2010). The budding yeast S. cerevisiae has been instrumental in understanding mtDNA organization and inheritance (Schatz, Haslbrunner and Tuppy 1964; Chen and Butow 2005). Yeast mtDNA is a circular 75–85 kbp molecule that encodes only a small fraction of genes required for mitochondrial function (Foury et al.1998). However, most mitochondrial proteins (Morgenstern et al.2017) are encoded by nuclear genes, synthesized in the cytosol and then targeted to the mitochondrial compartments. Thus, mitochondrial biogenesis and functions are under tight nuclear control, through the so-called anterograde regulation of gene expression. This involves signaling pathways that coordinate gene transcription to finely tune metabolic requirements with nutritional and environmental cues. Importantly, a whole-cell genomic profiling approach has revealed that mitochondrial and cytosolic translation are rapidly, dynamically and synchronously regulated by the nuclear genome to orchestrate the timely synthesis of OXPHOS complexes (Couvillion et al.2016). Therefore, nuclear-encoded mitochondrial protein import machinery, which has been extensively reviewed elsewhere (Schmidt, Pfanner and Meisinger 2010; Harbauer et al.2014; Neupert 2015; Wiedemann and Pfanner 2017), represents an important branch of the anterograde regulation that facilitates the nuclear control of mitochondrial activities.
Saccharomyces cerevisiae is a so-called petite-positive yeast, meaning that it can survive the elimination of mtDNA to form ‘petite’ colonies. Saccharomyces cerevisiae cells can proliferate in the absence of OXPHOS as long as a fermentable carbon source is made available for ATP synthesis via the glycolytic pathway. This property makes this yeast a powerful tool for dissecting the anterograde pathways that are required for mtDNA replication and transcription, mitochondrial protein synthesis and metabolite transport (Contamine and Picard 2000; Chen 2013; Chen and Clark-Walker 2018). On the other hand, most yeast species are petite-negative. In this case, loss of mtDNA is lethal. The use of these yeasts (e.g. Kluyveromyces lactis) has enabled investigators to establish the critical factors such as the inner membrane potential as a key determinant of cell survival upon mitochondrial damage (Chen and Clark-Walker 1993, 1995, 1996, 2000).
On the other hand, environmental changes trigger intracellular stress responses, which may disturb mitochondrial structure and/or function. To maintain cell homeostasis, damaged mitochondria relay signals through retrograde, as opposed to anterograde, communication pathways that drive specific nuclear gene transcription patterns in response to stress. If the mitochondrial stress is severe, retrograde communication can even promote the elimination of damaged mitochondria through a selective form of autophagy, named mitophagy (Kanki, Furukawa and Yamashita 2015; Wei, Liu and Chen 2015). It is worth noting that among the pathophysiologically relevant pathways that can easily be modeled in yeast are those regulating cell demise. Indeed, mammalian cells and yeast share some commons mechanisms of cell death. The use of yeast helped to uncover and establish factors and pathways involved in mammalian apoptosis and other controlled cell death subroutines [for Refs see Carmona-Gutierrez et al. (2018)].
Here we will focus on the genes, proteins and mechanisms identified in yeast that regulate cell homeostasis through a complex network of multi-directional signaling pathways among mitochondria, the cytoplasm and the nucleus. Specifically, we cover our current understanding of how these pathways are integrated to regulate transcription and maintain cell-wide proteostasis and metabolic homeostasis.
MITOCHONDRIA–NUCLEUS COMMUNICATION THROUGH ANTEROGRADE TRANSCRIPTIONAL REGULATION
Saccharomyces cerevisiae, being a facultative aerobe, is able to adapt its metabolism, i.e. fermentation versus respiration, according to the nature of available carbon substrates. When glucose is present as the sole carbon source, glycolysis and fermentation prevail. This phenomenon is named carbon catabolite repression that inhibits the transcription of a large set of genes involved in mitochondrial respiratory metabolism (Gancedo 1998; Schuller 2003; Conrad et al.2014; Kayikci and Nielsen 2015). In the presence of alternative carbon sources or when glucose is exhausted in the medium, as during the diauxic shift, carbon catabolite repression is relieved and mitochondrial respiration is activated to consume non-fermentable and non-glucose carbon substrates. Thus, it is apparent that nutrient selection and availability strongly affect mitochondrial biogenesis and activity. In this frame, fine-tuning of gene transcription allows rapid metabolic adaptations and appropriate growth under changing environments. The highly conserved nutrient-sensing pathways along the cyclic AMP (cAMP)-Protein Kinase A (PKA), Target of Rapamycin (TOR)-Sch9 and Snf1-Mig1 axis, play central roles in the regulation of mitochondrial biogenesis and activity, and consequently of cell growth and proliferation (Fig. 1).
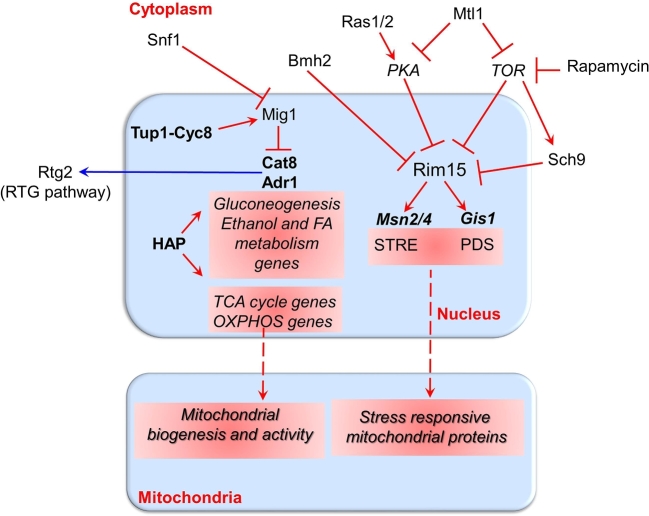
Figure 1.
Anterograde communication pathways regulating gene transcription in yeast. Key yeast genes and proteins involved in the transcriptional regulatory network of signaling pathways among the nucleus, the cytoplasm and mitochondria (see text for details). The nucleus regulates biosynthesis and function of mitochondria through anterograde pathways (red lines), which are also sensitive to carbon and nitrogen sources. Glucose and preferred nitrogen sources activate PKA and TOR kinases and inhibit Snf1. Rim15 and of Mig1 are regulated by shuttling between nucleus and cytoplasm. Anterograde regulation control the transcription of genes encoding proteins destined to mitochondria (red dashed lines). Nuclear genes highlighted in red are upregulated by the bolded transcription factors located above them. Cat8 and Adr1 can also regulate activation of RTG-dependent retrograde communication pathway (see Fig. 2, blue line).
cAMP-PKA and TOR-Sch9 pathways
Mitochondrial processes are tightly integrated with nutrient signaling pathways in order to enable rapid modulation of mitochondrial functions upon fluctuations in the intra- and extracellular environment. In turn, cells adapt cellular metabolism to changes in mitochondrial structure and function (Chandel 2014; Yun and Finkel 2014). PKA and TOR play the prominent role in determining how cells respond to the nutritional state. Consequently, these two proteins strongly influence both mitochondrial biogenesis and activity (Fig. 1). Important links have been established between Ras/cAMP/PKA signaling, nutritional sensing, mitochondrial biogenesis and ROS production. Ras1 and Ras2 proteins control adenylate cyclase activity and PKA is a highly conserved cAMP-dependent kinase that has a wide range of cellular effects centered on nutrient metabolism. In response to glucose signal, hyperactivation of PKA has been shown to induce transcriptional changes that reduce mitochondrial biogenesis, repress the electron transport chain (ETC), inhibit stress response mechanisms and lead to a regulated form of cell death (Leadsham and Gourlay 2010). PKA hyperactivation is associated with down-regulation of certain genes encoding components of the ETC, such as both external mitochondrial NADH dehydrogenases, NDE1 and NDE2, SDH2, a component of succinate dehydrogenase, two complex III subunits, CYC1, COX4, a subunit of cytochrome c oxidase and a component of ATP synthase. In addition, the expression of a number of regulatory components of the ETC was also reduced, including COQ2, which is required for ubiquinone biosynthesis; CYT2, an enzyme required for cytochrome c maturation; COX10 and SCO1, both required for Cytochrome c Oxidase formation; and NCA3 which regulates expression of the mitochondrially encoded ATP synthase subunits 6 and 8. The transcriptional repression of these mitochondrial genes is mediated by HAP4 (see below), SOK2 and SKO1, since their deletion suppresses PKA-dependent loss of mitochondrial function and production of ROS.
Under optimal growth conditions, i.e. high glucose availability and absence of cellular stress, PKA and TOR promote protein synthesis and ribosome biogenesis, as well as cell cycle progression (Loewith and Hall 2011). This is associated with reduced mitochondrial respiration that correlates with the reduced ETC component expression discussed above. PKA and TOR activities are reduced on non-fermentable carbon sources and under environmental conditions requiring cell cycle arrest. This reduction is concomitant with an increase in respiratory function. Mtl1 is critical for one branch of environment sensing upstream of PKA and TOR (Fig. 1). The absence of Mtl1, a member of the cell wall integrity pathway that acts as a sensor for oxidative stress and glucose starvation, severely impairs respiration and mitochondrial activation in the diauxic shift caused by downregulation of TORC1-Sch9 and PKA signaling pathways (Petkova et al.2010; Sundaram et al.2015). Therefore nutrient sensing involving MTL1 induces environment-dependent metabolic changes through TORC1, Sch9 and PKA signaling pathways.
A correlation between TOR signaling and mitochondrial activity has also been established (Pan et al.2011). Yeast strains with reduced TOR activity have enhanced mitochondrial ETC activity during growth that is efficiently coupled to ATP production. This metabolic alteration was also shown to increase mitochondrial membrane potential and superoxide production that was proposed to provide an adaptive signal enhancing survival in stationary phase, a condition where TOR activity is inhibited. Superoxide production downstream of reduced TOR was also proposed to extend chronological life span in these yeast strains, which also display reduction in cytosolic protein synthesis (see below). The same link between oxidative stress and reduced TOR signaling was found in yeast cells lacking the conserved mitochondrial protease Oma1. During logarithmic growth, transient accumulation of high levels of ROS contributes to the attenuation of TOR signaling in oma1Δ cells. Reduced signaling through the TOR-downstream effector Rim15 (Fig. 1), but not Sch9 or Rtg1 (see below) attenuates Msn2/Msn4-dependent gene expression thus compromising antioxidant defense mechanisms (Bohovych et al.2016). Indeed, oxidative stress responsive genes, such as CTT1, encoding the cytosolic catalase or SOD2, encoding the mitochondrial superoxide dismutase, are regulated by Msn2p/Msn4p (Werner-Washburne et al.1993; Herman 2002; Fabrizio et al.2003; Gray et al.2004). On the other hand, combined physiological and gene expression analysis has revealed transcriptional activation of both stress responsive MSN2/MSN4-target genes and genes involved in mitochondrial metabolism during starvation suggesting that stress response and aerobic metabolism are transcriptionally coordinated. It has been hypothesized that this could be due to TOR downregulation (Petti et al.2011).
In yeast, TOR inactivation by either nitrogen starvation or rapamycin treatment results in growth arrest that is associated with physiological changes, which are characteristic of stationary phase cells. These include G1 cell cycle arrest, repression of general transcription and mRNA translation, induction of stress response genes and synthesis of glycogen and trehalose (Werner-Washburne et al.1993; Jacinto and Hall 2003). Thus, stressed cells share transcriptional similarities with diauxic shift and stationary-phase cells, and the TORC1-Rim15-Msn2/Msn4 axis plays an important role in the metabolic remodeling through nucleus–mitochondria communications. Rim15, which is a common downstream effector of PKA and TOR pathways, plays a primary role in integrating nutrient signals for general transcription reprogramming (Fig. 1).
TOR regulates the nucleocytoplasmic distribution of Rim15 directly or indirectly through the yeast AGC kinase Sch9, homologue of mammalian S6 kinase 1 and protein kinase B (PKB/Akt) (Pedruzzi et al.2003; Powers 2007; Swinnen et al.2014) (Fig. 1). PKA negatively regulates Rim15 by causing its exclusion from the nucleus through phosphorylation. This results in the transcriptional inhibition of a wide array of stress-inducible genes and stationary-phase related genes, whose expression is controlled by the two partially redundant transcription factors Msn2/Msn4 and by Gis1, respectively (Fig. 1) (Reinders et al.1998; De Virgilio 2012). In particular, Msn2/4 regulate the transcription of a wide array of stress-response and tolerance genes, such as SOD2, that contain a STRE-element in their promoter and are expressed during respiratory growth and in stationary phase (Martinez-Pastor et al.1996). Interestingly, Msn2 transcriptional activity is sensitive to environmental signals and controlled at multiple levels, including phosphorylation, dephosphorylation, nuclear localization and degradation (Sadeh et al.2011). Gis1 is involved in the transition from diauxic shift to stationary phase and promotes the expression of genes with postdiauxic shift elements in their promoter (Lenssen et al.2002, 2005).
Independent of the growth phase, mitochondrial dysfunction due to the lack of mtDNA alters phosphorylation of certain factors, including Rim15 and Msn2, thus interfering with metabolic reprogramming associated with acute glucose starvation (Friis and Schultz 2016). This suggests a retrograde mechanism whereby mitochondrial dysfunction affects the transcription of nuclear-encoded mitochondrial genes. Mitochondrial retrograde regulation is addressed in subsequent sections.
Overall, these evidences indicate that integration of signals transmitted via at least three key nutrient-sensory kinases, i.e. TOR, PKA and Sch9 fine-tunes Rim15-controlled developmental processes. Crucial for nuclear import of Rim15 is the release of its binding to the 14-3-3 protein Bmh2 in the cytoplasm occurring through the de-phosphorylation of a specific threonine residue after inactivation of either TORC1 or the nutrient-signaling Pho80–Pho85 cyclin–cyclin‐dependent kinase complex (Wanke et al.2005). As described, Rim15-dependent metabolic reprogramming has important anterograde effects on mitochondrial function.
Snf1-Mig1 pathway
Snf1, the yeast orthologue of mammalian AMP-activated protein kinase (AMPK), together with Rim15 is a positive regulator of transcription of genes involved in the transition into stationary phase through the diauxic shift (Fig. 1). It can be considered a cellular energy sensor for generation and preservation of energy and a master regulator of metabolism in response to glucose limitation and other environmental stresses (Hedbacker and Carlson 2008). The SNF1 complex controls transcription of those genes mainly related to the utilization of alternative carbon sources, gluconeogenesis, glyoxylate cycle, salt stress and heat shock. Relevantly, the SNF1 complex plays an indirect role on mitochondrial functions through post-translational modulation of high energy-demand processes, such as protein synthesis and lipid metabolism, and stimulation of mitochondrial fatty acid oxidation (Sanz, Viana and Garcia-Gimeno 2016). SNF1-mediated transcription occurs through several mechanisms, including phosphorylation of transcriptional activators and repressors, chromatin modification and interference with preinitiation complex assembly. Mig1 is one of the best-studied targets of SNF1; when it is phosphorylated and retained in the cytoplasm, i.e. under low glucose conditions, the expression of hundreds of genes involved in the use of alternate carbon sources is permitted. Vice versa, Mig1 in a dephosphorylated state is localized in the nucleus and can act as a transcriptional repressor of these genes, usually in association with the corepressor Ssn6 (Cyc8)-Tup1 (Treitel and Carlson 1995). The localization of Mig1p is highly sensitive to glucose fluctuations in the medium as judged by rapid shuttling cytoplasm–nucleus in response to glucose concentration (De Vit, Waddle and Johnston 1997) (Fig. 1).
Another downstream target of Snf1p is Adr1, which is activated during the diauxic shift when glucose is exhausted or under low glucose conditions (Schuller 2003). The Snf1-Adr1 axis controls the expression of mitochondrial genes involved in ethanol and glycerol utilization and β-oxidation of fatty acids (Ratnakumar and Young 2010), leading to increased ethanol and fatty acid metabolism under low glucose conditions.
When glucose is absent, SNF1 complex phosphorylates and activates Cat8, Sip4 and Rds2 transcription factors allowing the expression of gluconeogenic genes (Vincent and Carlson 1999) (Fig. 1). CAT8 transcription is also inhibited by Mig1 and activated by the heteromultimeric HAP2/3/4/5 (HAP) complex. The HAP complex is a master transcriptional regulator of mitochondrial respiration and metabolism. It controls the expression of a vast array of mitochondrial proteins, including cytochrome c, TCA cycle enzymes and ETC complex subunits, as well as some putative transcriptional activators of unknown function (Buschlen et al.2003). Although the regulatory network activating the HAP complex during the diauxic shift (DeRisi, Iyer and Brown 1997) is not yet fully elucidated, Rds2 was shown to up-regulate HAP4, which encodes the limiting and activating subunit of the heteromeric transcriptional regulator [for Refs see Turcotte et al. (2010)]. Heme may also play a role in HAP complex activation during the diauxic shift. Interestingly, recent evidences indicate that the glucose-mediated repression of respiration in yeast is at least partly due to the low cellular heme level. Increased heme synthesis, even under conditions of glucose repression, activated Hap1p and the HAP complex and induced transcription of HAP4, leading to a switch from fermentation to respiration (Zhang et al.2017).
In the context of anterograde regulation, it is worth mentioning that Snf1 negatively regulates PKA-dependent transcription (Nicastro et al.2015). Snf1 itself as well as active Ras and certain components of cAMP/PKA, such as Ira2, Cyr1 and Bcy1, may localize to mitochondria under stress conditions (Belotti et al.2012; Strogolova et al.2012; Amigoni, Martegani and Colombo 2013; Galello, Moreno and Rossi 2014), although their intra-mitochondrial function is unknown.
MITOCHONDRIA–NUCLEUS COMMUNICATION THROUGH RETROGRADE TRANSCRIPTIONAL REGULATION
Mitochondria-to-nucleus communication, termed retrograde signaling, is in general, but not necessarily, activated by mitochondrial dysfunction, such as alterations in mitochondrial membrane potential, mutations in TCA cycle or OXPHOS genes, and complete loss or partial deletions of mtDNA. These defects, which can be sharpened by physiological changes due to environmental factors, elicit an adaptive transcriptional response to compensate for mitochondrial dysfunction thereby restoring metabolic fitness. The mitochondrial retrograde signaling is a multifaceted phenomenon whose complexity has evolved in parallel with the complexity of organisms and is, to some extent, conserved from fungi to plants to humans (Liu and Butow 2006; Ng et al.2014; Quiros, Mottis and Auwerx 2016). Retrograde signaling was first described in a model of yeast petite cells lacking mtDNA (Parikh et al.1987), in which it specifically regulates carbon and nitrogen metabolism under sub-optimal energy-production conditions. The retrograde (RTG) genes are the main positive regulators of yeast mitochondrial retrograde signaling and have been characterized in detail (Fig. 2). RTG1 and RTG3 encode for the subunits of a heterodimeric transcription factor activating RTG target gene expression (Jia et al.1997). RTG2, coding for a cytoplasmic protein with an N-terminal ATP-binding domain, acts as a sensor of mitochondrial dysfunction and regulates Rtg1/3 localization from the cytoplasm to the nucleus (Liu et al.2003). Multiple positive and negative regulators control the RTG pathway. Mks1 bound to functionally redundant 14-3-3 proteins Bmh1/2 represses RTG pathway, which is activated by Rtg2 binding to Mks1 (Liu et al.2003). Alternatively, Mks1 is subject to SCFGrr1-dependent ubiquitination and degradation (Liu et al.2005) (Fig. 2).
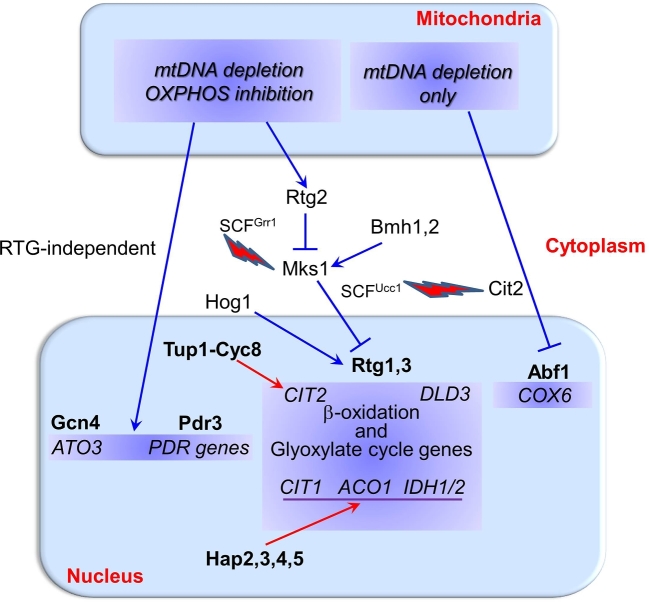
Figure 2.
Retrograde communication pathways regulating gene transcription in yeast. Key yeast genes and proteins involved in the transcriptional regulatory network of signaling pathways from mitochondria to the cytoplasm and the nucleus (see text for details). Mitochondrial dysfunction due to mtDNA depletion or OXPHOS enzyme complex inhibition can trigger retrograde pathways (blue lines), which activate-nuclear transcription to induce cell adaptation (see text for details). Dysfunctional mitochondria can activate different retrograde transcriptional responses that can be regulated by either RTG genes or alternative transcription factors and regulators, such as Abf1, Gcn4 and Pdr3. Hog1 can regulate RTG-dependent target genes. Nuclear genes highlighted in blue are upregulated by the bolded transcription factors located above them. Certain RTG-pathway target genes can also be regulated by anterograde transcription factors (see Fig. 1, red lines). The level of both retrograde target-gene products (e.g. Cit2) and regulators (e.g. Mks1) is under proteasomal degradation control, as indicated by the lightning bolts associated with the SCF ubiquitin ligase complexes.
Transcriptome profiling of yeast grown in raffinose showed that respiratory deficiency due to mtDNA depletion, but not inhibition of mitochondrial ATP synthesis per se, induces a suite of genes associated with both peroxisomal biogenesis and anaplerotic pathways. Thus, RTG-dependent transcriptional rewiring portends a metabolic reprogramming based on the activation of anaplerotic reactions that appear to alleviate the plausible interruption of TCA cycle and the ensuing decreased production of biosynthetic intermediates, including acetyl CoA and citrate, necessary for production of glutamate as a source of nitrogen (Epstein et al.2001). Interestingly enough, all rtg mutants, in which the retrograde pathway is inactive, are glutamate auxotrophs (Liao and Butow 1993; Jia et al.1997). RTG signaling is also induced in response to inhibition of amino acid biosynthetic enzymes, such as His3 and Gln1, apparently due to the central role of the tricarboxylic acid cycle in generating the carbon backbone in the biosynthesis of amino acids (Giannattasio et al.2005). It is of note that a microarray analysis in glucose-grown ρ° cells revealed an upregulation of genes encoding mitochondrial biogenesis factors and mitochondrial proteins, such as mitochondrial ribosomal proteins as well as a downregulation of ribosomal genes (Traven et al.2001). Thus, RTG pathway acts differently, depending on the carbon source used.
Rapamycin treatment, which inhibits the TORC1 complex mimicking nitrogen starvation, can activate RTG-dependent signaling (Komeili et al.2000). Lst8, a component of TORC1 complex, is a negative regulator of the RTG pathway (Liu et al.2001). Moreover, transcription of the genes encoding mitochondrial proteins functioning in the TCA cycle and OXPHOS as well as mitochondrial ribosomal proteins, is induced in sch9Δ cells and this seems tightly connected to the lifespan extension and stress resistance phenotype (Lavoie and Whiteway 2008). Recall, Sch9 is a key downstream effector of TORC1. Interestingly, ρ0 mutants and pet mutants that are respiratory-deficient due to nuclear mutations, differently affect the phosphorylation state of Sch9, suggesting that the induction of this pathway in response to mitochondrial stress is due, at least in part, to reduced TORC1 activity (Kawai et al.2011). However, it is apparent that TOR and RTG signaling can act through two separate branches in the regulation of RTG-dependent gene transcription (Giannattasio et al.2005).
CIT2 and DLD3, encoding the peroxisomal citrate synthase and d-lactate dehydrogenase respectively, are prototypical RTG-dependent target genes (Fig. 2). In particular, CIT2 expression, which is increased up to 30-fold in the absence of mtDNA, facilitates a more efficient utilization of carbon via the transfer of metabolites, such as citrate, from the glyoxylate cycle to the TCA cycle. CIT2 transcription is also controlled by Tup1-Cyc8 complex (Conlan et al.1999; Schuller 2003) and requires the Snf1-dependent transcription factors ADR1 and CAT8 (Laera et al.2016). Since Snf1 controls ADR1 and CAT8 by modulating interactions between Mig1 and the co-repressor Tup1-Cyc8 complex (see above) (Papamichos-Chronakis, Gligoris and Tzamarias 2004), glucose likely modulates RTG-dependent retrograde target gene transcription through Snf1 kinase. Interestingly, Cit2 is also under control of proteasomal degradation by the ubiquitin ligase SCFUcc1 (Nakatsukasa et al.2015) (Fig. 1). CIT1, ACO1 and IDH1/2, catalyzing the first three steps of TCA cycle, are regulated by HAP complex in respiratory competent cells but are under the control of RTG genes in response to a reduction or loss of respiratory function (Liu and Butow 1999). Thus, not only mitochondrial dysfunction, but also nutritional status has an impact on mitochondrial RTG-dependent signaling. Given that Aco1 is essential for mtDNA maintenance, the RTG pathway therefore plays a role in coupling metabolic regulation with mtDNA inheritance (Chen et al.2005).
The RTG pathway is linked to many biological pathways
The RTG-dependent signaling seems to have a role in different cellular processes and cell-stress response pathways rather than metabolic remodeling sensu stricto (Jazwinski 2005). These include aging, regulated cell death, cellular quality control and osmotic stress resistance.
RTG signaling is implicated in the aging process. As mitochondrial dysfunction and oxidative stress accelerate during aging, activation of retrograde signaling, by genetic and environmental means, proved to correlate with increased replicative lifespan (RLS) (Kirchman et al.1999). A role of RTG2 in modulating genome stability and RLS was further supported by the observation that the production of extrachromosomal ribosomal-DNA (rDNA) circles that accumulate in both aging yeast cells and cells lacking mitochondrial DNA was suppressed by Rtg2p (Borghouts et al.2004). This connection between retrograde signaling and aging seems to be conserved also in higher organisms, such as Caenorabditis elegans (Chang, Shtessel and Lee 2015; Palikaras, Lionaki and Tavernarakis 2015). Interestingly, Ras2 is required for retrograde response and RLS (Kirchman et al.1999). The molecular details of TOR-RAS-RTG connections deserve deeper investigations.
Activation of RTG pathway also causes regulated cell death evasion in yeast cells grown in raffinose, which differently from glucose favors mitochondrial respiration (Guaragnella et al.2013). In this regard, it has been shown that simultaneous activation of RTG pathway and SNF1/AMPK target gene expression can signal metabolic control to either extend chronological lifespan in respiratory-deficient ageing yeast cells or decrease acetic acid-induced regulated cell death in derepressed, respiratory competent yeast cells, plausibly through modulation of acetyl-CoA levels (Friis et al.2014; Laera et al.2016).
RTG signaling may be connected to aging through yet another process, namely cellular quality control. Cellular quality control has emerged as a central player in aging and mitophagy, a selective form of autophagy that degrades mitochondria, and has been hypothesized to serve a quality control function to prevent or slow the accumulation of malfunctioning mitochondria (see above). Mitophagy has been linked to the RTG pathway through the conserved mitochondrial protein phosphatase homolog, Aup1, which is required for efficient stationary phase mitophagy in yeast (Tal et al.2007). Aup1 is required for Rtg3/Rtg1-dependent transcription either during growth on non-fermentable carbon source or in response to 3-aminotriazole, an inhibitor of histidine biosynthesis. On the other hand, Rtg3 is essential for stationary-phase mitophagy, suggesting that Aup1 functions to induce a developmental transition, mediated through the RTG signaling pathway, that allows or facilitates mitophagy (Journo, Mor and Abeliovich 2009).
The RTG-dependent retrograde signaling has also been shown to contribute to osmotic stress resistance. The Hog1 stress-activated protein kinase (SAPK), induced by exposure to external hyperosmolarity, can control nuclear accumulation of Rtg1 and Rtg3 transcription factors and is required for their binding to the chromatin (Ruiz-Roig et al.2012). The significance of the interaction between HOG and RTG pathway needs further investigations.
Finally, Rtg2 has been found to function as an essential component of histone acetyltransferase-coactivator SAGA-like (SLIK) complex, which plays a critical role in transcription by RNA polymerase, thus linking chromatin remodeling to the retrograde signaling pathway. Notably, at least a portion of Rtg2 in yeast was found chromatin bound at the promoter of the activated CIT2 gene, which correlates with promoter acetylation (Pray-Grant et al.2002).
RTG-independent retrograde signaling
Beyond the RTG-mediated retrograde response, the existence of RTG-independent retrograde signaling has been reported (Fig. 2). In particular, ATO3 has been described as the prototypical target gene of RTG-independent mitochondrial retrograde pathway. ATO3 is up-regulated in ρ° cells under the transcriptional control of the general activator GCN4 and the amino acid sensor system Ssy1-Ptr3-Ssy5 (Fig. 2). ATO3 encodes an ammonium outward transporter and its up-regulation should eliminate the excess ammonia that arises because of a potential defect in ammonia assimilation in mtDNA-lacking cells (Guaragnella and Butow 2003). ATO3 has been involved in cell differentiation within yeast colonies, which are an excellent model for the investigation of processes involved in the development of specific cell types. Two major cell types have been identified within a yeast colony, U cells in upper regions which have a stress-resistant and longevity phenotype and L cells in lower regions, showing features of starving cells and sensitive to stresses (Cap et al.2012; Vachova, Cap and Palkova 2012). In this context, the Mks1-Rtg pathway exhibited heterogeneity of regulation, which differentially affects the properties and fate of U and L cells through a variety of yet unidentified factors that sense altered mitochondria differently (Podholova et al.2016).
The lack of mtDNA but not the absence of respiration activates another mitochondrial retrograde intergenomic signaling pathway involving the Abf1 transcription factor (Fig. 2). This retrograde response to a mitochondrial genotypic defect is distinct from RTG pathway and its activation causes down-regulation of a set of nuclear genes including subunits of cytochrome c oxidase (Woo et al.2009). Another example of mitochondrial retrograde signaling independent of RTG genes has been described for a long-lived mutant strain that harbors a deletion of AFO1, which encodes for a protein of the large subunit of the mitochondrial ribosome (Heeren et al.2009). This signaling is dependent on active TORC1, suggesting potential communication with the classic RTG pathway.
AFO1 deletion mutants are not the only long-lived mitochondrial mutants that appear to induce RTG-independent retrograde signaling. Mitochondrial dysfunction in cells lacking SOV1, a member of the yeast mitochondrial translation control module, causes a Sir2-dependent RLS extension through reduced cAMP-PKA signaling (Caballero et al.2011) [also see commentary Chen (2011)]. These discoveries confirm that dysfunctional mitochondria can activate different transcriptional responses to adapt functional changes and cellular needs.
The expression of multidrug resistance genes has also been connected with mitochondrial function through a regulatory pathway involving the transcription factor Pdr3. In both mtDNA- or OXA1-lacking cells, a Pdr3-dependent increase in the pleiotropic drug resistance (PDR) has been observed (Hallstrom and Moye-Rowley 2000; Devaux et al.2002; Moye-Rowley 2005) (Fig. 2). We will revisit this observation in the following section, as Pdr3 activation in this mutant yeast may induce changes in cytosolic proteostasis as well. In view of the relation between mitochondrial dysfunction, tumorigenesis and chemoresistance in cancer cells (Guaragnella, Giannattasio and Moro 2014), relations among retrograde pathway and multidrug resistance of cancer cells need further studies.
A novel mitochondria retrograde signaling pathway not initiated by a general loss of respiratory activity has been observed in respiring, but not in fermenting yeast cells lacking the i-AAA protease, Yme1. YME1 deletion abolished peptide generation in the intermembrane space and led to the induction of nuclear genes with functions in mitochondrial gene expression and the biogenesis of the respiratory chain, suggesting a link between export of peptides generated by mitochondrial protein degradation and nuclear gene expression. Many of these genes appear to adjust the energy supply by OXPHOS and to ensure cell survival under respiratory growth conditions, although the extent of readjustment is rather moderate (Arnold et al.2006).
Potential mitochondrial signals that trigger retrograde signaling
Despite the deep knowledge of the molecular details of the RTG-dependent retrograde signaling, what are the exact triggers for the activation of each pathway remain to be established. Several candidates have been proposed that include ATP depletion and mitochondrial membrane potential dissipation (Miceli et al.2011; Zhang et al.2013). ATP could be a good potential candidate because it directly regulates the dynamic interaction between Rtg2 and Mks1 in a conserved manner (Zhang et al.2013). Indeed, the integrity of ATP binding domain is required for the interaction with Mks1 and there is a direct correlation between the degree of Mks1p phosphorylation and the extent of RTG target gene repression (Liu et al.2003). Interestingly, in support of a role for ATP as signaling molecule, a recent work reported that most of the residues required for retrograde signaling surround the ATP-binding loops of Rtg2 (Rios-Anjos et al.2017). However, ATP as a second messenger molecule is a highly controversial subject, because it is dynamically short lived and involved in many other processes in the cell.
Reactive oxygen species might be other potential retrograde-activating signals resulting from dysfunctional mitochondria. Recent evidences have shown that RTG-dependent mitochondria-to-nucleus signaling modulates redox activity, resulting in increased stress resistance (Guaragnella et al.2013; Torelli et al.2015). ROS signaling from the mitochondrion to the nucleus is also involved in chronological lifespan extension in non-dividing cells, and this requires TORC1 (Pan et al.2011) (see above).
As reported above, another possible retrograde signaling molecule could be represented by single aminoacids or short peptides released from mitochondria and derived from damaged or misfolded mitochondrial proteins. However, if they serve a signaling function either their amount must be sensed or specific peptides must be recognized and this is not fully supported by published data (Arnold et al.2006).
While the specific mitochondrial signals that activate yeast's retrograde pathways are yet to be elucidated, it should be noted that novel mechanisms mediating retrograde mitochondrial signals are identified in C. elegans. In worms, mitochondrial damage, including the accumulation of misfolded proteins (proteotoxic stress), activates the mitochondrial unfolded protein response (UPRmt) by organelle partitioning of activating transcription factor associated with stress (ATFS-1). ATFS-1, which contains both a mitochondrial targeting sequence and a nuclear localization sequence, redistributes from mitochondria to the nucleus upon mitochondrial proteotoxic stress due to reduced mitochondrial protein import. Once in the nucleus, it directs a transcriptional program that upregulates mitochondrial chaperones to restore mitochondrial proteostasis (Nargund et al.2012). Therefore, ATFS-1 is a mitochondria-derived retrograde signaling activator in C. elegans’ UPRmt. The UPRmt was first discovered in human cells more than 15 years ago by the pioneering work of the Hoogenraad group, and is highly conserved (Zhao et al.2002). However, the specific UPRmt activation signals derived from dysfunctional mitochondria in the mammalian system have not been clearly identified, despite the discovery of a mammalian ATFS-1 orthologue (Fiorese et al.2016). Current evidence suggests mammalian UPRmt is multifaceted and far more complex than previously anticipated (Shpilka and Haynes 2018). Although the existence of a proteostatic retrograde regulation of nuclear gene transcription similar to UPRmt has not been documented so far in yeast, in the next paragraph we describe the elaborate network of mitochondria–cytosol communications activated by proteotoxic stress, which has been unveiled so far in this model organism.
PROTEOSTATIC CROSSTALK BETWEEN MITOCHONDRIA AND THE CYTOSOL
Beyond the trafficking of antero- and retrograde transcription factors, mitochondria and the cytosolic proteome interact in complex ways (Fig. 3). Hints of this relationship originate in early studies showing that mitochondrial dysfunction induces transcriptional changes directed, in part, at preserving cytosolic proteostasis. More recently it was demonstrated that a diverse range of mitochondrial stresses kill cells by challenging cytosolic proteostasis with unimported mitochondrial precursor proteins, a mechanism named mitochondrial precursor overaccumulation stress (mPOS) (Wang and Chen 2015; Wrobel et al.2015; Coyne and Chen 2018). The mPOS model provides a direct link between mitochondria and cytosolic proteostasis, and cytosolic response pathways exist to mitigate mPOS (Wrobel et al.2015). However, the relationship between mitochondria and cytosolic proteostasis appears to be more complex than just mPOS and its response mechanisms. Paradoxically, mitochondria are reported to actively improve cytosolic proteostasis (Ruan et al.2017). Moreover, while mitochondria can both positively and negatively affect cytosolic proteostasis, cytosolic proteostasis can affect mitochondrial function in intricate ways as well. Improved cytosolic proteostasis can rescue mitochondrial degeneration (Wang et al.2008), and cytosolic protein aggregation can cause mitochondrial defects (Solans et al.2006). In this section, we review our current understanding of this multifaceted interplay that occurs both anterogradely (cytosol-to-mitochondria) and retrogradely (mitochondria-to-cytosol) in S. cerevisiae.
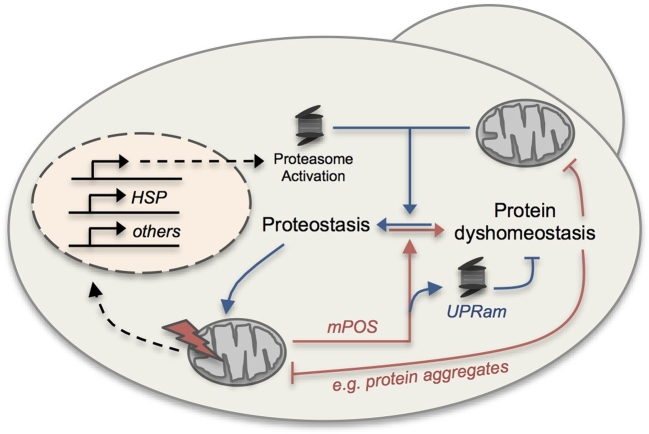
Figure 3.
Proteostatic crosstalk between mitochondria and the cytosol in yeast. A schematic of proteostatic crosstalk between mitochondria and the cytosol. Dysfunctional mitochondria (depicted with the red lightning bolt) can compromise cytosolic proteostasis via mitochondrial precursor overaccumulation stress (mPOS), which induces the unfolded protein response activated by mistargeting of proteins (UPRam) to improve cytosolic protein dyshomeostasis. Healthy mitochondria (without the lightning bolt) can benefit cytosolic proteostasis via mechanisms including increase in ATP supply, decrease in ROS production and the MAGIC pathway. Conversely, cytosolic proteostasis influences mitochondrial function. Beneficial and deleterious interactions are depicted by blue and red lines, respectively.
Hints of a mitochondria-cytosol crosstalk appear in early gene expression studies of cells with damaged mitochondria. Of course, mitochondria-cytosol crosstalk was extensively investigated in the context of the RTG-dependent retrograde pathway, as discussed above. However, in the search for retrograde pathways, gene expression studies of yeast with mitochondrial defects often showed transcriptional responses directed at improving cytosolic proteostasis, suggesting mitochondrial dysfunction directly affects the cytosolic proteome. For example, mtDNA depletion induces expression of the proteasome assembly factor, Spg5 (Epstein et al.2001). Consistent with this, Pdr3 is also preferentially expressed in ρ0 cells (Hallstrom and Moye-Rowley 2000; Devaux et al.2002). In addition to orchestrating the mitochondrial compromised import response (mitoCPR) (Weidberg and Amon 2018; discussed below), Pdr3 upregulates Rpn4 (Hahn, Neef and Thiele 2006), a transcriptional activator of the proteasome. Therefore, at least two transcriptional programs are triggered in ρ0 cells to activate the proteasome. The proteasome is responsible for the degradation of most cytosolic proteins. Altered expression of cytosolic heat shock proteins (HSPs) is also observed in ρ0 cells (Epstein et al.2001), which appears to be conserved in ρ0S. pombe (Guha et al.2011) and human cells (Behan, Doyle and Farrell 2005). HSPs are chaperones responsible for the refolding of damaged proteins under stress conditions. These cytosol-directed transcriptional changes represent a small fraction of those observed in ρ0 cells, perhaps explaining why they have received such little attention. However, the mPOS model provides a framework in which to understand these historical data, as proteasome activation and HSP expression would be predicted to relieve a mitochondria-induced cytosolic proteostatic burden.
As noted above, mitochondrial dysfunction can stress the cytosolic proteome, and one mechanism by which this occurs is mPOS wherein unimported mitochondrial precursor proteins challenge cytosolic proteostasis. mPOS can be caused by several different mitochondrial insults that do not directly target the mitochondrial protein import machinery. The prototypical mPOS inducer is protein misfolding in the inner mitochondrial membrane (IMM) (Wang and Chen 2015; Coyne and Chen 2018). The mechanism by which this occurs is unclear, though it may involve global IMM proteostatic stress and consequent destabilization of the mitochondrial protein translocases TIM22 and TIM23 located in the IMM (Liu, Wang and Chen 2015). Nevertheless, IMM protein misfolding causes the drastic accumulation and aggregation of mitochondrial precursor proteins in the cytosol, i.e. mPOS, which can ultimately kill cells.
A genetic screen for suppressors of mPOS-induced cell death revealed a pro-proteostatic network that decreases cytosolic protein synthesis and increases cytosolic protein chaperoning, degradation, and mRNA turnover (Wang and Chen 2015). Each gene in the network likely suppresses mPOS by reducing the cytosolic protein load. While some anti-degenerative genes likely only suppress mPOS when ectopically overexpressed, others may perform intrinsic anti-mPOS roles. There are at least three groups of genes in this category. The first group includes genes that are intrinsically upregulated upon mPOS. These include the ribosome-associated Gis2 and Nog2, which stimulate cap-independent translation (Gerbasi and Link 2007) and inhibit nuclear export of the 60S ribosomal subunit (Matsuo et al.2014) respectively. Not only are they intrinsically upregulated upon mPOS, likely by reduced protein turnover, but both can also ectopically suppress mPOS-induced cell death (Wang and Chen 2015). The mechanism by which Gis2 and Nog2 suppress mPOS is unclear, though we speculate that they stimulate the translation of specific stress-response mRNAs. The second group consists of cytosolic chaperones that can ectopically suppress mPOS-induced cell death and are also required for the cell viability in milder mPOS conditions. Ssb1 falls into this category, as it is a potent mPOS suppressor (Wang and Chen 2015) and is also required for viability in the ρ0 state (Dunn and Jensen 2003), which is an mPOS-inducing condition (Coyne and Chen 2018). These observations suggest that basal levels of Ssb1, and possibly other chaperones, function to contain mPOS when the stress is mild. Third, the proteasome assembly factors POC3 and POC4 are potent ectopic mPOS suppressors that also appear to respond to mPOS. Both genes are critical for the unfolded protein response activated by mistargeting of proteins (UPRam), wherein mistargeted mitochondrial proteins activate the proteasome to relieve the cytosolic protein burden (Wrobel et al.2015). Genetic ablation of either POC3 or POC4 prevents UPRam thereby sensitizing cells to growth inhibition caused by mistargeted mitochondrial proteins. The relationship between the UPRam and the proteasome-activating transcriptional changes discussed above is unclear.
Protein import defects elicit an additional mechanism to handle unimported mitochondrial precursors termed the mitochondrial compromised protein import response (mitoCPR) (Weidberg and Amon 2018). Upon import deficiency induced by various methods, Pdr3 is activated to drive the transcription of Cis1, which then physically associates with the translocase machinery on the outer membrane to facilitate the local proteasomal degradation of unimported proteins. It thus appears that this elegant pathway plays a role in preventing mPOS.
With the exceptions of the UPRam and mitoCPR, mechanistic investigations into intrinsic anti-mPOS pathways are lacking, and it is likely that additional pathways exist. One additional pathway demonstrated under mPOS conditions is the repression of global cytosolic translation (Wang and Chen 2015; Wrobel et al.2015), which remains poorly understood but may be induced by mitochondrial ROS (Topf et al.2018). Taken together, it is clear that cells have mechanisms for handling the cytosolic burden imposed by mPOS.
Mechanisms of mitochondria-induced cytosolic stress distinct from mPOS have also been documented. For example, mitochondria-derived oxidative stress induces the reversible disassembly of the 26S proteasome, which is likely to stress cytosolic proteostasis and may contribute to mitochondrial disease (Livnat-Levanon et al.2014; Segref et al.2014). This is in apparent conflict with the UPRam, which directs enhanced proteasome activity upon mitochondrial stress. One differentiating factor between proteasome disassembly and UPRam-inducing conditions is the nature of the mitochondrial stress; on the one hand, mitochondrial ROS is elevated and the proteasome is disassembled, while on the other hand, mitochondrial protein import is genetically impaired and the proteasome is activated. An interesting test of the relative dominance between these two pathways would be to measure proteasome assembly/activity under mitochondrial stress conditions that induce both mPOS and ROS production across varying intensities and durations.
In addition to perturbing cytosolic proteostasis, yeast mitochondria can actively improve cytosolic proteostasis. Perhaps the clearest demonstration of mitochondria as a double-edged sword for cytosolic proteostasis comes from an analysis of how mitochondrial translation fidelity affects the cytosol. While decreasing the accuracy of mitochondrial translation accelerates cytosolic protein aggregation in aging yeast cells, increasing translational accuracy reduces protein aggregation during aging (Suhm et al.2018). It was speculated that these effects are not related to mPOS, based on the observation that several nuclear-encoded mitochondrial proteins tested do not accumulate in the cytosol of cells with hypoaccurate mitochondrial translation. Similarly, it appears that the effects are also not due to the UPRam, as proteasomal activity remained unaffected in cells with both hyper- and hypoaccurate mitochondrial translation. More comprehensive studies are needed to capture potential proteostatic signals in the cytosol that reduce protein aggregation.
Other examples of mitochondrial function benefiting cytosolic proteostasis exist as well. A recent CRISPR-based genetic screen identified TIMM9, an intermembrane space chaperone that participates in the import and insertion of IMM proteins, as a potent suppressor of toxicity induced by cytosolic protein aggregation (Chen et al.2017). This may be indicative of an mPOS contribution to cytosolic protein aggregate toxicity, as improved mitochondrial protein import would limit potential mitochondrial protein contribution to cytosolic aggregates. Alternatively, TIMM9 suppression of cytosolic aggregate-induced toxicity may be related to the observation that aggregation-prone cytosolic proteins have been proposed to enter mitochondria for degradation by mitochondrial proteases, a process named Mitochondria As Guardian In Cytosol (MAGIC) (Ruan et al.2017). A beneficial effect of mitochondria on cytosolic protein aggregation is not limited to these observations. Inhibition of mitochondrial protein translation by chloramphenicol was shown to accelerate the clearance of cytosolic protein aggregates induced by heat shock (Suhm et al.2018). It is not clear how to interpret these observations; clearly, more work is needed to determine mechanistic details of how mitochondrial function can affect protein folding and aggregation in the cytosol.
Just as mitochondria affect cytosolic proteostasis, cytosolic proteostasis can affect mitochondria. Cytosolic proteostatic factors can benefit mitochondrial function by at least three mechanisms in yeast. First, the proteasome actively degrades misfolded outer mitochondrial membrane proteins damaged by ROS, a process termed mitochondria-associated degradation (MAD) (Heo et al.2010). MAD involves the mitochondrial translocation of Cdc48, a component of the ubiquitin/proteasome system known best for its role in degrading misfolded endoplasmic reticulum (ER) proteins. This ER mechanism is apparently repurposed for the degradation of mitochondrial proteins. Vms1p appears to be central for MAD, as its translocation to mitochondria under oxidative stress is required for the mitochondrial localization of Cdc48. Further, when Vms1 is mutated, ubiquitin-dependent mitochondrial protein degradation, mitochondrial respiratory function and cell viability are compromised. Interestingly enough, a cdc48 yeast mutant provided first evidences that cell death regulation is structurally and functionally conserved in yeast (Madeo, Frohlich and Frohlich 1997). For a discussion of how MAD relates to the oxidative stress-induced proteasome disassembly and the UPRam, the reader is referred elsewhere (Bragoszewski, Turek and Chacinska 2017).
The second mechanism by which cytosolic proteostatic factors benefit mitochondria also involves Vms1, but it is independent of Vms1’s functional interaction with Cdc48 (Izawa et al.2017). This pathway protects mitochondria from intra-mitochondrial aggregation of polypeptides that stalled on the cytosolic ribosome. Typically, a given polypeptide that stalls on the ribosome is modified with C-terminal alanyl/threonyl sequences (CAT-tails) to facilitate ubiquitination and degradation by the proteasome (Shen et al.2015). However, some stalled mitochondria-destined polypeptides can engage the import machinery before the proteasome has sufficient time and access to degrade the modified peptides. These inaccessible CAT-tailed polypeptides then aggregate and disrupt mitochondrial function (Izawa et al.2017). Vms1 functions to prevent the CAT-tail modification of stalled mitochondrial polypeptides, possibly through its recently identified role as a tRNA hydrolase that liberates aberrant polypeptides from the stalled 60S ribosome (Zurita Rendon et al.2018). This protects mitochondria from the toxic effects of CAT-tail-induced aggregation (Izawa et al.2017). This pathway, termed mitochondrial ribosome-associated quality control (mitoRQC) occurs under basal cellular conditions. What is Vms1’s role under mitochondrial stress conditions is an interesting question to be addressed in the future.
There is a third mechanism by which cytosolic proteostasis benefits mitochondria, though the details are much less clear than with MAD and mitoRQC. Reducing cytosolic protein synthesis by genetic, pharmacological, or environmental inhibition rescues mitochondrial degeneration induced by IMM stress, as judged by a restoration of mitochondrial membrane potential (Wang et al.2008). Similarly, genetic and pharmacological inhibition of cytosolic protein synthesis was demonstrated to fully restore biogenesis and activity of the OXPHOS system in yeast with dysfunctional cardiolipin, an important IMM phospholipid (de Taffin de Tilques et al.2018). Perhaps relatedly, yeast strains with reduced TOR signaling, which decreases global cytosolic protein synthesis, have greater oxygen consumption and overall electron transport chain activity that is efficiently coupled to ATP production (Pan and Shadel 2009; Pan et al.2011), as reported above for the anterograde transcriptional regulation of mitochondria-to-nucleus communication. This effect may or may not be related to a TOR-dependent reduction in cytosolic protein synthesis, as reduced TOR signaling causes an increase in the number of OXPHOS complexes/mitochondrion, which would clearly benefit mitochondrial function. Nevertheless, that reduced cytosolic protein synthesis can benefit mitochondrial function may be conserved in worms, flies, human cells and mice (Liu and Lu 2010; Baker et al.2012; Peng et al.2015). One simplistic explanation would be that a reduction of global protein synthesis reduces the overall proteostatic burden in subcellular compartments including mitochondria.
Conversely, cytosolic proteostatic factors can impair mitochondrial function. Cytosolic expression of disease-associated proteins with expanded polyglutamine tracts causes cytosolic protein aggregation, mitochondrial respiratory defects and significant cytotoxicity (Solans et al.2006). Enhanced mitochondrial biogenesis can suppress the cytotoxicity in this model (Ocampo, Zambrano and Barrientos 2010). Therefore, cytosolic protein aggregates kill yeast cells through impaired mitochondrial respiration, adding another complicating layer to mitochondria–cytosol crosstalk.
Overall, how mitochondria–cytosol proteostatic crosstalk pathways are integrated under different healthy and stressed conditions remains a daunting mystery, and will likely be the focus of many future studies.
OUTLOOK
The picture emerging from 40 years of intense yeast research on the mechanisms by which mitochondria communicate with cytoplasm and nucleus to maintain cell homeostasis is a dynamic and complex network of intracellular signaling pathways. This is apparently achieved both through modulation of the transcription of different sets of genes depending on the metabolic conditions of the cell and through a crosstalk between the cytosolic and mitochondrial proteostatic networks. Accumulating experimental evidences also suggest that signaling and transcription machineries might be eventually regulated by intracellular levels of a limited set of key metabolites, such as acetyl CoA (Chen, Siewers and Nielsen 2012) and NAD+ (Guarente 2016). In this context, it is of note that the levels of these molecules and others reported in this review, including ATP, reactive oxygen species, intermediates of amino acids synthesis, short peptides, iron-sulfur clusters, heme and the precursor proteins that fail to be imported by mitochondria, may not be sensed exclusively by mitochondria. Thus, it has also been argued that these substances are recognized by the cytosolic sensors which transmit the signals to the nucleus leading to general, as opposed to mitochondria specific, transcriptional response (Knorre et al.2016). Furthermore, gene transcriptional control cannot be the exclusive mechanism through which mitochondria exert their homeostatic function. For instance, the ubiquitin proteasome system has been shown to directly regulate both the yeast mitochondrial RTG-dependent retrograde pathway (see above and Fig. 1) and other mitochondrial processes, as extensively reviewed (Escobar-Henriques and Langer 2014).
Intriguingly, the mPOS mechanism dictates that the cytosolic protein degradation machinery has limited capacity to degrade unimported mitochondrial precursor proteins. This suggests that mitochondrial proteins may be directly engaged in cellular signaling in the cytosol and potentially other organelles (Friedman et al.2018). The UPRam mechanism is an example of this, as unimported mitochondrial proteins were shown to activate the cytosolic proteasome (Wrobel et al.2015). In worms, the failed import of ATFS-1 is critical for UPRmt induction (Nargund et al.2012). It will be interesting to see how this field develops, as unimported mitochondrial proteins could affect a diverse range of signaling pathways in response to mitochondrial dysfunction, as revealed by studies in yeast reviewed here.
Recent advances, made primarily in budding yeast, have provided novel insights into the existence of distinct micro-domains between intracellular organelles, known as membrane contact sites that coordinate diverse activities, including mitochondrial dynamics and cell stress signaling pathways. We refer to recent comprehensive reviews on the topic describing the structure and function of mitochondrial contact sites with multiple organelles (Murley and Nunnari 2016; Phillips and Voeltz 2016). Here, it is important to note that yeast is once more emerging as an ideal platform for membrane contact site hunting and characterization (Eisenberg-Bord et al.2016). The array of data showing that dysfunctional mitochondria dramatically induce RTG-dependent biogenesis of peroxisomes (Epstein et al.2001) together with finding that cardiolipin deficiency can cause Rtg2-mediated vacuolar defects (Chen et al.2008), suggest that there are multiple pathways of crosstalk between these organelles in yeast.
Except for a number of key regulatory factors involved in nutrient sensing and growth control, such as TOR, Sch9, PKA and Snf1, no direct orthologues were found so far of yeast anterograde- and retrograde-pathway regulators in other species, except for a number of mammalian functional homologues such as FOXOs, nuclear factor kB, ERα and Myc, as reviewed elsewhere (Hock and Kralli 2009; Guha and Avadhani 2013; Quiros, Mottis and Auwerx 2016). Despite this, yeast remains a preferred model organisms to explore eukaryotic cell biology, both due to the unique level of knowledge achieved on the molecular details of cellular processes of this unicellular organism, and to the high degree of functional conservation (Botstein and Fink 2011). Good evidence of this, for example, can be found in the apparent heterogeneity of RTG signaling in yeast colonies, which closely resembles the pleiotropic mitochondrial retrograde signaling in mammals, which includes a number of parallel regulatory events occurring under different conditions and in different cells (Podholova et al.2016).
It is important to stress that most studies have been performed under different environmental conditions with different yeast cell types. Therefore, it is difficult to establish general interconnection among the so-far identified anterograde and retrograde pathways that regulate mitochondria–cytosol–nucleus cross-talk at both transcription and proteostasis level, to define the key signaling molecules, and to decipher which pathways are homeostatic and which are pathological. The clarification of these aspects and understanding how cells adapt to changes in mitochondrial function in yeast will help to address relevant issues of mitochondria–cytosol–nucleus crosstalk in lifespan, cancer metabolism and growth control in higher eukaryotes.
Acknowledgements
We thank Dr. Sonia Colombo for critical reading of the manuscript and helpful discussions. The authors declare the absence of conflict of interest.
FUNDING
This work was supported by the NIH grants AG023731 and AG047400 to XJC.
Conflict of interest. None declared.
REFERENCES
- Amigoni L, Martegani E, Colombo S. Lack of HXK2 induces localization of active ras in mitochondria and triggers apoptosis in the yeast Saccharomyces cerevisiae. Oxid Med Cell Longev 2013;2013:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Wagner-Ecker M, Ansorge W et al. Evidence for a novel mitochondria-to-nucleus signalling pathway in respiring cells lacking i-AAA protease and the ABC-transporter Mdl1. Gene 2006;367:74–88. [DOI] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T et al. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet 2012;8:e1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behan A, Doyle S, Farrell M. Adaptive responses to mitochondrial dysfunction in the rho degrees Namalwa cell. Mitochondrion 2005;5:173–93. [DOI] [PubMed] [Google Scholar]
- Belotti F, Tisi R, Paiardi C et al. Localization of Ras signaling complex in budding yeast. Biochim Biophys Acta 2012;1823:1208–16. [DOI] [PubMed] [Google Scholar]
- Bohovych I, Kastora S, Christianson S et al. Oma1 links mitochondrial protein quality control and TOR signaling to modulate physiological plasticity and cellular stress responses. Mol Cell Biol 2016;36:2300–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J et al. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics 2004;166:765–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Fink GR. Yeast: an experimental organism for 21st century biology. Genetics 2011;189:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragoszewski P, Turek M, Chacinska A. Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system. Open Biol 2017;7:170007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem 2017;292:12754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschlen S, Amillet JM, Guiard B et al. The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comp Funct Genomics 2003;4:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A, Ugidos A, Liu B et al. Absence of mitochondrial translation control proteins extends life span by activating sirtuin-dependent silencing. Mol Cell 2011;42:390–400. [DOI] [PubMed] [Google Scholar]
- Cap M, Stepanek L, Harant K et al. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell 2012;46:436–48. [DOI] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Bauer MA, Zimmermann A et al. Guidelines and recommendations on yeast cell death nomenclature. Microb Cell 2018;5:4–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS. Mitochondria as signaling organelles. BMC Biol 2014;12:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Shtessel L, Lee SS. Collaboration between mitochondria and the nucleus is key to long life in Caenorhabditis elegans. Free Radic Biol Med 2015;78:168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tarsio M, Kane PM et al. Cardiolipin mediates cross-talk between mitochondria and the vacuole. MBoC 2008;19:5047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ. The search for nonconventional mitochondrial determinants of aging. Mol Cell 2011;42:271–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ. Mechanism of homologous recombination and implications for aging-related deletions in mitochondrial DNA. Microbiol Mol Biol Rev 2013;77:476–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD. Mutations in MGI genes convert Kluyveromyces lactis into a petite-positive yeast. Genetics 1993;133:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD. Specific mutations in alpha- and gamma-subunits of F1-ATPase affect mitochondrial genome integrity in the petite-negative yeast Kluyveromyces lactis. EMBO J 1995;14:3277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD. The mitochondrial genome integrity gene, MGI1, of Kluyveromyces lactis encodes the beta-subunit of F1-ATPase. Genetics 1996;144:1445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD. The petite mutation in yeasts: 50 years on. Int Rev Cytol 2000;194:197–238. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet 2005;6:815–25. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD. Unveiling the mystery of mitochondrial DNA replication in yeasts. Mitochondrion 2018;38:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Wang X, Kaufman BA et al. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 2005;307:714–7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Siewers V, Nielsen J. Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PLoS One 2012;7:e42475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Farzadfard F, Gharaei N et al. Randomized CRISPR-Cas transcriptional perturbation screening reveals protective genes against alpha-synuclein toxicity. Mol Cell 2017;68:247–257.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan RS, Gounalaki N, Hatzis P et al. The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J Biol Chem 1999;274:205–10. [DOI] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN et al. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 2014;38:254–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev 2000;64:281–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G et al. Synchronized mitochondrial and cytosolic translation programs. Nature 2016;533:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne LP, Chen XJ. mPOS is a novel mitochondrial trigger of cell death - implications for neurodegeneration. FEBS Lett 2018;592:759–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taffin de Tilques M, Lasserre JP, Godard F et al. Decreasing cytosolic translation is beneficial to yeast and human Tafazzin-deficient cells. Microb Cell 2018;5:220–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C. The essence of yeast quiescence. FEMS Microbiol Rev 2012;36:306–39. [DOI] [PubMed] [Google Scholar]
- De Vit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. MBoC 1997;8:1603–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 1997;278:680–6. [DOI] [PubMed] [Google Scholar]
- Devaux F, Carvajal E, Moye-Rowley S et al. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett 2002;515:25–8. [DOI] [PubMed] [Google Scholar]
- Dunn CD, Jensen RE. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics 2003;165:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Shai N, Schuldiner M et al. A tether is a tether is a tether: Tethering at membrane contact sites. Dev Cell 2016;39:395–409. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Waddle JA, Hale WT et al. Genome-wide responses to mitochondrial dysfunction. MBoC 2001;12:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M, Langer T. Dynamic survey of mitochondria by ubiquitin. EMBO Rep 2014;15:231–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Liou LL, Moy VN et al. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 2003;163:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin YF et al. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol 2016;26:2037–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F, Roganti T, Lecrenier N et al. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 1998;440:325–31. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kannan M, Toulmay A et al. Lipid homeostasis is maintained by dual targeting of the mitochondrial PE biosynthesis enzyme to the ER. Dev Cell 2018;44:261–270.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis RM, Glaves JP, Huan T et al. Rewiring AMPK and mitochondrial retrograde signaling for metabolic control of aging and histone acetylation in respiratory-defective cells. Cell Rep 2014;7:565–74. [DOI] [PubMed] [Google Scholar]
- Friis RMN, Schultz MC. Attenuation of transcriptional and signaling responses limits viability of rho(0)Saccharomyces cerevisiae during periods of glucose deprivation. Biochim Biophys Acta 2016;1860:2563–75. [DOI] [PubMed] [Google Scholar]
- Galello F, Moreno S, Rossi S. Interacting proteins of protein kinase A regulatory subunit in Saccharomyces cerevisiae. J Proteomics 2014;109:261–75. [DOI] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev 1998;62:334–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbasi VR, Link AJ. The myotonic dystrophy type 2 protein ZNF9 is part of an ITAF complex that promotes cap-independent translation. Mol Cell Proteomics 2007;6:1049–58. [DOI] [PubMed] [Google Scholar]
- Giannattasio S, Liu Z, Thornton J et al. Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J Biol Chem 2005;280:42528–35. [DOI] [PubMed] [Google Scholar]
- Goldenthal MJ, Marin-Garcia J. Mitochondrial signaling pathways: a receiver/integrator organelle. Mol Cell Biochem 2004;262:1–16. [DOI] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC et al. "Sleeping Beauty": Quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 2004;68:187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaragnella N, Butow RA. ATO3 encoding a putative outward ammonium transporter is an RTG-independent retrograde responsive gene regulated by GCN4 and the Ssy1-Ptr3-Ssy5 amino acid sensor system. J Biol Chem 2003;278:45882–7. [DOI] [PubMed] [Google Scholar]
- Guaragnella N, Giannattasio S, Moro L. Mitochondrial dysfunction in cancer chemoresistance. Biochem Pharmacol 2014;92:62–72. [DOI] [PubMed] [Google Scholar]
- Guaragnella N, Zdralevic M, Lattanzio P et al. Yeast growth in raffinose results in resistance to acetic-acid induced programmed cell death mostly due to the activation of the mitochondrial retrograde pathway. Biochim Biophys Acta 2013;1833:2765–74. [DOI] [PubMed] [Google Scholar]
- Guarente L. CELL METABOLISM. The resurgence of NAD(+). Science 2016;352:1396–7. [DOI] [PubMed] [Google Scholar]
- Guha M, Avadhani NG. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion 2013;13:577–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Lopez-Maury L, Shaw M et al. Transcriptional and cellular responses to defective mitochondrial proteolysis in fission yeast. J Mol Biol 2011;408:222–37. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Neef DW, Thiele DJ. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol Microbiol 2006;60:240–51. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Moye-Rowley WS. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J Biol Chem 2000;275:37347–56. [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A et al. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab 2014;19:357–72. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci 2008;13:2408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeren G, Rinnerthaler M, Laun P et al. The mitochondrial ribosomal protein of the large subunit, Afo1p, determines cellular longevity through mitochondrial back-signaling via TOR1. Aging 2009;1:622–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Livnat-Levanon N, Taylor EB et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell 2010;40:465–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK. Stationary phase in yeast. Curr Opin Microbiol 2002;5:602–7. [DOI] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol 2009;71:177–203. [DOI] [PubMed] [Google Scholar]
- Izawa T, Park SH, Zhao L et al. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell 2017;171:890–903.e18. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Hall MN. Correction: TOR signalling in bugs, brain and brawn. Nat Rev Mol Cell Biol 2003;4:117–26. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene 2005;354:22–7. [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J et al. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol 1997;17:1110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journo D, Mor A, Abeliovich H. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem 2009;284:35885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Furukawa K, Yamashita S. Mitophagy in yeast: Molecular mechanisms and physiological role. Biochim Biophys Acta 2015;1853:2756–65. [DOI] [PubMed] [Google Scholar]
- Kawai S, Urban J, Piccolis M et al. Mitochondrial genomic dysfunction causes dephosphorylation of Sch9 in the yeast Saccharomyces cerevisiae. Eukaryot Cell 2011;10:1367–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayikci O, Nielsen J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res 2015;15:fov068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY et al. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 1999;152:179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorre DA, Sokolov SS, Zyrina AN et al. How do yeast sense mitochondrial dysfunction? Microb Cell 2016;3:532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Wedaman KP, O'Shea EK et al. Mechanism of metabolic control. J Cell Biol 2000;151:863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laera L, Guaragnella N, Zdralevic M et al. The transcription factors ADR1 or CAT8 are required for RTG pathway activation and evasion from yeast acetic acid-induced programmed cell death in raffinose. Microb Cell 2016;3:621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W. The energetics of genome complexity. Nature 2010;467:929–34. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Whiteway M. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot Cell 2008;7:1127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadsham JE, Gourlay CW. cAMP/PKA signaling balances respiratory activity with mitochondria dependent apoptosis via transcriptional regulation. BMC Cell Biol 2010;11:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen E, Oberholzer U, Labarre J et al. Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol Microbiol 2002;43:1023–37. [DOI] [PubMed] [Google Scholar]
- Lenssen E, James N, Pedruzzi I et al. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation–via a newly identified Glc7/Bud14 type I protein phosphatase module–and TFIID promoter distribution. Mol Cell Biol 2005;25:488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 1993;72:61–71. [DOI] [PubMed] [Google Scholar]
- Liu S, Lu B. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS Genet 2010;6:e1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J et al. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 1996;86:147–57. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang X, Chen XJ. Misfolding of mutant adenine nucleotide translocase in yeast supports a novel mechanism of Ant1-induced muscle diseases. MBoC 2015;26:1985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol 1999;19:6720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet 2006;40:159–85. [DOI] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Epstein CB et al. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J 2001;20:7209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Spirek M, Thornton J et al. A novel degron-mediated degradation of the RTG pathway regulator, Mks1p, by SCFGrr1. Mol Biol Cell 2005;16:4893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Spirek M et al. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol Cell 2003;12:401–11. [DOI] [PubMed] [Google Scholar]
- Livnat-Levanon N, Kevei E, Kleifeld O et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep 2014;7:1371–80. [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011;189:1177–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol 1997;139:729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C et al. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 1996;15:2227–35. [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M et al. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 2014;505:112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli MV, Jiang JC, Tiwari A et al. Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Front Genet 2011;2:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern M, Stiller SB, Lubbert P et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep 2017;19:2836–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye-Rowley WS. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 2005;354:15–21. [DOI] [PubMed] [Google Scholar]
- Murley A, Nunnari J. The emerging network of mitochondria-organelle contacts. Mol Cell 2016;61:648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Nishimura T, Byrne SD et al. The ubiquitin ligase SCF(Ucc1) acts as a metabolic switch for the glyoxylate cycle. Mol Cell 2015;59:22–34. [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ et al. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 2012;337:587–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. A perspective on transport of proteins into mitochondria: a myriad of open questions. J Mol Biol 2015;427:1135–58. [DOI] [PubMed] [Google Scholar]
- Ng S, De Clercq I, Van Aken O et al. Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol Plant 2014;7:1075–93. [DOI] [PubMed] [Google Scholar]
- Nicastro R, Tripodi F, Gaggini M et al. Snf1 phosphorylates adenylate cyclase and negatively regulates protein kinase A-dependent transcription in Saccharomyces cerevisiae. J Biol Chem 2015;290:24715–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Zambrano A, Barrientos A. Suppression of polyglutamine-induced cytotoxicity in Saccharomyces cerevisiae by enhancement of mitochondrial biogenesis. FASEB J 2010;24:1431–41. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 2015;521:525–8. [DOI] [PubMed] [Google Scholar]
- Pan Y, Shadel GS. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging 2009;1:131–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A et al. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab 2011;13:668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Gligoris T, Tzamarias D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep 2004;5:368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R et al. The mitochondrial genotype can influence nuclear gene expression in yeast. Science 1987;235:576–80. [DOI] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E et al. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell 2003;12:1607–13. [DOI] [PubMed] [Google Scholar]
- Peng M, Ostrovsky J, Kwon YJ et al. Inhibiting cytosolic translation and autophagy improves health in mitochondrial disease. Hum Mol Genet 2015;24:4829–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova MI, Pujol-Carrion N, Arroyo J et al. Mtl1 is required to activate general stress response through Tor1 and Ras2 inhibition under conditions of glucose starvation and oxidative stress. J Biol Chem 2010;285:19521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti AA, Crutchfield CA, Rabinowitz JD et al. Survival of starving yeast is correlated with oxidative stress response and nonrespiratory mitochondrial function. Proc Natl Acad Sci USA 2011;108:E1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 2016;17:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Wallace DC, Burelle Y. The rise of mitochondria in medicine. Mitochondrion 2016;30:105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podholova K, Plocek V, Resetarova S et al. Divergent branches of mitochondrial signaling regulate specific genes and the viability of specialized cell types of differentiated yeast colonies. Oncotarget 2016;7:15299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. TOR signaling and S6 kinase 1: Yeast catches up. Cell Metab 2007;6:1–2. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ et al. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol 2002;22:8774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol 2016;17:213–26. [DOI] [PubMed] [Google Scholar]
- Ratnakumar S, Young ET. Snf1 dependence of peroxisomal gene expression is mediated by Adr1. J Biol Chem 2010;285:10703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Burckert N, Boller T et al. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev 1998;12:2943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Anjos RM, Camandona VL, Bleicher L et al. Structural and functional mapping of Rtg2p determinants involved in retrograde signaling and aging of Saccharomyces cerevisiae. PLoS One 2017;12:e0177090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan L, Zhou C, Jin E et al. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 2017;543:443–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Roig C, Noriega N, Duch A et al. The Hog1 SAPK controls the Rtg1/Rtg3 transcriptional complex activity by multiple regulatory mechanisms. Mol Biol Cell 2012;23:4286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh A, Movshovich N, Volokh M et al. Fine-tuning of the Msn2/4-mediated yeast stress responses as revealed by systematic deletion of Msn2/4 partners. Mol Biol Cell 2011;22:3127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P, Viana R, Garcia-Gimeno MA. AMPK in yeast: The SNF1 (sucrose non-fermenting 1) protein kinase complex. EXS 2016;107:353–74. [DOI] [PubMed] [Google Scholar]
- Schatz G, Haslbrunner E, Tuppy H. Deoxyribonucleic acid associated with yeast mitochondria. Biochem Biophys Res Commun 1964;15:127–32. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 2010;11:655–67. [DOI] [PubMed] [Google Scholar]
- Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 2003;43:139–60. [DOI] [PubMed] [Google Scholar]
- Segref A, Kevei E, Pokrzywa W et al. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metab 2014;19:642–52. [DOI] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 2015;347:75–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol 2018;19:109–20. [DOI] [PubMed] [Google Scholar]
- Solans A, Zambrano A, Rodriguez M et al. Cytotoxicity of a mutant huntingtin fragment in yeast involves early alterations in mitochondrial OXPHOS complexes II and III. Hum Mol Genet 2006;15:3063–81. [DOI] [PubMed] [Google Scholar]
- Strogolova V, Orlova M, Shevade A et al. Mitochondrial porin Por1 and its homolog Por2 contribute to the positive control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 2012;11:1568–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhm T, Kaimal JM, Dawitz H et al. Mitochondrial translation efficiency controls cytoplasmic protein homeostasis. Cell Metab 2018;27:1309–22. [DOI] [PubMed] [Google Scholar]
- Sun F, Cheng Y, Chen C. Regulation of heme biosynthesis and transport in metazoa. Sci China Life Sci 2015;58:757–64. [DOI] [PubMed] [Google Scholar]
- Sundaram V, Petkova MI, Pujol-Carrion N et al. Tor1, Sch9 and PKA downregulation in quiescence rely on Mtl1 to preserve mitochondrial integrity and cell survival. Mol Microbiol 2015;97:93–109. [DOI] [PubMed] [Google Scholar]
- Swinnen E, Ghillebert R, Wilms T et al. Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation in Saccharomyces cerevisiae. FEMS Yeast Res 2014;14:17–32. [DOI] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N et al. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem 2007;282:5617–24. [DOI] [PubMed] [Google Scholar]
- Topf U, Suppanz I, Samluk L et al. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat Commun 2018;9:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli NQ, Ferreira-Junior JR, Kowaltowski AJ et al. RTG1- and RTG2-dependent retrograde signaling controls mitochondrial activity and stress resistance in Saccharomyces cerevisiae. Free Radic Biol Med 2015;81:30–37. [DOI] [PubMed] [Google Scholar]
- Traven A, Wong JM, Xu D et al. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J Biol Chem 2001;276:4020–7. [DOI] [PubMed] [Google Scholar]
- Treitel MA, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA 1995;92:3132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B, Liang XB, Robert F et al. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res 2010;10:2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachova L, Cap M, Palkova Z. Yeast colonies: a model for studies of aging, environmental adaptation, and longevity. Oxid Med Cell Longev 2012;2012:601836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Carlson M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J 1999;18:6672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015;524:481–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zuo X, Kucejova B et al. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nat Cell Biol 2008;10:1090–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke V, Pedruzzi I, Cameroni E et al. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J 2005;24:4271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Liu L, Chen Q. Selective removal of mitochondria via mitophagy: distinct pathways for different mitochondrial stresses. Biochim Biophys Acta 2015;1853:2784–90. [DOI] [PubMed] [Google Scholar]
- Weidberg H, Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 2018;360:eaan4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC et al. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev 1993;57:383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem 2017;86:685–714. [DOI] [PubMed] [Google Scholar]
- Woo DK, Phang TL, Trawick JD et al. Multiple pathways of mitochondrial-nuclear communication in yeast: intergenomic signaling involves ABF1 and affects a different set of genes than retrograde regulation. Biochim Biophys Acta 2009;1789:135–45. [DOI] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 2015;524:485–8. [DOI] [PubMed] [Google Scholar]
- Yun J, Finkel T. Mitohormesis. Cell Metab 2014;19:757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Pracheil T, Thornton J et al. Adenosine triphosphate (ATP) is a candidate signaling molecule in the mitochondria-to-nucleus retrograde response pathway. Genes 2013;4:86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Bu P, Zeng J et al. Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J Biol Chem 2017;292:16942–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV et al. A mitochondrial specific stress response in mammalian cells. EMBO J 2002;21:4411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita Rendon O, Fredrickson EK, Howard CJ et al. Vms1p is a release factor for the ribosome-associated quality control complex. Nat Commun 2018;9:2197. [DOI] [PMC free article] [PubMed] [Google Scholar]