ABSTRACT
In mammals, the epithelial tissues of major salivary glands generate saliva and drain it into the oral cavity. For submandibular salivary glands (SMGs), the epithelial tissues arise during embryogenesis from naïve oral ectoderm adjacent to the base of the tongue, which begins to thicken, express SOX9 and invaginate into underlying mesenchyme. The developmental mechanisms initiating salivary gland development remain unexplored. In this study, we show that retinoic acid (RA) signaling activity at the site of gland initiation is colocalized with expression of retinol metabolic genes Rdh10 and Aldh1a2 in the underlying SMG mesenchyme. Utilizing a novel ex vivo assay for SMG initiation developed for this study, we show that RDH10 and RA are required for salivary gland initiation. Moreover, we show that the requirement for RA in gland initiation involves canonical signaling through retinoic acid receptors (RAR). Finally, we show that RA signaling essential for gland initiation is transduced specifically through RARα, with no contribution from other RAR isoforms. This is the first study to identify a molecular signal regulating mammalian salivary gland initiation.
KEY WORDS: Retinoic acid, Submandibular, Salivary gland, Embryo, RDH10, RAR, Mouse
Summary: This study, showing that retinoic acid regulates salivary gland initiation, is the first to identify a molecular signal directing transition of naïve oral epithelium toward salivary gland identity.
INTRODUCTION
Salivary gland dysfunction is a significant clinical problem, which negatively impacts oral health and quality of life for many individuals (Cassolato and Turnbull, 2003; Greenspan, 1996; Navazesh and Kumar, 2009; Sasportas et al., 2013; Villa et al., 2015). Developing therapies to regenerate damaged organs for patients will likely rely on insights gained from studies of embryonic development (Lombaert, 2017; Patel and Hoffman, 2014; Wei et al., 2007). During embryogenesis in mammals, salivary glands develop from reciprocal interactions between naïve oral epithelium and underlying mesenchyme. These tissue interactions are dynamic, with epithelium being instructive at one developmental stage and mesenchyme at another (Wells et al., 2013). Study of submandibular salivary gland (SMG) development in mice has revealed much about the genes and molecules involved in branching morphogenesis, differentiation and maintenance of progenitor pools (Knosp et al., 2015; Knox et al., 2010; Sequeira et al., 2010; Steinberg et al., 2005). However, knowledge of initiation events in salivary gland biogenesis is lacking and, to date, no genes or molecules regulating the initiation of the salivary gland epithelium have been identified.
In the mouse, SMG development begins between embryonic day (E) 11.0 and E11.5 as an epithelial thickening on either side of the lingual swellings (Tucker, 2007). This epithelium invaginates down into the mesenchyme to form an initial bud by E12.5. In subsequent stages, the epithelium undergoes branching morphogenesis, canalization and cell differentiation to create a mature arborized tissue that secretes most of the components of saliva.
The earliest known marker expressed in salivary gland epithelium is SOX9, which appears at ∼E11.0, when the oral epithelium begins to thicken (Chatzeli et al., 2017). Lineage tracing demonstrates that SOX9+ cells give rise to all epithelial cell types in the gland (Chatzeli et al., 2017). Signals responsible for activating the expression of SOX9 and, thus, specifying the population of early gland progenitors, are not yet known.
RA is a small lipid-soluble signaling molecule. It is the active metabolite of retinol, also known as vitamin A, and is essential for the morphogenesis of many organs and structures during embryonic development (Clagett-Dame and DeLuca, 2002; Duester, 2008; Metzler and Sandell, 2016; Niederreither and Dollé, 2008). RA has multiple modes of action, including nuclear functions and roles outside the nucleus (Al Tanoury et al., 2013; Iskakova et al., 2015). ‘Canonical’ RA signaling involves RA within the nucleus functioning as a ligand modulating the action of nuclear receptor transcription factors, specifically, members of the RAR family (RARα, RARβ and RARγ) and peroxisome proliferation-activated receptor (PPAR)-β/δ (Berry and Noy, 2009; Schug et al., 2007). The three distinct RAR isoforms have redundant activities and compensate for each other in some contexts (Ghyselinck et al., 1997; Manshouri et al., 1997), but perform isotype-specific activities in other settings (Duong and Rochette-Egly, 2011).
RA is metabolized from the precursor retinol by two successive oxidation reactions. First, retinol is reversibly converted to retinaldehyde primarily by the enzyme RDH10 (Sandell et al., 2007, 2012). The second reaction, oxidation of retinaldehyde to RA, is mediated by members of the aldehyde dehydrogenase 1A (ALDH1A) family (Niederreither and Dollé, 2008; Zhao et al., 1996). Owing to their role in the production of RA, RDH10 and ALDH1A family enzymes are crucial during embryonic development (Dupe et al., 2003; Sandell et al., 2012).
We and others have reported that RA signaling contributes to salivary gland development during branching morphogenesis (Abashev et al., 2017; DeSantis et al., 2017; Wright et al., 2015). We have also shown that RA has a role earlier in gland development, as evidenced by abnormal morphology of SMG initial buds in rare constitutive Rdh10 mutants that remain viable to E12.5 (Wright et al., 2015). We demonstrated that RA signaling is active within the mandibular portion of the first pharyngeal arch (PA1) in domains approximating the region of the initiating salivary glands before, and at the time of, gland initiation (Wright et al., 2015). Collectively, these observations suggest that RA, produced by RDH10-mediated retinol metabolism, is a crucial signal for initiating salivary gland development. However, no investigation has functionally assessed the role of RA, or any other molecular signal, in initiating salivary gland development. Here, we directly investigated the role of RDH10 and RA signaling in salivary gland initiation. We used a conditional in vivo genetic model and a novel ex vivo assay to show that RDH10 function and RA signaling regulate the initiation of SMG morphogenesis at the early stage of naïve oral ectoderm thickening, epithelial invagination and activation of SOX9. We also show that the action of RA in SMG development occurs via canonical signaling, mediated specifically by RARα. Thus, RA signaling is the first identified molecular mechanism regulating salivary gland initiation.
RESULTS
Rdh10 expression overlaps domains of RA signaling and precedes SMG initiation in developing mandibular arch
RA signaling does not occur uniformly throughout an embryo during development (Eichele and Thaller, 1987; Perz-Edwards et al., 2001; Rossant et al., 1991; Shimozono et al., 2013; Thaller and Eichele, 1987; Waxman and Yelon, 2011). We previously identified that RA signaling is present in the mandibular portion of PA1 at E10.5 and E11.5 in the region of SMG initiation (Wright et al., 2015). To investigate the relationship of mandibular arch tissues metabolizing retinol versus those where RA signaling is active, we compared the expression of Rdh10 with the distribution of RA signaling activity. We show here RA signaling data (Fig. 1D,H,J,L) similar to that we have previously reported (Wright et al., 2015) to facilitate comparison of domains of retinol metabolism with RA signaling.
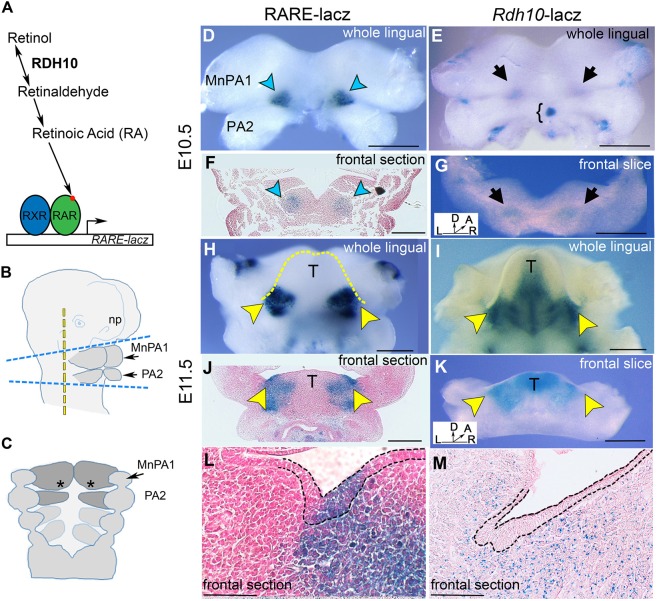
Fig. 1.
The retinol metabolism gene Rdh10 is expressed and RA signaling is active in developing mandible tissues before and during SMG initiation. (A) Diagram of RA metabolism and RA signaling. (B) Diagram of the cephalic region of the E10.5 mouse embryo with dashed lines to indicate the planes of dissection for whole-mount X-gal staining and explant culture. For explant culture, the tissues comprising the future mandible and adjacent neural tube, indicated as the area between the top and bottom blue dashed lines, were isolated and placed in a culture dish. For whole-mount X-gal staining, an additional cut was made, indicated by the vertical yellow dashed line. (C) Diagram of inner surface of the E10.5 PAs (asterisks indicate the region of prospective SMG initiation). (D,E) X-gal-stained E10.5 mandibular arch from RARE-lacZ transgenic embryos (D) and Rdh10βgeo embryos (E). RA signaling is indicated by blue arrowheads and Rdh10 expression is indicated by black arrows. (F,G) X-gal-stained frontal section or slice through the mandibular arch of reporter embryos at E10.5. (F) RARE-lacZ paraffin section. (G) Rdh10βgeo frontal tissue slice. Bracket indicates the thyroid primordium. (H,I) X-gal-stained E11.5 whole mandibular arch from RARE-lacZ transgenic embryos (H) and Rdh10βgeo embryos (I). Yellow dashed line outlines the lingual swellings/developing tongue. (J-L) X-gal-stained section through E11.5 SMG pre-bud region of RARE-lacZ embryo (J) and Rdh10βgeo embryo (K). (L) RARE-lacZ SMG pre-bud at E11.5 similar to that shown in J, but at a higher magnification (outline of epithelium shown by black dashed line). (M) Cryosection of Rdh10βgeo SMG pre-bud region at E11.5. Black dashed line outlines the oral epithelium. Yellow arrowheads indicate sites of SMG initiation. A, anterior; D, dorsal; L, left; MnPA1, mandibular portion of PA1; np, nasal prominence; R, right; T, tongue. Scale bars: 500 µm in D-K; 100 µm in L,M.
To compare the spatiotemporal distribution of Rdh10 expression and RA signaling within the developing mandibular arches, we utilized two different lacZ reporter mouse strains. Rdh10 expression was detected by X-gal staining tissues from mice carrying a Rdh10-lacZ knock-in reporter allele (Rdh10Bgeo) (Sandell et al., 2012), and RA signaling activity was visualized by X-gal staining tissues from mice with the RARE-lacZ transgene that detects canonical RA signaling (Rossant et al., 1991). The mandibular portions of PA1 along with second pharyngeal arches (PA2; shaded region in Fig. 1B), were isolated from E10.5 embryos and stained. At E10.5, Rdh10 expression and RA signaling were both detected in a bilateral pair of domains in posterior PA1, near the future site of fusion with PA2 (Fig. 1D,E). Frontal slices or sections through these tissues revealed that Rdh10 expression and RA signaling were both confined to the mesenchyme at this early stage (Fig. 1F,G). Visualizing the lingual surface of specimens that were 1 day older (E11.5) revealed that Rdh10 expression had expanded to include the midline of the tongue (Fig. 1I), whereas RA signaling remained as a bilateral pair of domains at the base of the developing tongue (Fig. 1H). Frontal slices or sections at E11.5 (Fig. 1J-M) revealed that Rdh10 expression remained restricted to the mesenchyme at this stage (Fig. 1K,M), whereas RA signaling expanded beyond the mesenchyme to include the thickened epithelium of the SMG pre-bud (Fig. 1J,L). Together, these data demonstrated that the metabolism of retinol, as assessed by Rdh10 expression, and RA signaling activity, as assessed by RARE-lacZ expression, occur in developing SMG tissues before and during initiation. At the time of initiation, production of the RA precursor retinaldehyde was restricted to the mesenchyme, whereas RA signaling occurred in both the underlying mesenchyme and the thickening epithelium.
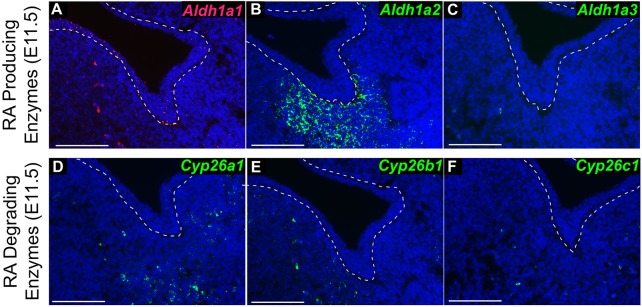
To fully characterize the distribution of RA metabolism and catabolism in tissues of the initiating SMG, we evaluated expression of Aldh1a family genes encoding RA-producing enzymes and Cyp26 family genes encoding RA-degrading enzymes. Analysis of E11.5 embryos by RNAscope RNA in situ hybridization (Wang et al., 2012) revealed that Aldh1a1 was expressed at a low level throughout the surface epithelium, with some expression in the mesenchyme (Fig. 2A). Aldh1a2 was expressed robustly in the SMG mesenchyme, but was not present in the epithelium (Fig. 2B). Aldh1a3 was not detected in mandibular tissues (Fig. 2C). Both Cyp26a1 and Cyp26b1 were sparsely expressed in the mesenchyme, whereas Cyp26c1 was not detected in mandibular tissues (Fig. 2D-F). These data indicate that ALDH1A2 is the predominant enzyme producing RA in the vicinity of SMG initiation.
Fig. 2.
The retinaldehyde metabolism gene Aldh1a2 is expressed in the mesenchyme of the SMG pre-bud at E11.5. (A-F) RNA in situ hybridization for Aldh1a1 (A), Aldh1a2 (B), Aldh1a3 (C), Cyp26a1 (D), Cyp26b1 (E) and Cyp26c1 (F) in frontal sections through the E11.5 SMG pre-bud (n=3 sections each). Oral epithelium is outlined by a dashed line. Scale bars: 100 µm.
Stage-specific inactivation of Rdh10 attenuates RA signaling in developing mandible
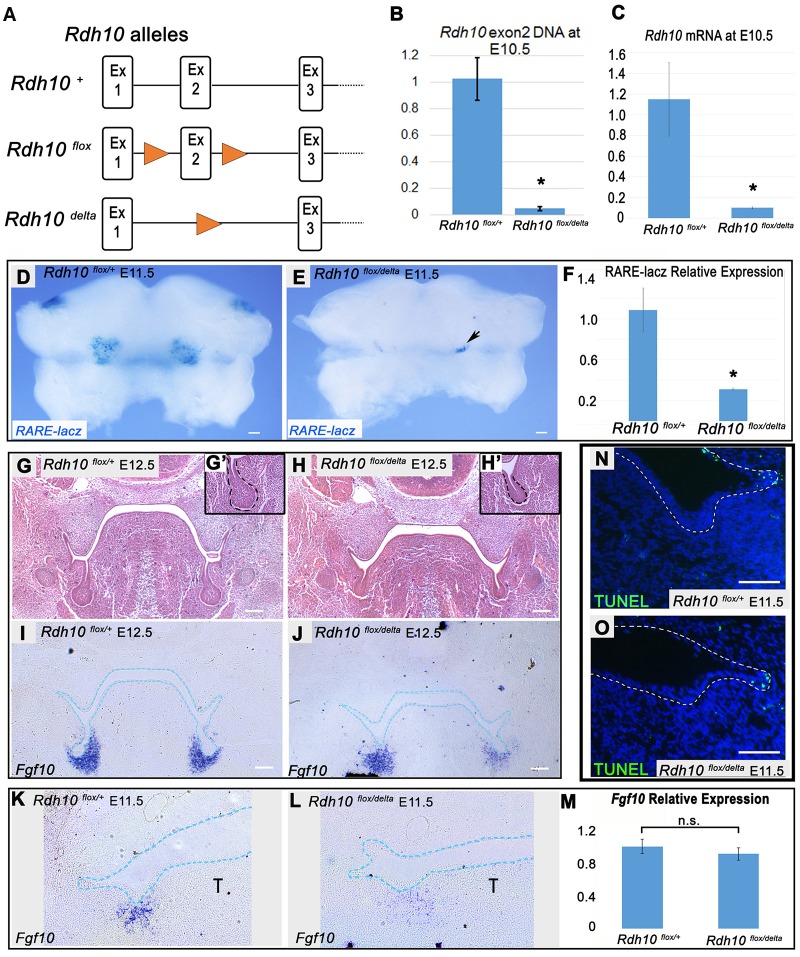
Owing to its essential role in retinol metabolism during embryogenesis, loss of RDH10 is usually embryonic lethal at or before E11.5 (Rhinn et al., 2011; Sandell et al., 2012). However, rare Rdh10−/− mutant embryos survive to E12.5 and we previously observed that SMG initial bud formation is abnormal in such embryos (Wright et al., 2015). The previous observations suggested that RDH10-mediated retinol metabolism is important for early gland development, but the overall dysmorphology of rare surviving E12.5 Rdh10−/− mutant embryos makes such phenotypes difficult to interpret. To better assess the role of RDH10 at initiation stages of SMG development, we utilized a tamoxifen-inducible Rdh10 knockout model (Kurosaka et al., 2017; Lenti et al., 2016; Sandell et al., 2012), which enables stage-specific inactivation of Rdh10. The genetic cross used for inactivation of Rdh10 involved a combination of Rdh10 mutant alleles (Fig. 3A). Rdh10+ represents the wild-type allele, Rdh10delta, a targeted knockout null allele in which Rdh10 Exon2 is deleted, and Rdh10flox, a floxed conditional allele of Rdh10 that is converted to Rdh10delta upon exposure to Cre recombinase (Sandell et al., 2012). Cre-mediated inactivation of the conditional Rdh10flox allele was accomplished using the tamoxifen-inducible Cre-ERT2 (Ventura et al., 2007).
Fig. 3.
Conditional inactivation of Rdh10 impairs SMG initial bud formation in vivo. (A) Rdh10 alleles used in conditional Rdh10 knockout cross. (B,C) qPCR of intact Rdh10 DNA (B) and Rdh10 RNA (C) from whole embryos at E10.5, 48 h after E8.5 tamoxifen administration, demonstrating the efficient conditional inactivation of Rdh10. (D,E) Whole-mount RARE-lacZ staining of E11.5 mandibles from Rdh10flox/+ control and Rdh10flox/delta conditional mutant embryos carrying the RARE-lacZ reporter transgene treated with tamoxifen at E8.5. Black arrow indicates a small region of residual RARE-lacZ staining in mutant embryos. (F) qPCR of RARE-lacZ gene expression from embryo bodies reveals that mutants retain 30% of the RA signaling activity (n=4 Rdh10flox/+;RARE-lacZ, n=2 Rdh10flox/delta;RARE-lacZ). *P<0.05. (G-H′) Frontal sections of E12.5 embryos stained with Hematoxylin and Eosin show defects in SMG initial gland bud formation in Rdh10flox/delta conditional mutants (H) relative to Rdh10flox/+ controls (G). Insets show higher magnification view of the initial bud (G′,H′). Dashed line outlines the oral epithelium. (I-L) Fgf10 RNA in situ hybridization of frontal sections through the SMG pre-bud of Rdh10flox/+ control (I) and Rdh10flox/delta conditional mutant (J) embryos at E12.5 (n=3 embryos each) and E11.5 (K,L) (n=3 embryos each) (blue dotted lines outline the epithelium). (M) qPCR of Fgf10 expression in whole E11.5 mandibular arches reveals no significant difference between control and Rdh10 conditional mutants (n=3 embryos each). (N,O) TUNEL staining of frontal sections through the SMG pre-bud at E11.5 reveals no difference in apoptosis in SMG initiation region between control (N) and Rdh10 conditional mutants (O) (n=3 embryos each). n.s., not significant; T, tongue. Scale bars: 100 µm.
For conditional inactivation of Rdh10 in embryos, timed matings were performed by crossing homozygous Rdh10flox/flox;Cre-ERT2/Cre-ERT2 mice with Rdh10delta/+ mice. In resulting litters, 50% of embryos were Rdh10+/flox ‘control’ embryos heterozygous for the wild-type allele of Rdh10, and 50% of embryos were Rdh10delta/flox ‘conditional mutants’ (Fig. S1A,B). All embryos had a single copy of Cre-ERT2. To inactivate the Rdh10 gene during development, a single dose of tamoxifen was administered via maternal oral gavage to activate Cre recombinase in embryos at E8.5. To measure the efficiency of gene inactivation, DNA from whole embryos was extracted at E10.5 (48 h post-tamoxifen treatment) from control Rdh10+/flox and conditional mutant Rdh10delta/flox embryos. To quantify the efficiency of Cre-mediated excision, qPCR was used to quantify Rdh10 exon2 (excised by Cre-recombinase) relative to Rdh10 exon3 (left intact by Cre-recombinase) by ΔCT measurement. The relative abundance of Rdh10 exon2 was then compared between control and conditional mutant embryos (n=3 embryos each) (Fig. 3B). Conditional mutant embryos retained 5% of the amount of Rdh10 exon2 relative to controls (P=0.001), indicating that Rdh10 gene inactivation at E10.5 was ∼95% efficient. To quantify the expression of any intact Rdh10 transcripts, mRNA was isolated from control and conditional mutant embryos at E10.5 (48 h post-tamoxifen treatment), and qPCR was performed for Rdh10 exon2 (n=3 embryos each) (Fig. 3C). In conditional mutant embryos, expression of intact Rdh10 mRNA was reduced to ∼10% of that of heterozygous control littermates (P=0.04).
To assess the level of RA signaling following Rdh10 inactivation, conditional mutant and control embryos carrying the RARE-lacZ reporter transgene were examined. Following treatment with tamoxifen at E8.5, mandibles from E11.5 embryos were isolated and X-gal stained. RARE-lacZ activity was noticeably reduced in mutant mandibles, indicating that conditional mutant embryos had a marked reduction in RA signaling relative to control littermates (Fig. 3D,E) (control n=4, mutant n=2 embryos). To quantify the reduction in RA signaling, qPCR was performed to measure lacZ expression in whole-embryo body tissues. By qPCR, RA signaling in the bodies of conditional mutant embryos was 30% of that of control littermates (Fig. 3F). The residual RA signaling activity could result from retinol metabolism by the small amount of remaining Rdh10 (Fig. 3B), from activity of an alternate RDH, or from metabolism of β-carotene, which can, under some conditions, reach an embryo from the maternal circulation (Wassef et al., 2013). These data demonstrated that conditional inactivation of Rdh10 attenuates, but does not eliminate, RA signaling in embryos at E11.5, when SMG morphogenesis is initiated.
Conditional inactivation of Rdh10 impairs early gland development
To determine whether RDH10 regulates early stages of SMG morphogenesis, we compared the formation of SMG initial buds in E12.5 Rdh10flox/+ control and Rdh10flox/delta conditional mutant embryos after tamoxifen treatment. Frontal paraffin sections through the mandible were stained with Hematoxylin and Eosin (n=3 embryos per genotype). The SMG initial buds of control Rdh10flox/+ embryos were well formed, each with a large rounded end bud and a distinct primary duct (Fig. 3G,G′). By contrast, the early initial bud of Rdh10flox/delta conditional mutant embryos was stunted, with a small, poorly formed end bud on a short stalk (Fig. 3H,H′). These data demonstrated that loss of RDH10 function impairs early SMG initial bud formation.
Given that the growth factor FGF10 is required for SMG morphogenesis beyond the pre-bud stage (Jaskoll et al., 2005), we examined Fgf10 expression in Rdh10flox/+ control and Rdh10flox/delta conditional mutant embryos by RNA in situ hybridization and qPCR. Analysis of Fgf10 RNA in situ-stained frontal paraffin sections at E11.5 and E12.5 revealed that Fgf10 was expressed in SMG mesenchyme of Rdh10flox/delta conditional mutant embryos (Fig. 3J,L), albeit at a somewhat reduced level and more diffuse pattern compared with Rdh10flox/+ control embryos (Fig. 3I,K) (n=3 per genotype). Quantitation of Fgf10 expression by qPCR on E11.5 mandibles showed that Fgf10 expression was unchanged in mutant tissue (Fig. 3M) (n=3 mandibles per genotype). These data indicated that the defect in initial SMG bud formation in Rdh10flox/delta conditional mutant embryos did not result from reduced expression of Fgf10.
We investigated the possibility that abnormal initial bud formation in mutants resulted from the elevated apoptotic cell death in SMG pre-bud tissues. Terminal deoxynucleotidyl transferase (dUTP) nick end labeling (TUNEL) analysis of E11.5 mandibles revealed sparse apoptotic cells in SMG epithelium and mesenchyme in both control and Rdh10flox/delta mutant embryos (Fig. 3N,O) (n=3 embryos per genotype). These data demonstrated that early SMG defects in mutant embryos do not result from elevated apoptosis. Condensation of the SMG mesenchyme appeared similar in the SMG of Rdh10flox/delta mutant embryos and of Rdh10flox/+ control embryos, as assessed by the ease of isolation and clearly defined perimeter of manually dissected glands at E13.5 (Fig. S2).
Novel assay for SMG initiation ex vivo
We next sought to develop an ex vivo assay to functionally test whether RA signaling regulates the transition of naïve oral epithelium into salivary gland epithelium. For these assays, pre-initiation mandibular arch tissues were cultured ex vivo using an air-interface culture system (Trowell, 1959) and SMG initiation was assessed by immunostaining and confocal microscopy.
Embryonic mandible tissues containing the mandibular component of PA1, PA2 and adjacent hindbrain (horizontal blue lines in Fig. 1B) were isolated from E10.5 embryos. Isolated tissue explants were placed with the interior lingual surface facing up onto filters at the air-liquid interface above a well filled with culture medium (Fig. 4A). Explants were cultured for 48 h (Fig. 4B), an interval paralleling the period of SMG initiation in vivo. To assess SMG epithelial invagination, tissues were whole-mount immunostained for E-cadherin (cadherin 1), cleared, and imaged by confocal microscopy through the entire thickness of the tissue (Fig. 4C,D). Rendering of the E-cadherin signal as a solid surface using Imaris image analysis software, and rotation of the 3D rendering 90° relative to the plane of growth, allows visualization of epithelial invagination (Fig. 4E,F). The extent of ex vivo SMG initiation was quantified using ImageJ or Imaris software to measure the volume of E-cadherin+ epithelium that had invaginated into underlying mesenchyme.
Fig. 4.
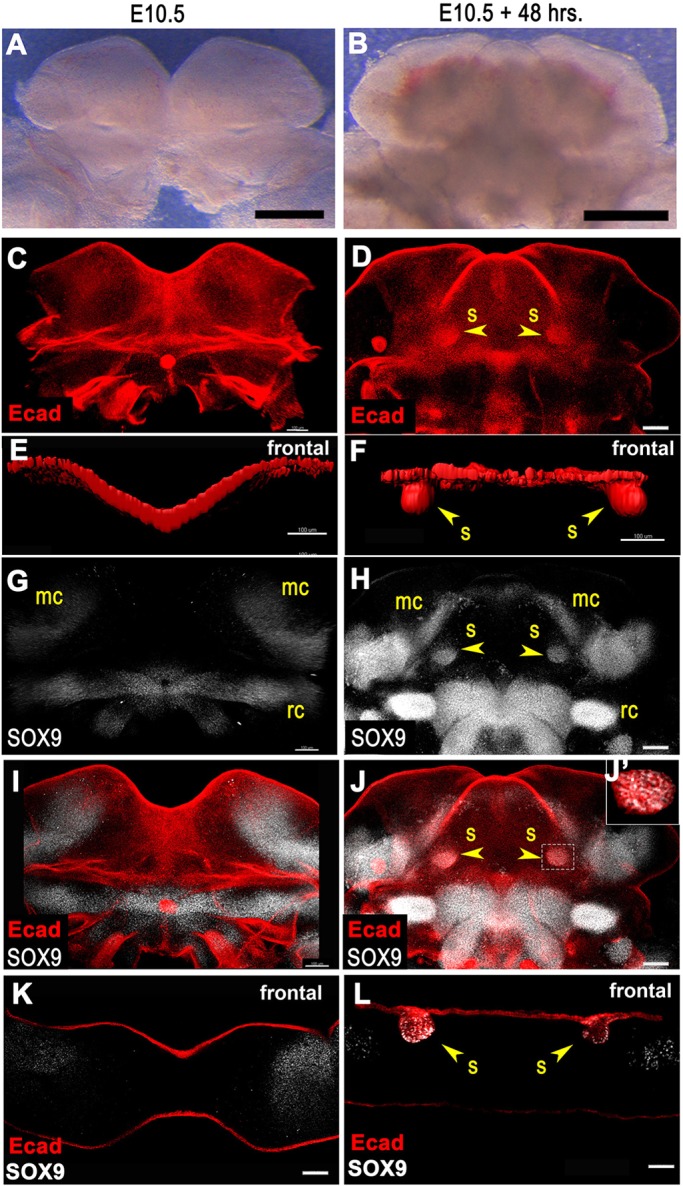
Novel ex vivo embryonic mandible culture system to assay SMG initiation. (A-D) E10.5 mandibular arch explants before (A) and after (B) 48 h of culture. (C,D) Mandibles before (C) and after (D) culture whole-mount immunostained for E-cadherin. Two dense spots of E-cadherin signal in cultured specimen correspond to sites of SMG initiation at the base of the tongue. (E,F) Surface rendering of E-cadherin immunofluorescence signal rotated to frontal view reveals SMG initial bud invaginated into underlying mesenchyme in cultured specimen (F). (G,H) Mandibles before (G) and after (H) culture whole-mount stained for SOX9. Two spots of SOX9 signals at the base of the tongue in cultured specimen correspond to initial SMG buds. (I,J) Merged whole-mount images with E-cadherin and SOX9. (J′) Single confocal optical slice shows regions of overlapping E-cadherin and SOX9 signals in the initiated SMG. (K,L) Frontal sections of mandibles co-stained for SOX9 and E-cadherin. (K) Optical frontal section of preculture mandible reveals no SOX9 staining in epithelium. (L) Frozen section of postculture mandible reveals two invaginated E-cadherin+ epithelial buds co-stained for SOX9. Yellow arrowheads indicate initiated SMG epithelial buds invaginated into underlying mesenchyme. mc, Meckel's cartilage; rc, Reichert's cartilage; s, submandibular gland initial bud. Scale bars: 500 µm in A,B; 100 µm in C-L.
Cultured explants were also immunostained for SOX9 to evaluate the differentiation of naïve oral epithelium into salivary epithelium (Fig. 4G-L). SOX9 marks cartilage progenitors in addition to salivary epithelium. In freshly isolated tissue explants, cartilage progenitor SOX9 was detected in the mesenchyme corresponding to regions of future cartilage development, but no SOX9 signal was detected in epithelium (Fig. 4G,I,K). After 48 h of culture, SOX9+ epithelium that co-stained with E-cadherin+ could be detected as epithelial buds that had invaginated downward from the surface into the underlying mesenchyme (Fig. 4H,J,L). The position of the bilateral pair of epithelial buds that had invaginated downward into the mesenchyme at the base of the tongue, and their SOX9+ status, indicated that these epithelial buds are the initiating SMGs.
The cultured mandible explant tissues preserve an in vivo-like expression pattern of Rdh10 and RA signaling (Fig. S3A,B). Supplementation of the culture medium with retinol, the metabolic precursor of retinaldehyde, and, ultimately, RA, resulted in enhanced RA signaling activity, demonstrating the explant tissues were able to metabolize retinol and respond to retinoid signaling in culture (Fig. S3C).
The Rdh10Bgeo allele used as a reporter for Rdh10 expression is a null mutation of Rdh10. To determine whether heterozygosity for Rdh10 in Rdh10Bgeo/+ explants impacts SMG initiation, we quantified SMG epithelium invagination in Rdh10+/+ versus Rdh10Bgeo/+ cultured mandible explants. No difference was detected in SMG initiation between Rdh10+/+ and Rdh10Bgeo/+ cultured explants (Fig. S4). Having established that heterozygosity for Rdh10 had no effect on SMG initiation, Rdh10Bgeo/+ and Rdh10+/+ embryos were subsequently used interchangeably for SMG explant culture experiments.
RDH10 is required for ex vivo SMG initiation
Our observation of early SMG defects in Rdh10flox/delta conditional mutant embryos (Fig. 3G,H) suggested that RDH10-mediated retinol metabolism is important for early SMG development. However, gland initiation was not completely abrogated in the in vivo Rdh10 conditional mutant model, possibly because the embryos in vivo retained a small amount of RA signaling (Fig. 3D-F). Given that the ex vivo cultured tissues responded to retinoids, we reasoned that the culture system could be used to elucidate the role of RDH10-mediated retinol metabolism and RA signaling in a reductive manner without the confounding influence of possible retinoid diffusion from other tissues or circulating retinoids or carotenoids.
Rdh10flox/+ control embryos and Rdh10flox/delta (or Rdh10flox/Bgeo) conditional mutant embryos were exposed to tamoxifen in vivo to inactivate the conditional floxed allele, as described above. Following tamoxifen treatment, mandibular arch tissues were isolated at E10.5 and cultured for 48 h. Whole-mount specimens were immunostained for E-cadherin and SOX9, and were imaged by confocal microscopy (Fig. 5A,B). All of the mandibles from Rdh10flox/+ control embryos exhibited robust SMG initiation, as assessed by invagination of the E-cadherin+ oral epithelium and by expression of SOX9 (n=3 mandibles) (Fig. 5A,A′,C,D). By contrast, Rdh10flox/delta conditional mutant mandibles completely failed to initiate SMG development (n=3 mandibles) (Fig. 5B,B′,C,D). These data demonstrated that SMG initiation requires RDH10 function.
Fig. 5.
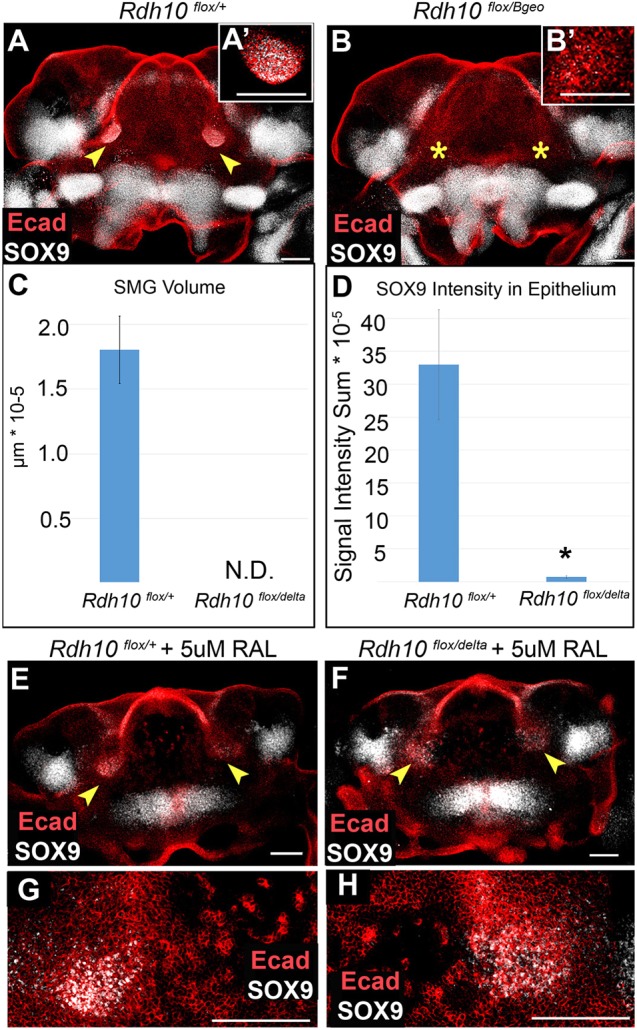
Conditional elimination of Rdh10 blocks ex vivo salivary gland initiation. (A,B) Mandibular explants from Rdh10flox/+ control and Rdh10flox/Bgeo conditional mutant embryos exposed to tamoxifen at E8.5 were cultured and whole-mount immunostained for E-cadherin and SOX9. SOX9+ epithelial buds are visible at the base of tongue in control mandibles, but are not detected in mutant mandibles. (A′,B′) Insets show higher magnification view of SMG region. (C) Gland volume assessed by measuring E-cadherin+ invaginated epithelium. Mutant mandible explants have no detectable SMG epithelium. Average volume and standard error bars are shown for each group (n=6). (D) SOX9 epithelial immunofluorescence was quantified by Imaris. Average intensity sum and standard error bars are shown for each group (n=6). *P<0.05. (E-H) Mandibles were cultured with 5 µM supplemental retinaldehyde, followed by immunostain. SOX9+ epithelial domains are detected in both Rdh10flox/+ control embryos (E,G) and (Rdh10flox/delta) conditional mutant embryos (F,H) (n=3 embryos each). (G,H) Higher magnification views of SOX9+ regions in E,F. Arrowheads indicate epithelial invagination into underlying mesenchyme, showing gland initiation; yellow asterisks indicate the absence of epithelial invagination. N.D., not detected; RAL, retinaldehyde. Scale bars: 100 µm.
RDH10 metabolizes retinol to retinaldehyde (Wu et al., 2002), and is the primary enzyme to do so as the first step in production of RA in embryos (Sandell et al., 2007). To determine whether the requirement for RDH10 in gland initiation results from its known enzymatic function in producing retinaldehyde, we tested whether adding exogenous retinaldehyde to cultured mutant mandibles could rescue the defect in SMG initiation. Whereas no SMG initiation was detected in the absence of retinaldehyde in mutant mandibles (Fig. 5A-D), supplementation with retinaldehyde rescued initiation in cultured mutant mandible explants, as assessed by the presence of SOX9+ epithelial cells at the base of the tongue (Fig. 5F,H). In retinaldehyde-supplemented explants, the SOX9+ expression domains were somewhat diffuse in both control and mutant tissues (Fig. 5E-H), relative to the tight domains in unsupplemented control samples, suggesting that localized production of retinaldehyde by RDH10 activity has a role in spatially restricting the SMG initiation domain.
SMG initiation is regulated by canonical RA signaling through RARs
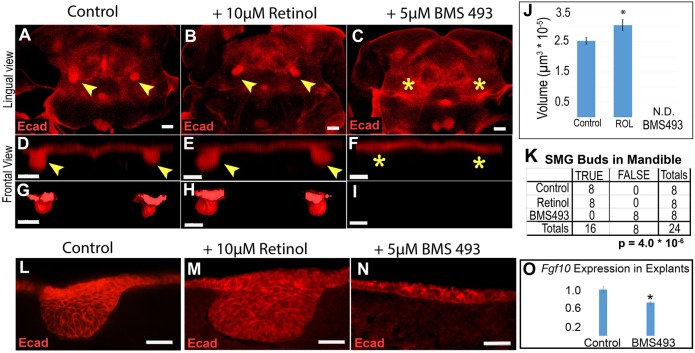
RA can function by a variety of mechanisms, including canonical signaling via RARs. We sought to determine whether SMG initiation was regulated by canonical RA signaling through RARs or by some other mechanism. Thus, we examined SMG initiation in the ex vivo mandible explant assay under conditions of abundant RA signaling, or when canonical RA signaling was completely blocked. To examine SMG initiation when RA signaling was robust, we added the metabolic precursor retinol to the culture medium, which enhanced RA signaling in a manner reflecting the in vivo pattern (Fig. S3C). To examine SMG initiation when canonical RA signaling was blocked, we added the pharmacological pan-RAR inhibitor BMS493 (Klein et al., 1996) to the culture medium. Mandibular arches from E10.5 embryos were cultured for 48 h with medium containing either 10 µM retinol, 5 µM BMS493, or vehicle dimethyl sulfoxide (DMSO) as a control (n=4 mandibles for each condition). Treatment with BMS493 or retinol did not impact the overall development of mandibular explants, because the cultured tissues continued to develop a visible tongue (Fig. S5). At the end of the culture period, explants were whole-mount stained for E-cadherin and imaged by confocal microscopy (Fig. 6A-I). ImageJ software was used to assess the presence or absence of SMG invaginated buds and to quantify gland bud volume (Fig. 6J,K). Imaris 3D image analysis software was used to render the E-cadherin signal as a solid surface to aid visualization (Fig. 6G-I).
Fig. 6.
SMG initiation is enhanced by addition of retinol and blocked by pharmacological inhibitor of RARs. (A-C) Mandibular explants were cultured with either vehicle control (A), 10 µM retinol (B) or 5 µM BMS493 (C), followed by whole-mount E-cadherin immunostaining and confocal microscopy. Bilateral patches of dense E-cadherin staining at base of tongue in control and retinol samples, but not in BMS493 samples, indicates that SMG initiation is blocked by RAR inhibitors. (D-I) Volume projections (D-F) and surface rendering (G-I) through the region of SMG gland initiation (rotated 90° from image plane for frontal view) are shown. (J) SMG epithelium volume quantitation indicates that retinol treatment increases initial bud size, whereas treatment with RAR inhibitor blocks initiation. Bar chart shows the average volume and standard error for each group (n=8 glands from four mandibles for each condition). (K) Contingency table for the presence or absence of initiated glands (TRUE versus FALSE) for each prospective site in the experiment (n=8 possible sites in four mandibles for each condition). (L-N) E-cadherin-stained frozen sections of cultured mandible explants reveal that inhibition of RAR blocks epithelial thickening (N). (O) qPCR of Fgf10 RNA in mandibles cultured on control or BMS493-treated medium reveals that treatment with RAR inhibitor causes a mild reduction in Fgf10 expression (n=3 mandibles). Yellow arrowheads indicate epithelial invagination into underlying mesenchyme, yellow asterisks indicate absence of epithelial invagination. *P<0.05. Scale bars: 100 µm.
All explants grown on control medium or retinol-treated medium initiated SMG development and had detectable epithelial buds (Fig. 6A,B,D,E,G,H,K). Addition of retinol to the culture medium caused a small but significant increase in the size of initiated SMG buds (Fig. 6J) (P=0.006). By contrast, explants cultured on medium containing BMS493 completely failed to initiate SMG development. No buds were observed in any mandibles treated with BMS493, indicating a significant absence of gland initiation (P=4.0*10−6, Fisher's exact test) (Fig. 6C,F,I,K). In BMS493-treated samples, Fgf10 expression was reduced to ∼70% of that in control explants, as quantified by qPCR (Fig. 6O). These data demonstrated that the presence of retinol enhanced SMG initiation and the action of retinoids on SMG initiation was mediated by canonical RA signaling through RARs.
To evaluate whether RA signaling is required to induce the earliest morphological manifestation of SMG initiation, the thickening of the oral epithelium, mandible specimens were cultured and processed for frozen-section immunostaining in frontal orientation (Fig. 6L-N) (n=3 mandibles per condition). For tissues grown on control medium or retinol-treated medium, E-cadherin staining revealed robust initial bud formation (Fig. 6L,M). By contrast, mandibles cultured on BMS493-treated medium failed to form any thickened epithelium in the SMG region (Fig. 6N). These data showed that the earliest stage of epithelial thickening in SMG initiation is regulated by canonical RA signaling through RARs.
Having identified that RA signaling through RARs is required for morphological initiation of SMG development, we next examined whether RA signaling is likewise needed to activate the expression of SOX9. Thus, we examined SOX9 in explants cultured with retinol or BMS493 by co-immunostaining whole-mount specimens for E-cadherin and SOX9. In all treatment groups, nonepithelial SOX9 signal was detected in mesenchymal domains lateral to the base of the tongue, consistent with the presence of SOX9 in cartilage progenitors (Fig. 7A-C″′). In control or retinol-cultured mandible explants, a strong epithelial SOX9 signal was detected coinciding with the SMG epithelial invaginations at the base of the tongue (Fig. 7A-B″′). In contrast to the control and retinol-treated explants, mandible explants cultured with BMS493 had a barely detectable epithelial SOX9 signal in the putative SMG initiation domain (Fig. 7C-C′″). Quantification of the SOX9 immunofluorescent signal revealed that epithelial SOX9 expression in BMS493-treated specimens was 0.6% of that of SOX9 expression in epithelial buds of mandibles grown on control medium (Fig. 7D) (P=0.002). These data demonstrated that the inhibition of canonical RA signaling by treatment with BMS493 blocks activation of SOX9 in oral epithelium. Thus, in addition to promoting morphological initiation of SMG development, active RA signaling through RARs is required for activation of a molecular marker of salivary gland initiation, SOX9.
Fig. 7.
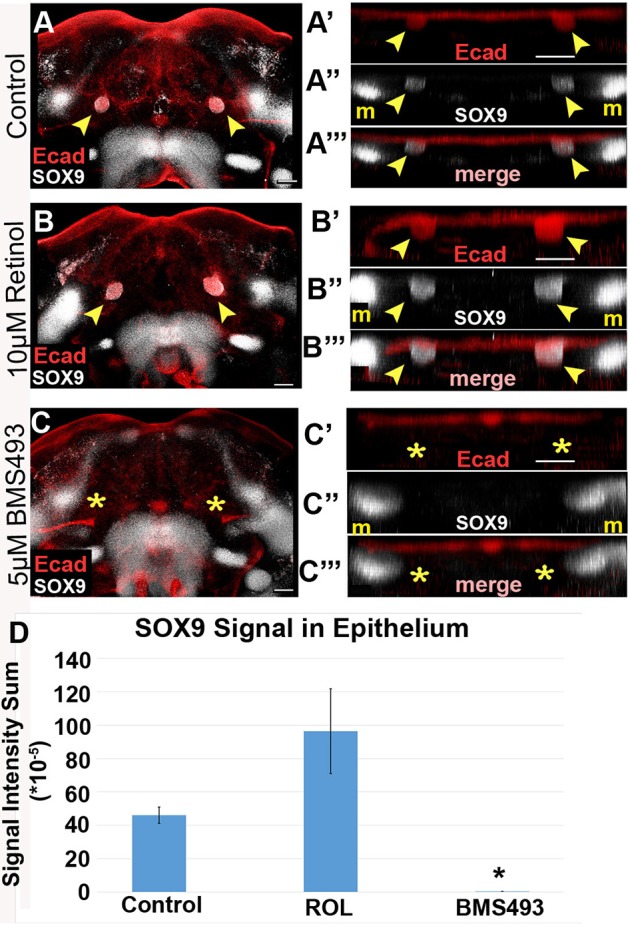
Treatment with a pan-RAR inhibitor blocks differentiation into SOX9+ salivary epithelium. (A-C‴) Mandibular explants were cultured on either 10 µM retinol, 5 µM BMS493, or vehicle control medium, whole-mount immunostained for E-cadherin and SOX9, and imaged by confocal microscopy. (A′-C′) Volume projections of E-cadherin staining in SMG initiation region, rotated for frontal view, show a bilateral pair of epithelial invaginations samples grown on control and retinol medium, but not on BMS493 medium. Control and retinol treated samples have a bilateral pair of SOX9+ expression domains that colocalize with the invaginated epithelium (A″,A‴,B″,B‴). The epithelial SOX9 expression domains are absent in BMS493-treated samples (C″,C‴). (D) SOX9 expression colocalized with epithelial buds was quantified by the intensity sum of the immunofluorescence signal. Average and standard error bars for each group are shown (n=4). Yellow arrowheads indicate epithelium invaginated into underlying mesenchyme, yellow asterisks indicate the absence of epithelial invagination. *P<0.05. m, Meckel's cartilage progenitors. Scale bars: 100 µm.
RA signal required for SMG initiation is transduced through RARα
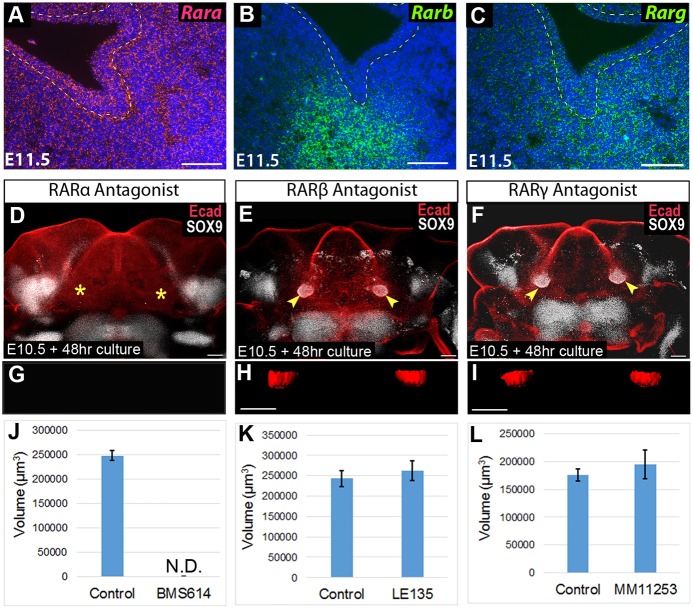
Having established that SMG initiation requires canonical RA signaling through RARs, we next investigated whether a specific RAR isoform was responsible for transducing the SMG initiation signal. To determine which RARs were present in early initiating SMG tissues, we used RNAScope RNA in situ hybridization to examine the transcription of the genes encoding each isoform during gland initiation. At E11.5, Rara was ubiquitously expressed throughout the oral tissues, Rarb was specifically localized to the SMG mesenchyme, and Rarg was expressed in epithelium and mesenchyme around the initiation SMG (Fig. 8A-C).
Fig. 8.
RA signaling required for gland initiation is mediated specifically by RARα, whereas other RAR isoforms are dispensable. (A-C) RNAscope in situ hybridization for Rara (A), Rarb (B) and Rarg (C) on frontal sections through the SMG pre-bud at E11.5. (D-F) E10.5 mandibular arch cultures were whole-mount stained for E-cadherin and SOX9 after treatment with an inhibitor for Rara (D), Rarb (E) or Rarg (F). (G-I) Surface rendering of representative SMG buds are shown for each condition (rotated 90° along the x-axis for frontal view). (J-L) Volume of invaginated gland buds for each treatment group versus vehicle control as quantified with ImageJ. Bar charts show the average and standard error bars for each group (n=4 for BMS614, n=6 for LE135 and MM11253). Yellow arrowheads indicate epithelial invagination into underlying mesenchyme, revealing gland initiation, yellow asterisks indicate the absence of epithelial invagination. N.D., not detected. Scale bars: 100 µm.
We next tested the effect of pharmacological inhibitors specific for each RAR isoform on ex vivo SMG initiation. RARα is inhibited by BMS614 (Germain et al., 2009), RARβ is inhibited by LE135 (Li et al., 1999) and RARγ is inhibited by MM11253 (Le et al., 2000). Each isotype-specific inhibitor was used at a final concentration of 5 µM. Mandibular arches were cultured on medium containing one of the pharmacological inhibitors, or on control medium containing the vehicle DMSO. Following culture, explant tissues were whole-mount immunostained for E-cadherin and SOX9, and were imaged by confocal microscopy.
Explants cultured on medium containing the RARα-specific antagonist BMS614 had no detectable SMG initiation (Fig. 8D,G,J). In these RARα-inhibited samples, no detectable SOX9 staining was observed in the region of gland initiation (Fig. 8D), and no gland bud could be detected (Fig. 8G,J) (n=4 glands). In contrast to the RARα inhibitor, which blocked salivary gland initiation, inhibitors specific for RARβ or RARγ did not disrupt ex vivo SMG development. Mandibles cultured with the RARβ inhibitor (LE135) and RARγ inhibitor (MM11253), initiated SMG development equivalent to tissues grown on control medium. For these latter specimens, SOX9 expression was activated in the epithelium (Fig. 8E,F), and the volume of the initial bud was similar to that of control tissues (Fig. 8H,I,K,L). These data demonstrated that SMG initiation requires RA signaling mediated exclusively by RARα.
DISCUSSION
This is the first study to identify a molecular signal directing initiation of salivary gland development. We demonstrated that retinol metabolism occurs in a pattern overlapping with, but not identical to, RA signaling in mandibular tissues before and during SMG initiation (Fig. 1). At E11.5, Rdh10 expression, marking sites where retinol is converted to retinaldehyde, and Aldh1a2 expression, marking sites of retinaldehyde conversion to RA, occurred exclusively in the mesenchyme (Fig. 1K,M, Fig. 2B), whereas RA signaling extended into overlying epithelium (Fig. 1J,L). Using a conditional genetic model of RA deficiency (Fig. 3), in conjunction with a novel ex vivo mandible explant assay (Fig. 4), we showed that RDH10-mediated retinol metabolism was crucial for SMG initiation (Fig. 5) and, using a pharmacological RAR inhibitor to block transduction of RAR signaling, we demonstrated that RAR signaling activity was required to direct the oral epithelium to initiate thickening and invagination into the underlying mesenchyme (Fig. 6), and to activate expression of the salivary gland marker SOX9 (Fig. 7). Furthermore, using isotype-specific inhibitors, we identified that the crucial RA signal needed for salivary gland initiation was mediated exclusively by RARα, with no contribution from RARβ or RARγ (Fig. 8). Together, these findings reveal that RA, produced by localized RDH10-mediated metabolism of retinol to retinaldehyde, signals through RARα to induce initiation of SMG development.
Addition of exogenous retinol ex vivo produced larger glands but did not produce any ectopic glands (Fig. 6). This is most likely because of the localized expression of retinol metabolic enzymes (Fig. 2), which restrict production of RA to regions of gland development.
Interactions between epithelium and mesenchyme orchestrate salivary gland development at growth and branching stages of morphogenesis (Grobstein, 1953a,b; Kratochwil, 1969; Kusakabe et al., 1985; Tucker, 2007; Tyler and Koch, 1977) and at earlier stages, including initiation (Wells et al., 2013). Retinoid molecules, which are lipophilic, can diffuse between cells and form gradients (Eichele and Thaller, 1987; Schilling et al., 2012; Shimozono et al., 2013; Thaller and Eichele, 1987). Our data showed that Rdh10 and Aldh1a2, which together encode enzymes responsible for the first and second steps in the conversion of retinol to RA, are both expressed in pre-SMG mandibular mesenchyme (Figs 1K,M, and 2B). These data suggested that diffusion of RA from mesenchyme into overlying epithelium is one tissue interaction that regulates salivary gland initiation. Such tissue interactions, based on differential sites of RA production and RA action, might parallel those that have been documented during early kidney development (Rosselot et al., 2010).
Initiation could be induced by RA signaling directly within oral epithelium, or by activity within the underlying mesenchyme. We previously showed that RA signaling acts directly within epithelial cells at the pseudoglandular stage of epithelial branching morphogenesis (Abashev et al., 2017; DeSantis et al., 2017). The fact that RA signaling is active within the epithelial cells of the initial pre-bud stage, SMG, suggests that RA acts directly within naïve oral epithelium to direct initiation. However, the presence of RA signaling in underlying mesenchyme also suggests that RA regulates SMG initiation indirectly. Future studies are needed to identify direct target genes of RA signaling and their localization during SMG initiation.
We also investigated the possibility that RA impacts SMG initiation by influencing the expression of Fgf10, the product of which is essential for SMG development at later stages (Chatzeli et al., 2017; Entesarian et al., 2005; Jaskoll et al., 2005; Lombaert and Hoffman, 2010; Milunsky et al., 2006; Ohuchi et al., 2000; Teshima et al., 2016; Wells et al., 2013). We found that abnormal initial bud formation in Rdh10 conditional mutant embryos (Fig. 3H,H″) was not associated with significant changes in Fgf10 expression (Fig. 3M), indicating that the early SMG phenotypes do not result from a loss of Fgf10. Similarly, treatment of mandible explants with the pan-RAR inhibitor BMS493 completely blocked SMG initiation, but caused only a mild reduction in Fgf10 expression (Fig. 6O). The modest reduction in Fgf10 in BMS493-treated samples could result from the lack of SMG epithelium, which controls the expression of Fgf10 in the underlying mesenchyme (Wells et al., 2013). That RA exerts its effect on SMG initiation independent of Fgf10, is consistent with the observation that the first epithelial invagination at SMG initiation does not require Fgf10 (Jaskoll et al., 2005).
RARs regulate gene transcription by interacting with different proteins, including co-activators and co-repressors. We demonstrated that SMG initiation was blocked in explants cultured with two different pharmacological inhibitors of RA signaling; the pan-RAR inhibitor BMS493, and the RARα isotype-specific inhibitor BMS614. BMS493, an ‘inverse agonist’, increases the affinity of RARs for co-repressors, leading to active repression of gene transcription. BMS614, a ‘neutral antagonist’, displaces co-activators but does not affect the recruitment of co-repressors to RAR (le Maire et al., 2010). The fact that both molecules had the same impact on SMG initiation shows that gland initiation can be inhibited either by an increase in co-repressor binding or loss of co-activators.
RARα null mutant mice have been generated previously (Lufkin et al., 1993), but a salivary gland phenotype has not been reported. A salivary gland phenotype in RARα null mutants might have been overlooked or, alternatively, RARα null mutants might have normal salivary glands owing to compensation by other RAR isoforms. Compensation between different RAR isoforms has been previously demonstrated for other structures and organs (Lohnes et al., 1994), and we showed here that both RARβ and RARγ are also expressed in the early salivary gland (Fig. 8B,C).
Here, we also demonstrated that expression of SOX9, the earliest known marker of SMG cell fate (Chatzeli et al., 2017), is dependent upon RA signaling. RA has been shown to regulate expression of Sox9 in other contexts. An agonist for RARα upregulates Sox9 in a breast cancer cell line (Afonja et al., 2002), and treatment with RA increases the expression of Sox9 in F9 teratocarcinoma cells (Laursen et al., 2013). However, the current study is the first to show that endogenous levels of RA signaling influence Sox9 expression in the context of normal tissue development. How RA regulates Sox9 is currently not known. No RAR-responsive cis-regulatory elements have been identified in Sox9 promoter DNA. RA might influence expression of Sox9 indirectly, or it might regulate Sox9 by direct action through an unidentified distant enhancer.
Current efforts to develop regenerative therapies for patients with salivary gland damage have mainly focused on the identification and isolation of stem cell or progenitor cell populations residing in adult glands. Manipulation of tissue-resident stem and/or progenitor populations might hold potential benefit for some patients. The current study suggests that retinoid signaling is one molecular pathway that could be manipulated to promote salivary epithelial identity.
MATERIALS AND METHODS
Mice
Mouse strains used in this paper are presented with official names in Table S1. Rdh10 mutant and conditional mutant alleles, Rdh10Bgeo, Rdh10flox and Rdh10delta, have been previously described (Sandell et al., 2012). The Rdh10Bgeo/+ strain was on a C57BL/6N background, as originally derived from embryonic stem cells obtained from the trans-NIH Knockout Mouse Project (KOMP) Repository, a NCRR-NIH supported strain repository (www.komp.org). For the experiments in this study, Rdh10flox/flox and Rdh10delta/+ mice were bred extensively to FVB/NJ mice, such that their background was mixed with a significant contribution from FVB/NJ mice. Additional mouse strains used were FVB/NJ, C57BL/6, RARE-lacZ and Cre-ERT2, all obtained from Jackson Laboratories and maintained at the University of Louisville.
For timed breeding, the day of the vaginal plug was considered to be E0.5. All experiments involving mice were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee at the University of Louisville.
Tamoxifen-induced conditional mutation
To induce conditional inactivation of the Rdh10flox allele in embryos, time-mated pregnant dams were administered tamoxifen plus progesterone. The progesterone was added to avoid spontaneous abortion (Nakamura et al., 2006). Each pregnant dam was given a single dose of 5 mg tamoxifen+1 mg progesterone in 250 µl of corn oil by oral gavage. The solution was administered between 09:00 h and 11:00 h on E8.5. The tamoxifen+progesterone solution was prepared as follows: 20 mg/ml tamoxifen (Sigma-Aldrich, T5648), 40 µl/ml 100% ethanol and 1 mg/ml progesterone (Sigma-Aldrich, P3972) were dissolved in corn oil (VWR, 700000-136) by incubating the solution at 56°C and vortexing every 15-30 min until clear. Solution was aliquoted into single-use tubes and stored at −20°C. Solution was used within 2 weeks.
X-gal staining of lacZ reporter embryos
Tissue from embryos containing either the RARE-lacZ or Rdh10Bgeo reporter were stained with an X-gal substrate as previously described (Wright et al., 2015). For whole-mount tissue staining, tissue was fixed in 2% paraformaldehyde/0.2% glutaraldehyde for 45-90 min on ice, incubated with Rise Solution A (Millipore, BG-6-B) for 30 min at room temperature, and then incubated with Rise Solution B (Millipore, BG-7-B) for 15 min at 37°C. Tissue was then placed in X-gal solution [Stain Base Solution (Millipore, BG-8-C)+1 mg/ml X-gal (Sigma-Aldrich, B4252)] overnight in a light-protected area. For paraffin sectioning of whole-mount stained specimens, RARE-lacZ embryos were whole-mount stained as described above, and then processed and embedded in paraffin. After sectioning, slides were counter stained with Nuclear Fast Red (VWR, JTS635-1). LacZ expression in Rdh10βgeo embryos was very weak relative to the strong expression in RARE-lacZ embryos, and did not withstand the paraffin-embedding procedure. Therefore, to detect lacZ expression signal in a frontal section view of Rdh10βgeo embryos, tissues were stained after slicing or cryosectioning. Tissue slices were prepared using a McIlwain tissue chopper.
Mandible ex vivo culture for SMG initiation
Mandible tissue explants comprising the mandibular component of PA1 plus the entire PA2 along with the adjacent hindbrain region were dissected out of E10.5 embryos. The mandible explants were plated with the interior surface facing up onto filter disks (Whatman Nucleopore, 13 mm, 0.1 μm pore size: VWR) supported on medium surface by silicone gaskets (CultureWell Gaskets, Grace Biolabs). The culture medium was DMEM/F12 with 100 U/ml penicillin, 100 μg/ml streptomycin, 150 μg/ml Vitamin C and 50 μg/ml transferrin. Tissue was cultured in a humidified incubator at 37°C with 5% CO2/ 95% air. For RAR inhibition experiments, cultures were treated with 5 µM of either BMS493, BMS614, LE135 or MM 11253 (Tocris, #3509, #3660, #2021 and #3822, respectively). All chemical inhibitors were suspended in DMSO. For supplementation with retinol (vitamin A), retinol (Sigma-Aldrich, #95144) suspended in ethanol was added to the culture medium at a final concentration of 10 µM. For supplementation with retinaldehyde, 5 µM retinaldehyde (Sigma-Aldrich, #R2500) suspended in DMSO was added to the culture medium at a final concentration of 5 µM. In each case, control samples were cultured with an equal volume of the vehicle DMSO or ethanol added. Culture media was changed daily.
Immunostaining
Immunostaining of frozen sections and whole-mount tissues was performed as previously described (Abashev et al., 2017). Primary antibodies used were anti-E-cadherin (BD Biosciences #610182; 1:50), and anti-SOX9 (Abcam, #185966; 1:200). Fluorescently conjugated secondary antibodies Alexa Fluor 546, or Alexa Fluor 660 (Invitrogen), were used at 1:300.
Imaging and analysis
Confocal imaging of whole-mount tissue was performed using an Olympus MPE FV1000 equipped with a far-red laser. Volume measurements were performed in ImageJ using the 3D Objects Counter plugin. Epithelial-specific SOX9 signal was measured using IMARIS (Bitplane AG). To quantify the SOX9 immunofluorescent signal specifically within SMG (omitting non-SMG signal), invaginating SMG epithelial buds were isolated based on a surface-rendered E-cadherin signal. Once non-SMG signal was excluded, the SOX9 immunostain Signal Intensity Sum was recorded for each sample. Statistical analysis was performed using the Student’s t-test.
In situ hybridization
RNA in situ hybridization for Fgf10 on paraffin sections was performed as previously described (Elliott et al., 2018).
RNA in situ hybridization for RA metabolic enzymes as well as RAR isoforms was performed using RNAscope Multiplex Fluorescent Assay v2 (Cat. # 323110), from Advanced Cell Diagnostics, on paraffin sections according to the manufacturer’s instructions. Probes were used to hybridize to Aldh1a1 (491321-C2), Aldh1a2 (447391), Aldh1a3 (501201), Cyp26a1 (468911), Cyp26b1 (454241), Cyp26c1 (481861), RARα (463081-C2), RARβ (463101) and RARγ (463111). Probes were fluorescently labeled using Opal Fluorophores 520 (FP1487001KT) or 570 (FP1488001KT) from PerkinElmer. Images were acquired on a Zeiss Axio Imager.A1 mounted with a Zeiss Axiocam MRc5.
Apoptosis assay
TUNEL staining was performed on paraffin sections using the In Situ Cell Death Detection Kit, Fluorescein (Cat. # 11684795910) from Sigma-Aldrich as per the manufacturer’s instructions.
qPCR
RNA was extracted from tissue using the RNeasy Micro kit (Qiagen), and converted to cDNA using the SuperScript III Reverse Transcriptase kit (Invitrogen). Primers used for qPCR amplification are listed in Table S2. The calculated efficiency for each primer pair was 100-108% (data not shown).
All qPCR was performed on an ABI 7500, using SYBR Select Master Mix (Thermo Fisher Scientific). For data analysis, both Gapdh and Actb were used as normalization control genes. Relative expression was quantified using the ΔΔCT method (Livak and Schmittgen, 2001), and the Student’s t-test was used to determine statistical significance.
Supplementary Material
Acknowledgements
The authors thank the University of Louisville School of Dentistry, Department of Oral Immunology and Infectious Diseases for helpful discussions, and the use of core equipment and resources.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.A.M., L.L.S.; Methodology: M.A.M., L.L.S.; Validation: M.A.M.; Formal analysis: M.A.M.; Investigation: M.A.M., S.R., K.H.E., R.M.F., N.Q.H.T.; Resources: L.L.S.; Data curation: M.A.M.; Writing - original draft: M.A.M.; Writing - review & editing: M.A.M., S.A.B., M.L., L.L.S.; Visualization: M.A.M., L.L.S.; Supervision: S.A.B., L.L.S.; Project administration: L.L.S.; Funding acquisition: M.A.M., S.A.B., L.L.S.
Funding
This work was supported by the National Institute of Dental and Craniofacial Research (M.A.M. is supported by F32DE027277; R15DE025960 to L.L.S.; R01DE023804 to S.A.B.; R56 DE2246706 to M.L.) and the National Institutes of Health (T32GM105526 to K.H.E.). Confocal imaging was supported by National Institute of General Medicine (GM10350). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.164822.supplemental
References
- Abashev T. M., Metzler M. A., Wright D. M. and Sandell L. L. (2017). Retinoic acid signaling regulates Krt5 and Krt14 independently of stem cell markers in submandibular salivary gland epithelium. Dev. Dyn. 246, 135-147. 10.1002/dvdy.24476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonja O., Raaka B. M., Huang A., Das S., Zhao X., Helmer E., Juste D. and Samuels H. H. (2002). RAR agonists stimulate SOX9 gene expression in breast cancer cell lines: evidence for a role in retinoid-mediated growth inhibition. Oncogene 21, 7850-7860. 10.1038/sj.onc.1205985 [DOI] [PubMed] [Google Scholar]
- Al Tanoury Z., Piskunov A. and Rochette-Egly C. (2013). Vitamin A and retinoid signaling: genomic and nongenomic effects. J. Lipid Res. 54, 1761-1775. 10.1194/jlr.R030833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. C. and Noy N. (2009). All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol. Cell. Biol. 29, 3286-3296. 10.1128/MCB.01742-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassolato S. F. and Turnbull R. S. (2003). Xerostomia: clinical aspects and treatment. Gerodontology 20, 64-77. 10.1111/j.1741-2358.2003.00064.x [DOI] [PubMed] [Google Scholar]
- Chatzeli L., Gaete M. and Tucker A. S. (2017). Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development 144, 2294-2305. 10.1242/dev.146019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clagett-Dame M. and DeLuca H. F. (2002). The role of vitamin A in mammalian reproduction and embryonic development. Annu. Rev. Nutr. 22, 347-381. 10.1146/annurev.nutr.22.010402.102745E [DOI] [PubMed] [Google Scholar]
- DeSantis K. A., Stabell A. R., Spitzer D. C., O'Keefe K. J., Nelson D. A. and Larsen M. (2017). RARα and RARγ reciprocally control K5+ progenitor cell expansion in developing salivary glands. Organogenesis 13, 125-140. 10.1080/15476278.2017.1358336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. (2008). Retinoic acid synthesis and signaling during early organogenesis. Cell 134, 921-931. 10.1016/j.cell.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong V. and Rochette-Egly C. (2011). The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim. Biophys. Acta 1812, 1023-1031. 10.1016/j.bbadis.2010.10.007 [DOI] [PubMed] [Google Scholar]
- Dupe V., Matt N., Garnier J.-M., Chambon P., Mark M. and Ghyselinck N. B. (2003). A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. USA 100, 14036-14041. 10.1073/pnas.2336223100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichele G. and Thaller C. (1987). Characterization of concentration gradients of a morphogenetically active retinoid in the chick limb bud. J. Cell Biol. 105, 1917-1923. 10.1083/jcb.105.4.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott K. H., Millington G. and Brugmann S. A. (2018). A novel role for cilia-dependent sonic hedgehog signaling during submandibular gland development. Dev. Dyn. 247, 818-831. 10.1002/dvdy.24627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entesarian M., Matsson H., Klar J., Bergendal B., Olson L., Arakaki R., Hayashi Y., Ohuchi H., Falahat B., Bolstad A. I. et al. (2005). Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nat. Genet. 37, 125-128. 10.1038/ng1507 [DOI] [PubMed] [Google Scholar]
- Germain P., Gaudon C., Pogenberg V., Sanglier S., Van Dorsselaer A., Royer C. A., Lazar M. A., Bourguet W. and Gronemeyer H. (2009). Differential action on coregulator interaction defines inverse retinoid agonists and neutral antagonists. Chem. Biol. 16, 479-489. 10.1016/j.chembiol.2009.03.008 [DOI] [PubMed] [Google Scholar]
- Ghyselinck N. B., Dupe V., Dierich A., Messaddeq N., Garnier J. M., Rochette-Egly C., Chambon P. and Mark M. (1997). Role of the retinoic acid receptor beta (RARbeta) during mouse development. Int. J. Dev. Biol. 41, 425-447. [PubMed] [Google Scholar]
- Greenspan D. (1996). Xerostomia: diagnosis and management. Oncology 10, 7-11. [PubMed] [Google Scholar]
- Grobstein C. (1953a). Inductive epitheliomesenchymal interaction in cultured organ rudiments of the mouse. Science 118, 52-55. 10.1126/science.118.3054.52 [DOI] [PubMed] [Google Scholar]
- Grobstein C. (1953b). Morphogenetic interaction between embryonic mouse tissues separated by a membrane filter. Nature 172, 869-870. 10.1038/172869a0 [DOI] [PubMed] [Google Scholar]
- Iskakova M., Karbyshev M., Piskunov A. and Rochette-Egly C. (2015). Nuclear and extranuclear effects of vitamin A. Can. J. Physiol. Pharmacol. 93, 1065-1075. 10.1139/cjpp-2014-0522 [DOI] [PubMed] [Google Scholar]
- Jaskoll T., Abichaker G., Witcher D., Sala F. G., Bellusci S., Hajihosseini M. K. and Melnick M. (2005). FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC Dev. Biol. 5, 11 10.1186/1471-213X-5-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E. S., Pino M. E., Johnson A. T., Davies P. J. A., Nagpal S., Thacher S. M., Krasinski G. and Chandraratna R. A. S. (1996). Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J. Biol. Chem. 271, 22692-22696. 10.1074/jbc.271.37.22692 [DOI] [PubMed] [Google Scholar]
- Knosp W. M., Knox S. M., Lombaert I. M. A., Haddox C. L., Patel V. N. and Hoffman M. P. (2015). Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Dev. Cell 32, 667-677. 10.1016/j.devcel.2015.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox S. M., Lombaert I. M. A., Reed X., Vitale-Cross L., Gutkind J. S. and Hoffman M. P. (2010). Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science 329, 1645-1647. 10.1126/science.1192046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil K. (1969). Organ specificity in mesenchymal induction demonstrated in the embryonic development of the mammary gland of the mouse. Dev. Biol. 20, 46-71. 10.1016/0012-1606(69)90004-9 [DOI] [PubMed] [Google Scholar]
- Kurosaka H., Wang Q., Sandell L., Yamashiro T. and Trainor P. A. (2017). Rdh10 loss-of-function and perturbed retinoid signaling underlies the etiology of choanal atresia. Hum. Mol. Genet. 26, 1268-1279. 10.1093/hmg/ddx031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe M., Sakakura T., Sano M. and Nishizuka Y. (1985). A pituitary-salivary mixed gland induced by tissue recombination of embryonic pituitary epithelium and embryonic submandibular gland mesenchyme in mice. Dev. Biol. 110, 382-391. 10.1016/0012-1606(85)90097-1 [DOI] [PubMed] [Google Scholar]
- Laursen K. B., Mongan N. P., Zhuang Y., Ng M. M., Benoit Y. D. and Gudas L. J. (2013). Polycomb recruitment attenuates retinoic acid-induced transcription of the bivalent NR2F1 gene. Nucleic Acids Res. 41, 6430-6443. 10.1093/nar/gkt367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Q., Dawson M. I., Soprano D. R. and Soprano K. J. (2000). Modulation of retinoic acid receptor function alters the growth inhibitory response of oral SCC cells to retinoids. Oncogene 19, 1457-1465. 10.1038/sj.onc.1203436 [DOI] [PubMed] [Google Scholar]
- le Maire A., Teyssier C., Erb C., Grimaldi M., Alvarez S., de Lera A. R., Balaguer P., Gronemeyer H., Royer C. A., Germain P. et al. (2010). A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor. Nat. Struct. Mol. Biol. 17, 801-807. 10.1038/nsmb.1855 [DOI] [PubMed] [Google Scholar]
- Lenti E., Farinello D., Yokoyama K. K., Penkov D., Castagnaro L., Lavorgna G., Wuputra K., Sandell L. L., Tjaden N. E. B., Bernassola F. et al. (2016). Transcription factor TLX1 controls retinoic acid signaling to ensure spleen development. J. Clin. Invest. 126, 2452-2464. 10.1172/JCI82956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hashimoto Y., Agadir A., Kagechika H. and Zhang X. (1999). Identification of a novel class of retinoic acid receptor beta-selective retinoid antagonists and their inhibitory effects on AP-1 activity and retinoic acid-induced apoptosis in human breast cancer cells. J. Biol. Chem. 274, 15360-15366. 10.1074/jbc.274.22.15360 [DOI] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔC(T) method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lohnes D., Mark M., Mendelsohn C., Dolle P., Dierich A., Gorry P., Gansmuller A. and Chambon P. (1994). Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development 120, 2723-2748. [DOI] [PubMed] [Google Scholar]
- Lombaert I. M. A. and Hoffman M. P. (2010). Epithelial stem/progenitor cells in the embryonic mouse submandibular gland. Front. Oral Biol. 14, 90-106. 10.1159/000313709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert I. M. A. (2017). Implications of salivary gland developmental mechanisms for the regeneration of adult damaged tissues. In Salivary Gland Development and Regeneration (ed. S. Cha), pp. 3-22. Cham: Springer. [Google Scholar]
- Lufkin T., Lohnes D., Mark M., Dierich A., Gorry P., Gaub M. P., LeMeur M. and Chambon P. (1993). High postnatal lethality and testis degeneration in retinoic acid receptor alpha mutant mice. Proc. Natl. Acad. Sci. USA 90, 7225-7229. 10.1073/pnas.90.15.7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manshouri T., Yang Y., Lin H., Stass S. A., Glassman A. B., Keating M. J. and Albitar M. (1997). Downregulation of RAR alpha in mice by antisense transgene leads to a compensatory increase in RAR beta and RAR gamma and development of lymphoma. Blood 89, 2507-2515. [PubMed] [Google Scholar]
- Metzler M. A. and Sandell L. L. (2016). Enzymatic metabolism of vitamin A in developing vertebrate embryos. Nutrients 8, E812 10.3390/nu8120812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky J. M., Zhao G., Maher T. A., Colby R. and Everman D. B. (2006). LADD syndrome is caused by FGF10 mutations. Clin. Genet. 69, 349-354. 10.1111/j.1399-0004.2006.00597.x [DOI] [PubMed] [Google Scholar]
- Nakamura E., Nguyen M.-T. and Mackem S. (2006). Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 235, 2603-2612. 10.1002/dvdy.20892 [DOI] [PubMed] [Google Scholar]
- Navazesh M. and Kumar S. K. (2009). Xerostomia: prevalence, diagnosis, and management. Compend. Contin. Educ. Dent. 30, 326-328. [PubMed] [Google Scholar]
- Niederreither K. and Dollé P. (2008). Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 9, 541-553. 10.1038/nrg2340 [DOI] [PubMed] [Google Scholar]
- Ohuchi H., Hori Y., Yamasaki M., Harada H., Sekine K., Kato S. and Itoh N. (2000). FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 277, 643-649. 10.1006/bbrc.2000.3721 [DOI] [PubMed] [Google Scholar]
- Patel V. N. and Hoffman M. P. (2014). Salivary gland development: a template for regeneration. Semin. Cell Dev. Biol. 25-26, 52-60. 10.1016/j.semcdb.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz-Edwards A., Hardison N. L. and Linney E. (2001). Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev. Biol. 229, 89-101. 10.1006/dbio.2000.9979 [DOI] [PubMed] [Google Scholar]
- Rhinn M., Schuhbaur B., Niederreither K. and Dolle P. (2011). Involvement of retinol dehydrogenase 10 in embryonic patterning and rescue of its loss of function by maternal retinaldehyde treatment. Proc. Natl. Acad. Sci. USA 108, 16687-16692. 10.1073/pnas.1103877108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Zirngibl R., Cado D., Shago M. and Giguere V. (1991). Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5, 1333-1344. 10.1101/gad.5.8.1333 [DOI] [PubMed] [Google Scholar]
- Rosselot C., Spraggon L., Chia I., Batourina E., Riccio P., Lu B., Niederreither K., Dolle P., Duester G., Chambon P. et al. (2010). Non-cell-autonomous retinoid signaling is crucial for renal development. Development 137, 283-292. 10.1242/dev.040287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L. L., Lynn M. L., Inman K. E., McDowell W. and Trainor P. A. (2012). RDH10 oxidation of Vitamin A is a critical control step in synthesis of retinoic acid during mouse embryogenesis. PLoS ONE 7, e30698 10.1371/journal.pone.0030698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L. L., Sanderson B. W., Moiseyev G., Johnson T., Mushegian A., Young K., Rey J.-P., Ma J.-X., Staehling-Hampton K. and Trainor P. A. (2007). RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 21, 1113-1124. 10.1101/gad.1533407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasportas L. S., Hosford D. N., Sodini M. A., Waters D. J., Zambricki E. A., Barral J. K., Graves E. E., Brinton T. J., Yock P. G., Le Q.-T. et al. (2013). Cost-effectiveness landscape analysis of treatments addressing xerostomia in patients receiving head and neck radiation therapy. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 116, e37-e51. 10.1016/j.oooo.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling T. F., Nie Q. and Lander A. D. (2012). Dynamics and precision in retinoic acid morphogen gradients. Curr. Opin. Genet. Dev. 22, 562-569. 10.1016/j.gde.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug T. T., Berry D. C., Shaw N. S., Travis S. N. and Noy N. (2007). Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129, 723-733. 10.1016/j.cell.2007.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira S. J., Larsen M. and DeVine T. (2010). Extracellular matrix and growth factors in salivary gland development. Front. Oral Biol. 14, 48-77. 10.1159/000313707 [DOI] [PubMed] [Google Scholar]
- Shimozono S., Iimura T., Kitaguchi T., Higashijima S. and Miyawaki A. (2013). Visualization of an endogenous retinoic acid gradient across embryonic development. Nature 496, 363-366. 10.1038/nature12037 [DOI] [PubMed] [Google Scholar]
- Steinberg Z., Myers C., Heim V. M., Lathrop C. A., Rebustini I. T., Stewart J. S., Larsen M. and Hoffman M. P. (2005). FGFR2b signaling regulates ex vivo submandibular epithelial cell proliferation and branching morphogenesis. Development 132, 1223-1234. 10.1242/dev.01690 [DOI] [PubMed] [Google Scholar]
- Teshima T. H. N., Lourenco S. V. and Tucker A. S. (2016). Multiple Cranial Organ Defects after Conditionally Knocking Out Fgf10 in the Neural Crest. Front. Physiol. 7, 488 10.3389/fphys.2016.00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller C. and Eichele G. (1987). Identification and spatial distribution of retinoids in the developing chick limb bud. Nature 327, 625-628. 10.1038/327625a0 [DOI] [PubMed] [Google Scholar]
- Trowell O. A. (1959). The culture of mature organs in a synthetic medium. Exp. Cell Res. 16, 118-147. 10.1016/0014-4827(59)90201-0 [DOI] [PubMed] [Google Scholar]
- Tucker A. S. (2007). Salivary gland development. Semin. Cell Dev. Biol. 18, 237-244. 10.1016/j.semcdb.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Tyler M. S. and Koch W. E. (1977). In vitro development of palatal tissues from embryonic mice. III. Interactions between palatal epithelium and heterotypic oral mesenchyme. J. Embryol. Exp. Morphol. 38, 37-48. [PubMed] [Google Scholar]
- Ventura A., Kirsch D. G., McLaughlin M. E., Tuveson D. A., Grimm J., Lintault L., Newman J., Reczek E. E., Weissleder R. and Jacks T. (2007). Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661-665. 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- Villa A., Connell C. L. and Abati S. (2015). Diagnosis and management of xerostomia and hyposalivation. Ther. Clin. Risk Manag. 11, 45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Flanagan J., Su N., Wang L.-C., Bui S., Nielson A., Wu X., Vo H.-T., Ma X.-J. and Luo Y. (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22-29. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassef L., Spiegler E. and Quadro L. (2013). Embryonic phenotype, beta-carotene and retinoid metabolism upon maternal supplementation of beta-carotene in a mouse model of severe vitamin A deficiency. Arch. Biochem. Biophys. 539, 223-229. 10.1016/j.abb.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman J. S. and Yelon D. (2011). Zebrafish retinoic acid receptors function as context-dependent transcriptional activators. Dev. Biol. 352, 128-140. 10.1016/j.ydbio.2011.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Larsen M., Hoffman M. P. and Yamada K. M. (2007). Self-organization and branching morphogenesis of primary salivary epithelial cells. Tissue Eng. 13, 721-735. 10.1089/ten.2006.0123 [DOI] [PubMed] [Google Scholar]
- Wells K. L., Gaete M., Matalova E., Deutsch D., Rice D. and Tucker A. S. (2013). Dynamic relationship of the epithelium and mesenchyme during salivary gland initiation: the role of Fgf10. Biol. Open 2, 981-989. 10.1242/bio.20135306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. M., Buenger D. E., Abashev T. M., Lindeman R. P., Ding J. and Sandell L. L. (2015). Retinoic acid regulates embryonic development of mammalian submandibular salivary glands. Dev. Biol. 407, 57-67. 10.1016/j.ydbio.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B. X., Chen Y., Chen Y., Fan J., Rohrer B., Crouch R. K. and Ma J. X. (2002). Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Invest. Ophthalmol. Vis. Sci. 43, 3365-3372. [PubMed] [Google Scholar]
- Zhao D., McCaffery P., Ivins K. J., Neve R. L., Hogan P., Chin W. W. and Dräger U. C. (1996). Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur. J. Biochem. 240, 15-22. 10.1111/j.1432-1033.1996.0015h.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.