Abstract
Lotus japonicus is a well-studied nodulating legume and a model organism for the investigation of plant-microbe interactions. The majority of legume transcriptome studies have focused on interactions with compatible symbionts, whereas responses to non-adapted rhizobia and pathogenic bacteria have not been well-characterized. In this study, we first characterized the transcriptomic response of L. japonicus to its compatible symbiont, Mesorhizobium loti R7A, through RNA-seq analysis of various plant tissues. Early symbiotic signaling was largely Nod factor-dependent and enhanced within root hairs, and we observed large-scale transcriptional reprogramming in nodule primordia and mature nitrogen-fixing nodules. We then characterized root transcriptional responses to a spectrum of L. japonicus interacting bacteria ranging from semi-compatible symbionts to pathogens. M. loti R7A and the semi-compatible strain Sinorhizobium fredii HH103 showed remarkably similar responses, allowing us to identify a small number of genes potentially involved in differentiating between fully and semi-compatible symbionts. The incompatible symbiont Bradyrhizobium elkanii USDA61 induced a more attenuated response, but the weakest response was observed for the foliar pathogen Pseudomonas syringae pv. tomato DC3000, where the affected genes also responded to other tested bacteria, pointing to a small set of common bacterial response genes. In contrast, the root pathogen Ralstonia solanacearum JS763 induced a pronounced and distinct transcriptomic pathogen response, which we compared to the results of the other treatments. This comparative analysis did not support the concept that an early defense-like response is generally evoked by compatible rhizobia during establishment of symbiosis.
Keywords: symbiosis, nodulation, RNA-seq, pathogen, legume, nitrogen fixation, plant-microbe interaction, rhizobia
Introduction
Lotus japonicus develops determinate root nodules in association with its compatible rhizobia Mesorhizobium loti. Establishment of an effective nitrogen-fixing symbiosis requires molecular communication to ensure compatibility and coordinate the developmental processes of rhizobial infection and nodule organogenesis with legume LysM-receptor kinases playing an integral role in these processes (Madsen et al., 2010; Oldroyd et al., 2011; Kelly et al., 2017a). Nod factor (NF) is the key signal molecule produced by rhizobia (Long, 1996) and is perceived by a NF receptor complex, which in Lotus consists of NFR1, NFR5 and SYMRK (Madsen et al., 2003; Radutoiu et al., 2003; Broghammer et al., 2012; Antolin-Llovera et al., 2014). Perception of compatible NF results in rapid physiological and transcriptional responses in the host (Desbrosses and Stougaard, 2011; Oldroyd, 2013). An additional level of compatibility scrutiny in Lotus occurs through the perception of rhizobial exopolysaccharides (EPSs) by the EPR3 receptor (Kelly et al., 2013; Kawaharada et al., 2015, 2017b).
Significant resources have been established to assist with investigations of L. japonicus. The genome sequence is available (Sato et al., 2008) as is an extensive LORE1 retrotransposon mutant resource consisting of 150,000 lines that provides potential mutants for an estimated 90% of active protein coding genes (Malolepszy et al., 2016). A Lotus japonicus gene expression atlas (LjGEA) was established that incorporated and extended on available Lotus transcriptome data in response to rhizobia and abiotic stresses (Verdier et al., 2013). All of these genomic resources can now be accessed through Lotus Base (Mun et al., 2016). Comparable transcriptomic resources are available for the indeterminate nodulating model legume Medicago truncatula (Benedito et al., 2008) and additional significant transcriptome data for this host was provided by the specific analysis of root hairs, which revealed a role for auxin signaling in infection thread (IT) formation (Breakspear et al., 2014).
Transcriptome responses of L. japonicus to compatible rhizobia and arbuscular mycorrhiza form the bulk of host transcriptional data available, while responses to incompatible rhizobia and pathogens have been less well-defined. Microarray analysis has been performed on two L. japonicus ecotypes that show differing leaf phenotypes when challenged with Pseudomonas syringae pv. tomato DC3000 and the response to pathogenic fungal exudates has also been investigated, identifying potential defense related genes (Bordenave et al., 2013; Giovannetti et al., 2015).
The interplay between symbiotic and pathogenic responses in legumes has largely been investigated through co-inoculation experiments, which revealed a negative impact of pathogens and defense eliciting compounds on symbiotic efficiency (Lopez-Gomez et al., 2012; Chen et al., 2017). Analysis of a cDNA array of expressed sequence tags from L. japonicus at various time points after inoculation with M. loti TONO indicated that an initial defense-like transcriptional response in L. japonicus is subsequently dampened through symbiotic signaling. The identified defense-like genes encode hyper-sensitive related proteins, pathogenesis-related (PR) proteins, and proteins associated with phytoalexin biosynthesis and cell wall modification (Kouchi et al., 2004).
In this study, a comprehensive symbiotic transcriptomic response of L. japonicus to its compatible symbiont M. loti R7A and a spectrum of interacting bacteria was obtained through RNA-seq analysis, revealing distinct transcriptional responses and challenging the concept that an early defense-like response is evoked by compatible rhizobia in L. japonicus during the establishment of symbiosis.
Materials and Methods
Plant Material and Growth Conditions
Lotus japonicus ecotype Gifu (Handberg and Stougaard, 1992) was used as the wild-type plant. Seed sterilization and plant-growth setups were as previously described (Kawaharada et al., 2015). For tissue-specific analysis of transcriptome responses to M. loti R7A, plants were grown at 21°C with a 16 h day and 8 h night cycle. For the analysis of transcriptome responses to a spectrum of interacting bacteria, plants were grown at 25°C with a 16 h day and 8 h night cycle. Plant growth plates, each containing 10 seedlings, were inoculated with 750 μL of OD600 = 0.02–0.05 bacterial suspensions along the length of the root. Purified M. loti R7A NF was obtained as previously described (Rodpothong et al., 2009) and used at 10-8 M for plant inoculation.
Bacterial Strains
Bacterial strains used in this study are listed in Supplementary Table 1. All strains were cultured at 28°C. M. loti R7A, Bradyrhizobium elkanii USDA61 and Sinorhizobium fredii HH103 were cultured in yeast mannitol broth (YMB) (Vincent, 1970), Pseudomonas syringae pv. tomato DC3000 in NYG medium (Daniels et al., 1984) and Ralstonia solanacearum JS763 in BGT medium (BG medium plus 0.005% tetrazolium chloride) (Saile et al., 1997). Antibiotics were added to media as required at the following concentrations: polymyxin, 50 μg mL-1; rifampicin, 50 μg mL-1; kanamycin, 50 μg mL-1.
Plant Phenotyping
Phenotypic responses of L. japonicus to the spectrum of interacting bacteria was determined though observation of plants at weekly intervals following inoculation. Symbiotic phenotypes at 21 dpi were observed using a Zeiss Discovery V8 stereomicroscope. Disease responses, root-browning and wilting symptoms, of L. japonicus to R. solanacearum JS763 were analyzed at 5 and 29 dpi.
Plant Tissue Harvesting
Whole root samples were harvested by removing the root tip (∼3 mm), to minimize hormone and meristematic activity specific to the root tip, and then collecting roots in tubes immersed in liquid nitrogen. Each whole root sample consisted of 10 roots. Root hairs were harvested using a previously described method (Sauviac et al., 2005). Roots were separated from shoots and root tips were removed. Ten roots were transferred to 50 mL Falcon tubes immersed in liquid nitrogen and after 1 min were twice vortexed for 15 s, tubes were returned to liquid nitrogen between vortexing. Falcon tubes were gently tapped to collect root hairs in the bottom and tweezers were used to remove roots stripped of root hairs. The process was then repeated with another 10 roots and a total of 100 roots was processed for each sample. Nodule primordia were collected at 7 dpi using a scalpel to slice through the root above and below the developing primordia. Excised primordia were collected in tubes immersed in liquid nitrogen and each sample represents the total primordia collected from 30 roots. Nodules collected at 21 dpi were gently separated from the root using tweezers and collected in tubes immersed in liquid nitrogen. Each sample represents the total nodules collected from 30 roots.
RNA Isolation, Library Preparation, and Sequencing
Total RNA was isolated from whole root, nodule primordia, and nodule tissue samples using a NucleoSpin® RNA Plant kit (Macherey-Nagel) according to the manufacturer’s instructions. mRNA was directly isolated from root hair samples using a Dynabeads mRNA DIRECT kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA quality was assessed on an Agilent 2100 Bioanalyser and samples were sent to GATC-Biotech1 for library preparation and sequencing. Sequencing data is deposited at the Short Read Archive with the BioProject ID SRP127678. Sequencing data for roots and shoots 3 dpi with R7A was obtained previously (Munch et al., 2018) and is deposited at the Short Read Archive with the BioProject ID PRJNA384655.
Bioinformatics and Statistical Analysis
Reads were mapped to the L. japonicus v. 3.0 genome and differential gene expression was analyzed using CLC Genomics Workbench 9.5.3 (Qiagen). For each sample a minimum of 30 million reads were obtained with >90% of reads mapped to the reference genome. A criterion of fourfold change with a FDR p-value ≤ 0.05 was applied to determine differential gene expression. Results of the analysis are available on Lotus Base2 (Mun et al., 2016). Multidimensional scaling (MDS) analysis was performed in R (R Core Team, 2017) from total read count data using the script outlined in Supplementary Data 1. Heatmaps were generated in R using Heatmap and Heatmap3 functions (Zhao et al., 2014), an example script is provided in Supplementary Data 1. Venn diagrams were constructed using BioVenn (Hulsen et al., 2008). Detection of differentially expressed genes was also performed using the R package DEGseq (Wang et al., 2010) with a criterion of fourfold change with a FDR p-value ≤ 0.05 to examine the effects of combining replicates from different time points.
Results
Tissue-Specific Responses of Lotus japonicus to Its Compatible Symbiont Mesorhizobium loti R7A
A comprehensive transcriptome response of L. japonicus ecotype Gifu to its compatible symbiotic partner M. loti R7A was obtained through RNA-seq analysis of specific tissues at various stages of symbiosis. Initial interactions between rhizobia and the legume were investigated through the isolation of root hairs following R7A inoculation at 1 and 3 dpi. The NF-deficient strain R7AnodC was included to reveal the role of NF in these early responses, which was further examined through the isolation of purified NF-treated root hairs 2 days post-treatment. Root hair deformations of L. japonicus are evident as early as 24 h following inoculation with M. loti or treatment with purified NF (Radutoiu et al., 2003; Maekawa-Yoshikawa and Murooka, 2009). Transcriptome responses during nodule colonization were examined through the harvesting of 7 dpi nodule primordia that are in the early stages of colonization by R7A and mature nitrogen-fixing nodules that are fully colonized by R7A. RNA-seq of whole roots (minus root tips) and shoots collected at 3 dpi to provide a basis for tissue-specific responses was previously performed (Munch et al., 2018). For each condition examined, biological triplicate samples were harvested and total RNA was isolated. The reads were mapped to the L. japonicus v3.0 genome and all data is available from the Lotus Base resource website (Mun et al., 2016)2.
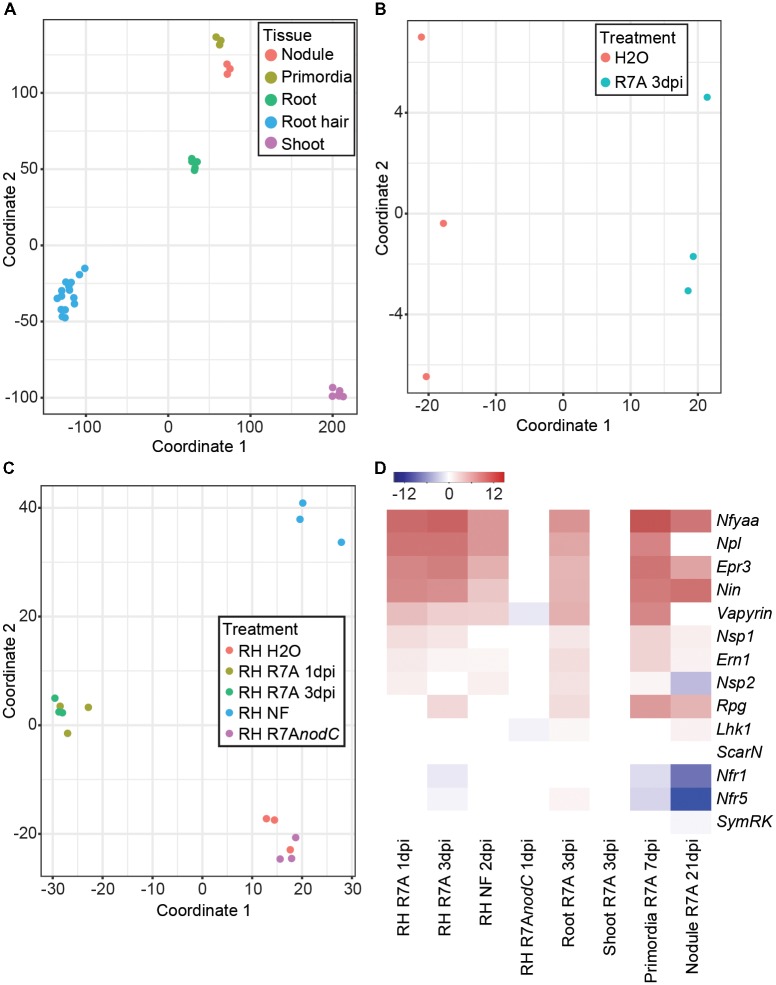
Multidimensional scaling analysis clearly separated the samples based on their tissue origin (Figure 1A). A closer examination of whole root samples revealed a clear shift following R7A inoculation (Figure 1B). Most root hair samples separated into distinct groups depending on their treatment (Figure 1C). H2O and R7AnodC samples clustered together, indicating that the majority of early root hair transcriptome responses to R7A are dependent on NF production. R7A treated root hairs formed a cluster with separation between the 1 and 3 dpi samples, while NF-treated root hairs formed a distinct cluster removed from the R7A inoculated samples.
FIGURE 1.
Lotus japonicus tissue-specific responses to the compatible symbiont Mesorhizobium loti R7A. (A) MDS plot of L. japonicus tissue samples. (B) MDS plot of whole root samples following treatment with H2O or R7A 3dpi. (C) MDS plot of all root hair samples following the indicated treatments. (D) Heatmap representation of log2 fold changes of various known symbiotic genes in tissues following treatment compared to H2O controls. NF, Nod factor; RH, root hairs.
The expression of a set of known symbiotic genes was examined in the different tissues in response to inoculation with R7A or NF treatment compared to H2O-treated controls (Figure 1D). No difference in gene expression was observed in shoots 3 dpi with R7A or root hairs 1 dpi with the R7AnodC mutant compared to their respective controls, 3 dpi H2O-treated shoots and 3 dpi H2O-treated root hairs. Similar expression patterns were observed for R7A and NF treated root hair and root samples compared to control 3 dpi H2O-treated root hairs. Symbiotic transcription factors, Nin (Schauser et al., 1999), NfyaA1 (Soyano et al., 2013), and Ern1 (Cerri et al., 2017; Kawaharada et al., 2017a; Yano et al., 2017) were strongly induced while Nsp1 and Nsp2 (Heckmann et al., 2006; Kawaharada et al., 2017a) showed only slight induction. The EPS receptor Epr3 (Kawaharada et al., 2015, 2017b), pectate lyase Npl (Xie et al., 2012) and Lotus homologs of Vapyrin (Murray et al., 2011) and Rpg (Arrighi et al., 2008) that were identified as required for IT formation in Medicago were all strongly induced. Notably, Rpg was not induced until 3 dpi with R7A. The receptors involved in NF perception, Nfr1, Nfr5, and Symrk (Stracke et al., 2002; Radutoiu et al., 2003) showed little change in expression other than a slight downregulation in root hairs 3 dpi.
Nodule primordia, compared to control 3 dpi H2O-treated roots, shared a similar expression pattern to root hair and root samples with the exception that the NF receptors Nfr1 and Nfr5 were downregulated. This downregulation of the NF receptors was stronger in the 21 dpi mature nodule samples compared to control 3 dpi H2O-treated roots. The Nsp2 transcription factor also showed downregulation in mature nodules and expression of Epr3, Npl, and Vapyrin was reduced compared to primordia, root hairs, and roots.
Early Symbiotic Responses Are Enhanced in Root Hairs and Largely Depend on Nod Factor Signaling
Predictably, significant overlap was observed in differentially expressed genes between root hairs harvested at 1 and 3 dpi compared to control 3 dpi H2O-treated root hairs and whole roots at 3 dpi compared to control 3 dpi H2O-treated whole roots (Figure 2A). Although expected symbiotic gene induction was observed in both tissues, the root hair samples showed enhanced transcriptional responses compared to whole roots (Figure 1D and Supplementary Figure 1). Among the most highly induced of the 93 genes identified as specifically differentially expressed within root hairs at 1 and 3 dpi compared to whole roots, we identified an aspartic protease and a calmodulin-binding protein as well as several blue copper proteins, expansins and pectinesterase/pectinesterase inhibitors (Supplementary Table 2). The root hair transcriptional changes observed in response to R7A are largely NF-dependent with root hairs inoculated with the NF-deficient R7AnodC strain sharing minimal overlap with R7A or NF treated samples (Figure 2B). Nodule primordia and mature nodules show a large transcriptional response compared to 3 dpi root samples with downregulation of genes more prominent than in other tissues investigated (Figure 2C).
FIGURE 2.
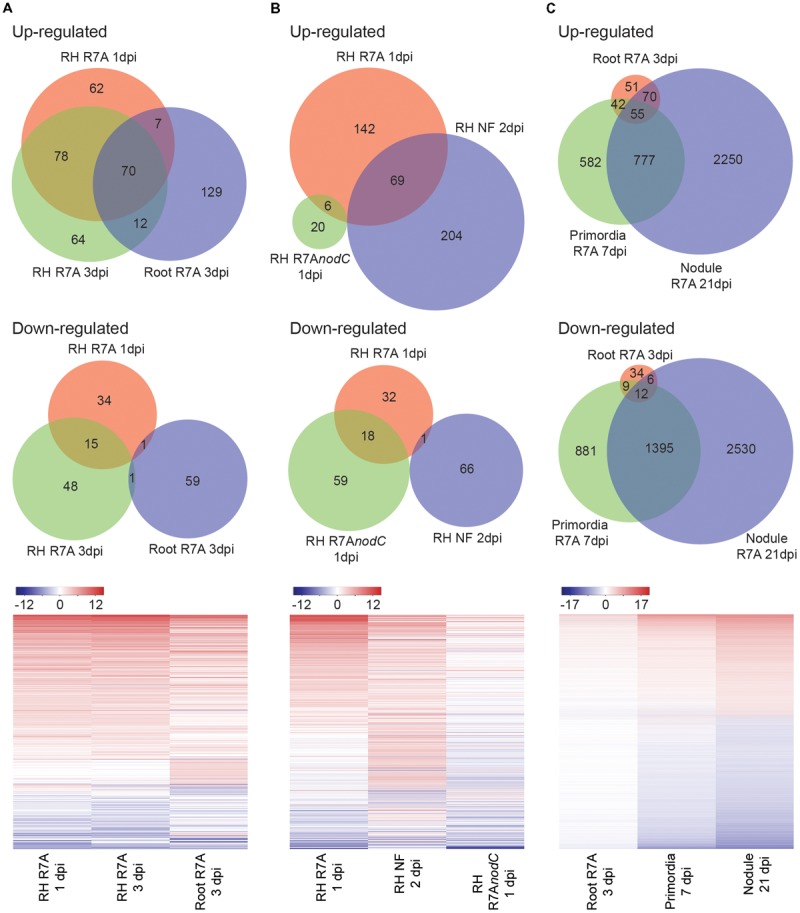
Lotus japonicus tissue-specific responses to compatible M. loti R7A and Nod factor. Venn diagram and heatmap representation of differential gene expression in the indicated conditions compared to H2O controls. Significant gene expression differences were designated based on a fourfold change in expression with a FDR p-value ≤ 0.05. (A) R7A induced gene expression in whole roots and root hairs. (B) NF-dependency of early root hair differential gene expression. (C) R7A induced gene expression in nodule primordia and mature nodules compared to whole roots. For heatmaps, Log2 fold change values were used. NF, Nod factor; RH, root hairs.
Altogether, the tissue transcriptome data of L. japonicus responses to R7A provided here represents an important resource for future research into this determinate nodulating model system.
Lotus japonicus Transcriptome Responses to a Spectrum of Interacting Bacteria
Transcriptional responses of L. japonicus to interacting bacteria representing a spectrum from compatible symbiont to pathogen were investigated to determine similarities or differences in the host transcriptome response to these diverse bacteria (Figure 3A). M. loti R7A (Ml R7A) represents an adapted symbiont that forms nitrogen-fixing nodules on L. japonicus. Sinorhizobium fredii HH103 (Sf HH103) is a broad host-range rhizobium that forms nitrogen-fixing nodules of some species of Lotus but induces uninfected primordia on L. japonicus (Acosta-Jurado et al., 2016) and (Figure 3B). Bradyrhizobium elkanii USDA61 (Be USDA61) is a symbiont of soybean that induces minimal responses on L. japonicus roots, with only an occasional root swelling near lateral root junctions observed (Figure 3B). Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) is a well-known foliar phytopathogen (Xin and He, 2013). We observed no responses to root inoculation with Pst DC3000 indicating that the strain is non-pathogenic to L. japonicus Gifu, as has been previously reported (Bordenave et al., 2013). Ralstonia solanacearum causes bacterial wilt in over 200 plant species and represents one of the most damaging bacterial pathogens in plants (Hayward, 1991). We observed that Rs JS763 causes brown discoloration of the roots from 3 dpi followed by discoloration at the base of the hypocotyl as well as chlorosis and wilting symptoms on leaves in L. japonicus Gifu (Figure 3C). These are typical plant disease symptoms due to R. solanacearum infection, confirming it to be a genuine root pathogen of L. japonicus, as previously described (Nagata et al., 2008).
FIGURE 3.
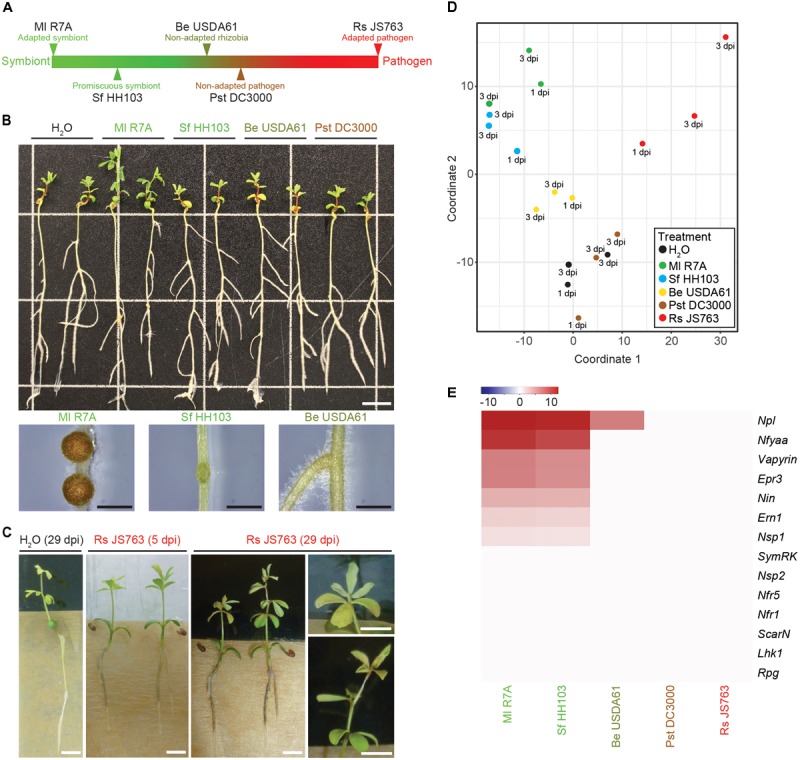
Lotus japonicus responses to a spectrum of interacting bacteria ranging from compatible symbiont to pathogen. (A) Schematic representation of the positioning of investigated bacteria on a spectrum from compatible symbiont to pathogen. (B) Phenotypes of L. japonicus 21 dpi with the indicated strains. Close up images show the differences in nodule development induced by Ml R7A, Sf HH103, and Be USDA61. Scale bars are 1 cm in the whole plant image and 1 mm in nodule images. (C) Representative images of root-browning and shoot wilting pathogenic phenotypes caused by Rs JS763 inoculation of L. japonicus at 5 and 29 dpi. Scale bars are 5 mm. (D) MDS plot of whole root samples following treatment with the indicated strains. (E) Heatmap representation of log2 fold changes of various known symbiotic genes following inoculation with the indicated strains compared to H2O controls.
Whole roots (minus root tips) treated with the spectrum of diverse bacterial strains or water were harvested at 1 and 3 dpi in biological duplicates. RNA isolation, Illumina sequencing and read mapping were performed as described above for the tissue samples. All data are available from the Lotus Base resource website (Mun et al., 2016)2. We detected outlier replicates for each inoculum and these were removed before proceeding with downstream analysis. The remaining three datasets, consisting of two 3 dpi and one 1 dpi samples, were combined to allow robust detection of differentially expressed genes. The effects of combining replicates from two 3 dpi and one 1 dpi samples were investigated using the R package DEGseq (Wang et al., 2010), which allows for differential gene expression analysis to be performed with fewer than three replicates. Differentially expressed genes were identified using the combined three replicates as well as using just the two 3 dpi replicates. A strong overlap in the genes identified between the three and two replicate analysis was observed. Almost all genes identified using the combined three replicates in our CLC genomics workbench analysis were also identified as differentially expressed in both the two and three replicate analyses using DEGseq, indicating that they likely represent genuine differentially regulated genes (Supplementary Data 2). However, genes oppositely regulated at 1 and 3 dpi or differentially expressed at only one time point may not have been detected due to the combined analysis of 1 and 3 dpi time points. We used the conservative lists of differentially expressed genes obtained from CLC genomics workbench in the downstream analyses. MDS analysis clustered Ml R7A and Sf HH103 together, Pst DC3000 clustered with H2O while Be USDA61 was slightly removed from them and Rs JS763 samples formed a less defined group removed from the other samples (Figure 3D).
To assess the symbiotic response induced by the diverse strains, expression of the same set of known symbiotic genes analyzed in the various tissue samples following inoculation with R7A or NF was examined through comparing bacteria inoculated root samples to control H2O-treated roots (Figure 3E). Ml R7A and Sf HH103 induced similar responses that are comparable to those obtained in analysis of the various tissues examined following R7A or NF treatment (Figure 1D) with the exception of Rpg expression, which was not induced in the Ml R7A and Sf HH103 samples. Only the Npl gene showed any expression change following inoculation with Be USDA61 and no response for any of the genes was observed following Pst DC3000 or Rs JS763 inoculation.
L. japonicus Responses to a Non-adapted Pathogen and Incompatible Rhizobia
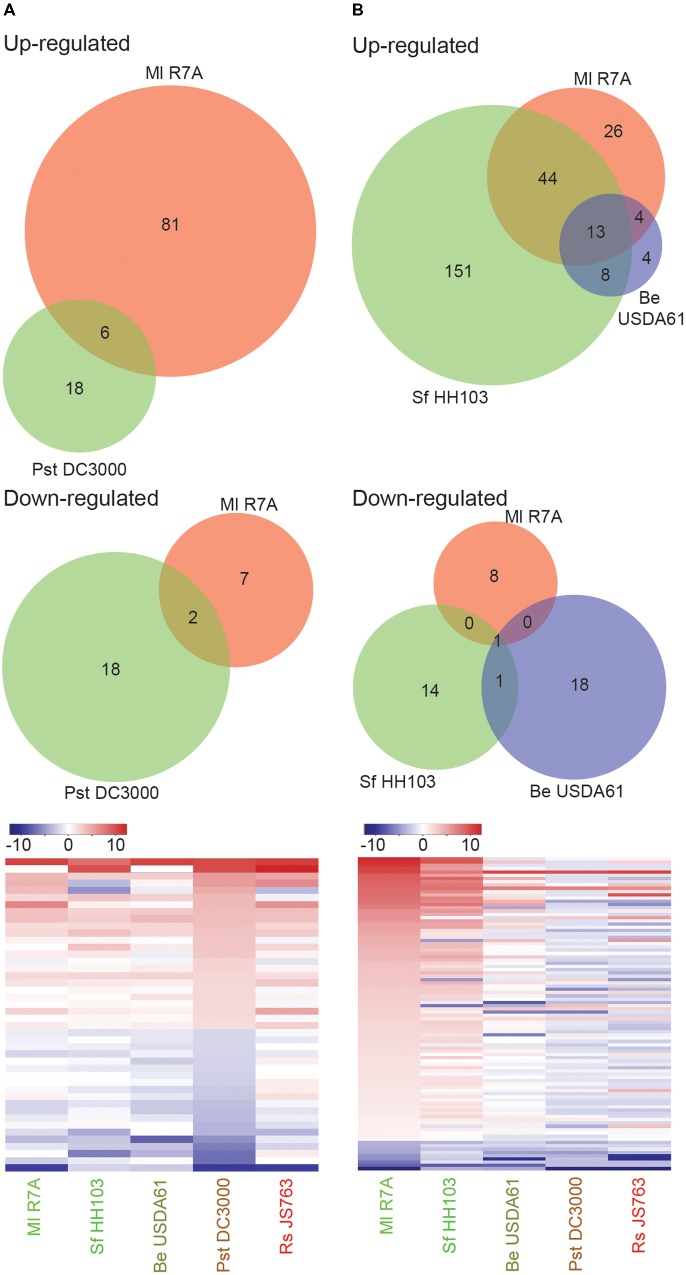
Pst DC3000 induced no discernible phenotypic responses on L. japonicus. Only a small set of genes (44) showed significant expression changes following Pst DC3000 treatment, 8 of which showed similar regulation following Ml R7A inoculation (Figure 4A and Supplementary Table 3). Similar expression of the identified Pst DC3000 differentially regulated genes was observed following inoculation with the spectrum of strains investigated, indicating that these genes may represent a common response to bacteria by L. japonicus (Figure 4A and Supplementary Table 3). Of interest in the identified genes are two leucine-rich repeat (LRR) receptor like proteins (Lj0g3v0095839 and Lj0g3v0331049), an unknown-conserved GYF-domain containing protein (Lj5g3v0101610) and an exportin-7 like protein (Lj1g3v2392240) that show upregulated expression in response to all of the diverse strains. LRR receptors represent the largest receptor family in plants and are involved in diverse developmental, defense and symbiotic processes (Torii, 2004). Exportin-7 proteins are reportedly involved in nuclear export processes (Mingot et al., 2004).
FIGURE 4.
Lotus japonicus root responses to non-adapted pathogenic bacteria and incompatible rhizobia. Venn diagram and heatmap representations of differential gene expression in the indicated conditions compared to H2O controls. Significant gene expression differences were designated based on a fourfold change in expression with a FDR p-value ≤ 0.05. (A) Non-adapted pathogen Pst DC3000 induced differential gene expression. (B) Compatible symbiont Ml R7A compared to incompatible rhizobia Sf HH103 and Be USDA61 differential gene expression. For heatmaps, Log2 fold change values were used.
Be USDA61 and Sf HH103 represent two rhizobial species that are incompatible for the establishment of nitrogen-fixing symbiosis with L. japonicus (Figure 3B). Significant gene expression changes were limited following Be USDA61 inoculation, whilst Sf HH103 inoculation induced a greater response and shared a large overlap with Ml R7A inoculation (Figure 4B).
Genes showing significant differential expression following Ml R7A inoculation but not Sf HH103 or Be USDA61 represent candidates involved in specifically promoting interaction with the compatible symbiont. The 34 genes identified that fit these criteria are listed in Supplementary Table 4. Of particular interest in this list are Lj5g3v2288900 that encodes a legume specific chalcone isomerase that has recently been shown to be involved in the biosynthesis of an Ml R7A NodD1 activating inducer, specifically within ITs (Kelly et al., 2017b) and Lj4g3v2365210 that encodes for the Nfy-B1 subunit, which forms part of the Nuclear Factor Y transcriptional factor known to be involved in nodulation (Soyano et al., 2013). Lj4g3v1983610 encodes an ENOD-like protein with a predicted cupredoxin domain. The protein shares 46% amino acid identity to ENOD16 of M. truncatula, which has been shown to be induced during symbiosis. No clear roles for ENOD16 have been demonstrated but it has been proposed that they may be involved in cell wall reorganization (Greene et al., 1998), which is required for IT development.
Interplay Between Symbiotic and Defense Responses in L. japonicus
Rs JS763 was shown to be a root pathogen of L. japonicus (Figure 3B). Rs JS763 inoculation resulted in a distinct transcription response, with 347 (277 upregulated and 70 downregulated) differentially expressed genes identified (Supplementary Data 3). Among the genes upregulated by Rs JS763 are five genes annotated as WRKY transcription factors (Lj0g3v0074419, Lj0g3v0244439, Lj1g3v1815540, Lj1g3v1787630, and Lj1g3v1220240). WRKY transcription factors belong to a large family of regulatory proteins involved in various plant processes but most notably in plant immunity against pathogen attack, including R. solanacearum infection (Mukhtar et al., 2008; Wang et al., 2013, 2014; Dang et al., 2014; Cai et al., 2015; Liu et al., 2017). Two 1-aminocyclopropane-1-carboxylate oxidase (ACO) 1-like genes (Lj2g3v0911860 and Lj6g3v0086450) were also induced. ACO proteins are involved in biosynthesis of ethylene, a plant hormone that is involved in many developmental and physiological responses including defense (Broekgaarden et al., 2015) and symbiosis (Reid et al., 2018). Ethylene signaling plays a critical role in bacterial wilt disease development in A. thaliana (Hirsch et al., 2002), M. truncatula (Moreau et al., 2014) and in tomato resistance to R. solanacearum (Zhang et al., 2004). In addition to inducing ethylene production by the host plant, R. solanacearum is also capable of producing ethylene itself (Weingart et al., 1999). Other defense related genes induced by Rs JS763 included pathogenesis-related proteins [such as PR1a: Lj6g3v2170740, beta-1,3-glucanase (PR-2): Lj0g3v0278459; PR10: Lj0g3v0286359; peroxidase 1-like: Lj0g3v0336889; two chitinases: Lj5g3v1961260 and Lj6g3v1078650] and several protease inhibitor proteins potentially involved in plant immunity (Lj0g3v0174139, Lj0g3v0174109, Lj2g3v0604160, Lj0g3v0265049, Lj5g3v1174410). Homologs of some of these genes were also reported as differentially expressed in response to R. solanacearum in M. truncatula according to proteomic and transcriptomic analyses (Yamchi et al., 2018). Interestingly, a gene annotated as a WAT1 (Walls Are Thin1)-related gene (Lj1g3v0913340) was downregulated in response to Rs JS763. In A. thaliana, inactivation of WAT1 gene, which is required for secondary cell-wall deposition, conferred broad-spectrum resistance to vascular pathogens, including R. solanacearum (Denance et al., 2013).
A transient defense response in L. japonicus roots during the early stages of symbiosis with compatible rhizobia had previously been reported (Kouchi et al., 2004). We investigated expression of the defense related genes identified by Kouchi et al. (2004) but did not observe comparable expression in response to symbiotic or pathogenic strains in our dataset (Figure 5A and Supplementary Data 4). Since symbiosis genes were consistently induced across all our experiments, we take this as an indication that the set of genes with defense-related annotations identified by Kouchi et al. (2004), are not generally associated with symbiotic or pathogenic interactions and our data does not support the notion that an early defense-like response is evoked by compatible rhizobia in the establishment of symbiosis with L. japonicus.
FIGURE 5.
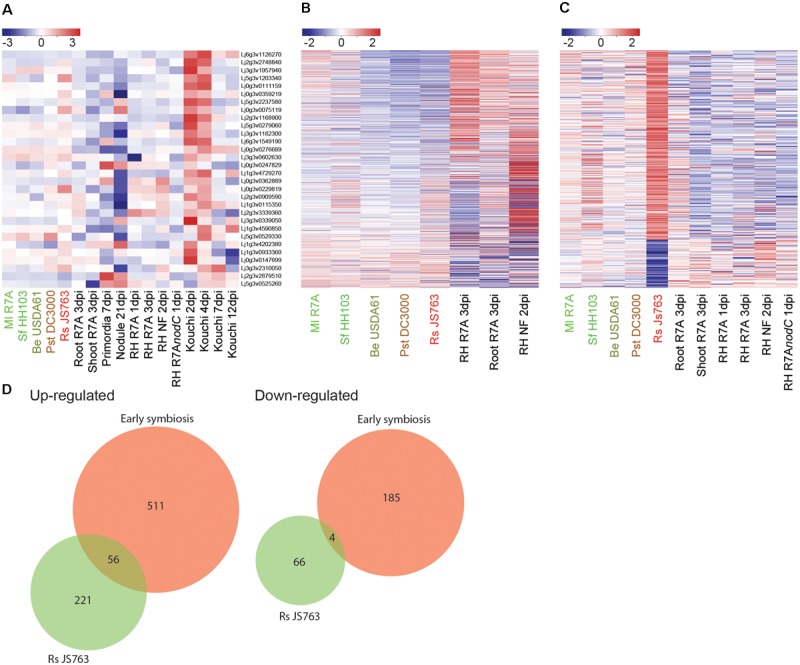
Interplay between symbiotic and defense transcriptional responses. (A) Log2 fold changes of potential defense associated genes identified as induced in the early stages of symbiosis by Kouchi et al. (2004) were examined across our datasets. (B) Expression patterns of 71 commonly differentially expressed genes in early symbiotic samples compiled from Ml R7A inoculated roots and root hairs was analyzed in the spectrum of interacting bacteria. (C) Rs JS763 differentially regulated genes were examined for expression changes in early symbiotic and diverse strain datasets. (D) Venn diagrams showing the overlap in gene expression between early symbiosis, corresponding to a compiled list of genes that were responsive to either Ml R7A or NF in whole roots or root hairs, and response to pathogenic Rs JS763. Differentially expressed genes were filtered by a minimum fourfold change and a FDR p-value ≤ 0.05.
It remains possible, however, that overlaps in gene expression responses could exist between infection-competent symbiotic and pathogenic bacteria. To further examine such potential similarities, a list of commonly differentially expressed genes in early symbiotic samples was compiled from Ml R7A inoculated roots and root hairs. Expression of the 71 genes was analyzed in the spectrum of interacting bacteria, including Rs JS763 (Figure 5B). In a complementary analysis, expression of the 347 differentially expressed genes following Rs JS763 inoculation was compared in response to the diverse strains (Figure 5C). Rs JS763 inoculated roots did not show similar gene expression patterns with the symbiotic samples in either of these analyses. However, some overlapping expression changes were observed between Rs JS763 responsive genes and a compiled list of early symbiotic genes that were responsive to either Ml R7A or NF in whole roots or root hairs (Figure 5D and Supplementary Table 5).
This overlap included the cytokinin oxidase Ckx3 (Lj5g3v0692300) that is involved in cytokinin breakdown during nodulation (Reid et al., 2016) and the sulfate transporter Sst1 (Lj2g3v0776860.1) that is required for nitrogen-fixation within nodules. In addition, putative defense-related genes identified were a PR10-like gene (Lj0g3v0286359), the WAT1-related gene (Lj1g3v0913340) and a gene encoding a salicylate O-methyltransferase-like protein (Lj6g3v0509430) that is involved in the biosynthesis of methyl salicylate which was found to participate in root responses against fungal pathogens (Ament et al., 2010; Boba et al., 2016). Further studies will be required to determine the roles of the symbiosis-related genes in the Lotus–Ralstonia interaction and of the putative defense genes in Lotus–rhizobium interactions. However, the identification of genes responding to both Ralstonia and rhizobium treatments does not necessarily indicate that symbiotic rhizobia are initially perceived as pathogenic by Lotus. In contrast, our data indicate that the pathogenic and symbiotic responses are well-separated as early as 1 dpi (Figure 3D).
Discussion
Lotus japonicus has been extensively utilized for the study of symbiotic interactions with rhizobial bacteria and mycorrhizal fungi. Analysis of the host transcriptional response during these interactions has been beneficial in developing our understanding of the molecular processes behind the symbiotic interactions and identifying potential genes important for these.
Transcriptome data collected from numerous microarray based studies of L. japonicus tissues at various stages of development or in association with biotic and abiotic treatments has been collated together in the LjGEA and is now accessible through Lotus Base2 (Sanchez et al., 2008, 2011; Guether et al., 2009; Hogslund et al., 2009; Diaz et al., 2010; Verdier et al., 2013; Mun et al., 2016). We have expanded on this through the generation of a comprehensive RNA-seq based transcriptome analysis of the response of L. japonicus to its compatible symbiont M. loti R7A. Previous studies in M. truncatula have highlighted the benefits of isolating root hairs to amplify early symbiotic transcriptome responses (Breakspear et al., 2014). In our analysis, early symbiotic signaling was similarly amplified in root hair samples compared to whole roots. Npl (Xie et al., 2012), Vapyrin (Murray et al., 2011), and Rpg (Arrighi et al., 2008) represent genes known to be required for IT development that were highly induced in our root hair samples. Additional genes identified as showing enhanced expression within root hairs represent potential candidates that may be involved in IT development. An interesting candidate from this analysis is the aspartic peptidase APN1 (Lj3g3v0950730.1) that was recently discerned to be expressed in nodules and demonstrated to play a crucial role in nodule functioning. apn1 mutant nodules fail to fix nitrogen and undergo early senescence in a M. loti strain-depended manner (Yamaya-Ito et al., 2017). Our identification of root hair enhanced expression of Apn1 compared to whole roots suggests that IT phenotypes of apn1 inoculated with R7A would be of interest to examine. As expected, early symbiotic signaling was largely NF-dependent and although considerable overlap was observed for differentially expressed genes following R7A or NF treatment, including most of the known symbiotic genes, there remains clear differences in the responses. A comparable difference in differentially expressed genes following NF treatment or S. meliloti inoculation was observed in M. truncatula (Breakspear et al., 2014). The observed differences in responses to purified NF compared to rhizobial inoculation may be due to the effects of other rhizobial factors, for example surface glycans, or differences in the concentration of NF roots are exposed to following rhizobial inoculation compared to purified NF treatment. Rhizobia produce an array of NF species and it is also possible that this induces differences in transcriptional responses compared to application of one particular purified NF species. Interestingly, Rpg induction was not observed in 2 dpi NF-treated or at 1 dpi with R7A, suggesting that Rpg induction is delayed compared to the other symbiotic genes investigated. This delay was not observed in M. truncatula, where Rpg induction was detected in roots at 1 dpi with S. meliloti or NF-treated root hairs (Arrighi et al., 2008; Breakspear et al., 2014). Nodule primordia and mature nodules exhibited large transcriptional reprogramming as has previously been observed for Lotus nodules (Takanashi et al., 2012). Highly upregulated genes include various transporters, auxin and cytokinin responsive genes as well as genes known to be required for nodule functioning such as the sulfate transporter Sst1 (Krusell et al., 2005) and homocitrate synthase Fen1 (Hakoyama et al., 2009).
Model legume resources, developed largely through the study of symbiotic interactions, have not been exploited to such an extent in interactions with diverse microbes. In this study, we identified distinct transcriptome responses to a spectrum of interacting bacteria ranging from symbiotic to pathogenic. The broad host-range rhizobium Sf HH103 forms small nodule primordia that remain uninfected on L. japonicus (Sandal et al., 2012; Acosta-Jurado et al., 2016). A remarkably similar early transcriptome response of L. japonicus to Ml R7A and Sf HH103 was observed, indicating that NF-induced signaling is comparable between the two strains. Both strains produce a variety of NFs with one notable difference being the substitution on the reducing terminal residue, which is generally an acetylfucosyl for Ml R7A NF and a methylfucosyl for Sf HH103 NF (Gil-Serrano et al., 1997; Rodpothong et al., 2009). Genes identified as potentially limiting the symbiotic capacity of Sf HH103 based on differential expression compared to Ml R7A include a legume specific chalcone isomerase, Nfy-B1 and an ENOD16 encoding gene. Be USDA61 induced only occasional root swellings on L. japonicus and a limited transcriptome response was observed, suggesting that the NF species produced by this rhizobial strain are not well-perceived by L. japonicus. We also detected minimal transcriptome responses to Pst DC3000 inoculation, which appeared to be non-pathogenic to L. japonicus roots. Differentially expressed genes identified from these non-symbioitic/non-pathogenic strains may represent a common response to bacteria as similar transcription responses were overserved across the spectrum of interacting bacteria.
Rs JS763 was characterized as a genuine pathogen of L. japonicus with typical disease symptoms observed following root inoculation and a clear and distinct root transcriptome response was detected, allowing us to investigate the interplay between pathogenic and symbiotic signaling. Transcriptome-based studies have previously suggested that an initial defense-like response to rhizobia is subsequently dampened through symbiotic signaling in soybean/B. japonicum, M. truncatula/S. meliloti, and L. japonicus/M. loti symbioses (Kouchi et al., 2004; Lohar et al., 2006; Libault et al., 2010). Kouchi et al. (2004) identified a set of potential defense-related genes that were induced at early time points before being suppressed at later stages of L. japonicus/M. loti interactions. We did not observe similar induction of the identified genes in our RNA-seq transcriptome analysis following M. loti R7A inoculation, indicating that the gene set is not reproducibly associated with symbiotic infection. Furthermore, only two of these genes (Lj0g3v0229819 and Lj5g3v1203340) encoding carboxylesterase 6-like and basic 7S globulin-like proteins respectively, showed a significant response following Rs JS763 inoculation in our analysis, suggesting that although the genes identified by Kouchi et al. (2004) have potential defense associated annotations they may not represent genes that are generally pathogen responsive in L. japonicus. It is possible that differences in host and M. loti genotypes as well as growth setups used in our analysis and by Kouchi et al. (2004) may be responsible for the differences in transcriptional responses observed.
We did however identify a small overlap between early time point Ml R7A responsive genes and Rs JS763 differentially expressed genes. The only known symbiotic genes identified as commonly transcriptionally regulated are the cytokinin oxidase Ckx3 and the sulfate transporter Sst1. Cytokinin is a key plant hormone and is tightly regulated in root development and symbiotic processes (Reid et al., 2017). Transcriptional changes in the expression of cytokinin related genes have previously been reported following R. solanacearum inoculation of M. truncatula (Moreau et al., 2014). SST1 functions during the later stages of symbiosis to transport sulfate from the plant to rhizobia where it is essential for protein synthesis, including nitrogenase biosynthesis (Krusell et al., 2005). It is unclear if a similar role is performed by SST1 in earlier stages of interactions with rhizobia or other bacteria. The only obvious potentially defense-related genes identified in the list of commonly induced genes were a PR10-like gene (Lj0g3v0286359), WAT1-related gene (Lj1g3v0913340) and a gene encoding a salicylate O-methyltransferase-like protein (Lj6g3v0509430). Lj6g3v0509430 is one of several salicylate O-methyltransferase-like proteins with high similarity encoded by L. japonicus and additional studies are required to determine if it is involved in salicylic acid responses.
Altogether, our analysis of transcriptome responses to a spectrum of interacting bacteria indicates that distinct transcriptome responses are observed in response to symbiotic and pathogenic bacteria and does not support the concept that an early defense-like response is generally evoked by compatible rhizobia in L. japonicus during the establishment of symbiosis.
Author Contributions
SK and TM performed the plant experiments, isolated RNA, and analyzed the sequencing data. SUA conceived and supervised the study. CB and JS devised the experiments and contributed resources. SK, CB, and SUA wrote the manuscript with input from all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank J. Luisetti for providing strain JS763 of R. solanacearum used in this work, Finn Pedersen for greenhouse assistance and Clive Ronson, John Sullivan, and Nicolai Maolanon for purified Nod factor.
Funding. The work was supported by the Danish National Research Foundation grant DNRF79 (JS) and a fellowship from the Institut Français du Danemark in the frame of the Blåtand Program (CB).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01218/full#supplementary-material
References
- Acosta-Jurado S., Rodriguez-Navarro D. N., Kawaharada Y., Perea J. F., Gil-Serrano A., Jin H., et al. (2016). Sinorhizobium fredii HH103 invades Lotus burttii by crack entry in a Nod factor-and surface polysaccharide-dependent manner. Mol. Plant Microbe Interact. 29 925–937. 10.1094/MPMI-09-16-0195-R [DOI] [PubMed] [Google Scholar]
- Ament K., Krasikov V., Allmann S., Rep M., Takken F. L., Schuurink R. C. (2010). Methyl salicylate production in tomato affects biotic interactions. Plant J. 62 124–134. 10.1111/j.1365-313X.2010.04132.x [DOI] [PubMed] [Google Scholar]
- Antolin-Llovera M., Ried M. K., Parniske M. (2014). Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 24 422–427. 10.1016/j.cub.2013.12.053 [DOI] [PubMed] [Google Scholar]
- Arrighi J. F., Godfroy O., De Billy F., Saurat O., Jauneau A., Gough C. (2008). The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. U.S.A. 105 9817–9822. 10.1073/pnas.0710273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito V. A., Torres-Jerez I., Murray J. D., Andriankaja A., Allen S., Kakar K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55 504–513. 10.1111/j.1365-313X.2008.03519.x [DOI] [PubMed] [Google Scholar]
- Boba A., Kostyn K., Kostyn A., Wojtasik W., Dziadas M., Preisner M., et al. (2016). Methyl salicylate level increase in lax after Fusarium oxysporum infection is associated with phenylpropanoid pathway activation. Front. Plant Sci. 7:1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenave C. D., Escaray F. J., Menendez A. B., Serna E., Carrasco P., Ruiz O. A., et al. (2013). Defense responses in two ecotypes of Lotus japonicus against non-pathogenic Pseudomonas syringae. PLoS One 8:e83199. 10.1371/journal.pone.0083199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26 4680–4701. 10.1105/tpc.114.133496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekgaarden C., Caarls L., Vos I. A., Pieterse C. M., Van Wees S. C. (2015). Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 169 2371–2379. 10.1104/pp.15.01020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broghammer A., Krusell L., Blaise M., Sauer J., Sullivan J. T., Maolanon N., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. U.S.A. 109 13859–13864. 10.1073/pnas.1205171109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Yang S., Yan Y., Xiao Z., Cheng J., Wu J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66 3163–3174. 10.1093/jxb/erv125 [DOI] [PubMed] [Google Scholar]
- Cerri M. R., Wang Q., Stolz P., Folgmann J., Frances L., Katzer K., et al. (2017). The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol. 215 323–337. 10.1111/nph.14547 [DOI] [PubMed] [Google Scholar]
- Chen T., Duan L., Zhou B., Yu H., Zhu H., Cao Y., et al. (2017). Interplay of pathogen-induced defense responses and symbiotic establishment in Medicago truncatula. Front. Microbiol. 8:973. 10.3389/fmicb.2017.00973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang F., Wang Y., She J., Lei Y., Liu Z., Eulgem T., et al. (2014). Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 150 397–411. 10.1111/ppl.12093 [DOI] [PubMed] [Google Scholar]
- Daniels M. J., Barber C. E., Turner P. C., Cleary W. G., Sawczyc M. K. (1984). Isolation of mutants of Xanthomonas campestris pv. campestris showing altered pathogenicity. J. Gen. Microbiol. 130 2447–2455. 10.1099/00221287-130-9-2447 [DOI] [Google Scholar]
- Denance N., Ranocha P., Oria N., Barlet X., Riviere M. P., Yadeta K. A., et al. (2013). Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73 225–239. 10.1111/tpj.12027 [DOI] [PubMed] [Google Scholar]
- Desbrosses G. J., Stougaard J. (2011). Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10 348–358. 10.1016/j.chom.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Diaz P., Betti M., Sanchez D. H., Udvardi M. K., Monza J., Marquez A. J. (2010). Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 188 1001–1013. 10.1111/j.1469-8137.2010.03440.x [DOI] [PubMed] [Google Scholar]
- Gil-Serrano A. M., Franco-Rodriguez G., Tejero-Mateo P., Thomas-Oates J., Spaink H. P., Ruiz-Sainz J. E., et al. (1997). Structural determination of the lipo-chitin oligosaccharide nodulation signals produced by Rhizobium fredii HH103. Carbohydr. Res. 303 435–443. 10.1016/S0008-6215(97)00189-4 [DOI] [PubMed] [Google Scholar]
- Giovannetti M., Mari A., Novero M., Bonfante P. (2015). Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Front. Plant Sci. 6:480. 10.3389/fpls.2015.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E. A., Erard M., Dedieu A., Barker D. G. (1998). MtENOD16 and 20 are members of a family of phytocyanin-related early nodulins. Plant Mol. Biol. 36 775–783. 10.1023/A:1005916821224 [DOI] [PubMed] [Google Scholar]
- Guether M., Balestrini R., Hannah M., He J., Udvardi M. K., Bonfante P. (2009). Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 182 200–212. 10.1111/j.1469-8137.2008.02725.x [DOI] [PubMed] [Google Scholar]
- Hakoyama T., Niimi K., Watanabe H., Tabata R., Matsubara J., Sato S., et al. (2009). Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature 462 514–517. 10.1038/nature08594 [DOI] [PubMed] [Google Scholar]
- Handberg K., Stougaard J. (1992). Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 2 487–496. 10.1111/j.1365-313X.1992.00487.x [DOI] [Google Scholar]
- Hayward A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29 65–87. 10.1146/annurev.py.29.090191.000433 [DOI] [PubMed] [Google Scholar]
- Heckmann A. B., Lombardo F., Miwa H., Perry J. A., Bunnewell S., Parniske M., et al. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142 1739–1750. 10.1104/pp.106.089508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J., Deslandes L., Feng D. X., Balague C., Marco Y. (2002). Delayed symptom development in ein2-1, an Arabidopsis ethylene-insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum. Phytopathology 92 1142–1148. 10.1094/PHYTO.2002.92.10.1142 [DOI] [PubMed] [Google Scholar]
- Hogslund N., Radutoiu S., Krusell L., Voroshilova V., Hannah M. A., Goffard N., et al. (2009). Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS One 4:e6556. 10.1371/journal.pone.0006556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T., De Vlieg J., Alkema W. (2008). BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. 10.1186/1471-2164-9-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y., James E. K., Kelly S., Sandal N., Stougaard J. (2017a). The ethylene responsive factor required for nodulation 1 (ERN1) transcription factor is required for infection-thread formation in Lotus japonicus. Mol. Plant Microbe Interact. 30 194–204. 10.1094/MPMI-11-16-0237-R [DOI] [PubMed] [Google Scholar]
- Kawaharada Y., Nielsen M. W., Kelly S., James E. K., Andersen K. R., Rasmussen S. R., et al. (2017b). Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun. 8:14534. 10.1038/ncomms14534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y., Kelly S., Nielsen M. W., Hjuler C. T., Gysel K., Muszynski A., et al. (2015). Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523 308–312. 10.1038/nature14611 [DOI] [PubMed] [Google Scholar]
- Kelly S., Radutoiu S., Stougaard J. (2017a). Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr. Opin. Plant Biol. 39 152–158. 10.1016/j.pbi.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Kelly S., Sullivan J., Kawaharada Y., Radutoiu S., Ronson C., Stougaard J. (2017b). Regulation of Nod factor biosynthesis by alternative NodD proteins at distinct stages of symbiosis provides additional compatibility scrutiny. Environ. Microbiol. 20 97–110. 10.1111/1462-2920.14006 [DOI] [PubMed] [Google Scholar]
- Kelly S. J., Muszynski A., Kawaharada Y., Hubber A. M., Sullivan J. T., Sandal N., et al. (2013). Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol. Plant Microbe Interact. 26 319–329. 10.1094/MPMI-09-12-0227-R [DOI] [PubMed] [Google Scholar]
- Kouchi H., Shimomura K., Hata S., Hirota A., Wu G. J., Kumagai H., et al. (2004). Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 11 263–274. 10.1093/dnares/11.4.263 [DOI] [PubMed] [Google Scholar]
- Krusell L., Krause K., Ott T., Desbrosses G., Kramer U., Sato S., et al. (2005). The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17 1625–1636. 10.1105/tpc.104.030106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Farmer A., Brechenmacher L., Drnevich J., Langley R. J., Bilgin D. D., et al. (2010). Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152 541–552. 10.1104/pp.109.148379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Liu Y., Tang Y., Chen J., Ding W. (2017). Overexpression of NtWRKY50 increases resistance to Ralstonia solanacearum and alters salicylic acid and jasmonic acid production in Tobacco. Front. Plant Sci. 8:1710. 10.3389/fpls.2017.01710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar D. P., Sharopova N., Endre G., Penuela S., Samac D., Town C., et al. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140 221–234. 10.1104/pp.105.070326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. R. (1996). Rhizobium symbiosis: nod factors in perspective. Plant Cell 8 1885–1898. 10.1105/tpc.8.10.1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gomez M., Sandal N., Stougaard J., Boller T. (2012). Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J. Exp. Bot. 63 393–401. 10.1093/jxb/err291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. 10.1038/nature02045 [DOI] [PubMed] [Google Scholar]
- Madsen L. H., Tirichine L., Jurkiewicz A., Sullivan J. T., Heckmann A. B., Bek A. S., et al. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1:10. 10.1038/ncomms1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa-Yoshikawa M., Murooka Y. (2009). Root hair deformation of symbiosis-deficient mutants of Lotus japonicus by application of Nod factor from Mesorhizobium loti. Microbes Environ. 24 128–134. 10.1264/jsme2.ME09103 [DOI] [PubMed] [Google Scholar]
- Malolepszy A., Mun T., Sandal N., Gupta V., Dubin M., Urbanski D., et al. (2016). The LORE1 insertion mutant resource. Plant J. 88 306–317. 10.1111/tpj.13243 [DOI] [PubMed] [Google Scholar]
- Mingot J. M., Bohnsack M. T., Jakle U., Gorlich D. (2004). Exportin 7 defines a novel general nuclear export pathway. EMBO J. 23 3227–3236. 10.1038/sj.emboj.7600338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Fromentin J., Vailleau F., Vernie T., Huguet S., Balzergue S., et al. (2014). The symbiotic transcription factor MtEFD and cytokinins are positively acting in the Medicago truncatula and Ralstonia solanacearum pathogenic interaction. New Phytol. 201 1343–1357. 10.1111/nph.12636 [DOI] [PubMed] [Google Scholar]
- Mukhtar M. S., Deslandes L., Auriac M. C., Marco Y., Somssich I. E. (2008). The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum. Plant J. 56 935–947. 10.1111/j.1365-313X.2008.03651.x [DOI] [PubMed] [Google Scholar]
- Mun T., Bachmann A., Gupta V., Stougaard J., Andersen S. U. (2016). Lotus Base: An integrated information portal for the model legume Lotus japonicus. Sci. Rep. 6:39447. 10.1038/srep39447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch D., Gupta V., Bachmann A., Busch W., Kelly S., Mun T., et al. (2018). The brassicaceae family displays divergent, shoot-skewed NLR resistance gene expression. Plant Physiol. 176 1598–1609. 10.1104/pp.17.01606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. D., Muni R. R., Torres-Jerez I., Tang Y., Allen S., Andriankaja M., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65 244–252. 10.1111/j.1365-313X.2010.04415.x [DOI] [PubMed] [Google Scholar]
- Nagata M., Murakami E., Shimoda Y., Shimoda-Sasakura F., Kucho K., Suzuki A., et al. (2008). Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus. Mol. Plant Microbe Interact. 21 1175–1183. 10.1094/MPMI-21-9-1175 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11 252–263. 10.1038/nrmicro2990 [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Murray J. D., Poole P. S., Downie J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45 119–144. 10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Gronlund M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. 10.1038/nature02039 [DOI] [PubMed] [Google Scholar]
- Reid D., Liu H., Kelly S., Kawaharada Y., Mun T., Andersen S. U., et al. (2018). Dynamics of ethylene production in response to compatible Nod factor. Plant Physiol. 176 1764–1772. 10.1104/pp.17.01371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D., Nadzieja M., Novak O., Heckmann A. B., Sandal N., Stougaard J. (2017). Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 175 361–375. 10.1104/pp.17.00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D. E., Heckmann A. B., Novak O., Kelly S., Stougaard J. (2016). CYTOKININ OXIDASE/DEHYDROGENASE3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiol. 170 1060–1074. 10.1104/pp.15.00650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodpothong P., Sullivan J. T., Songsrirote K., Sumpton D., Cheung K. W., Thomas-Oates J., et al. (2009). Nodulation gene mutants of Mesorhizobium loti R7A-nodZ and nolL mutants have host-specific phenotypes on Lotus spp. Mol. Plant Microbe Interact. 22 1546–1554. 10.1094/MPMI-22-12-1546 [DOI] [PubMed] [Google Scholar]
- Saile E., Mcgarvey J. A., Schell M. A., Denny T. P. (1997). Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87 1264–1271. 10.1094/PHYTO.1997.87.12.1264 [DOI] [PubMed] [Google Scholar]
- Sanchez D. H., Lippold F., Redestig H., Hannah M. A., Erban A., Kramer U., et al. (2008). Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J. 53 973–987. 10.1111/j.1365-313X.2007.03381.x [DOI] [PubMed] [Google Scholar]
- Sanchez D. H., Pieckenstain F. L., Szymanski J., Erban A., Bromke M., Hannah M. A., et al. (2011). Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS One 6:e17094. 10.1371/journal.pone.0017094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandal N., Jin H., Rodriguez-Navarro D. N., Temprano F., Cvitanich C., Brachmann A., et al. (2012). A set of Lotus japonicus Gifu x Lotus burttii recombinant inbred lines facilitates map-based cloning and QTL mapping. DNA Res. 19 317–323. 10.1093/dnares/dss014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakamura Y., Kaneko T., Asamizu E., Kato T., Nakao M., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15 227–239. 10.1093/dnares/dsn008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauviac L., Niebel A., Boisson-Dernier A., Barker D. G., De Carvalho-Niebel F. (2005). Transcript enrichment of Nod factor-elicited early nodulin genes in purified root hair fractions of the model legume Medicago truncatula. J. Exp. Bot. 56 2507–2513. 10.1093/jxb/eri244 [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. 10.1038/46058 [DOI] [PubMed] [Google Scholar]
- Soyano T., Kouchi H., Hirota A., Hayashi M. (2013). Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9:e1003352. 10.1371/journal.pgen.1003352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., et al. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. 10.1038/nature00841 [DOI] [PubMed] [Google Scholar]
- Takanashi K., Takahashi H., Sakurai N., Sugiyama A., Suzuki H., Shibata D., et al. (2012). Tissue-specific transcriptome analysis in nodules of Lotus japonicus. Mol. Plant Microbe Interact. 25 869–876. 10.1094/MPMI-01-12-0011-R [DOI] [PubMed] [Google Scholar]
- Torii K. U. (2004). Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234 1–46. 10.1016/S0074-7696(04)34001-5 [DOI] [PubMed] [Google Scholar]
- Verdier J., Torres-Jerez I., Wang M., Andriankaja A., Allen S. N., He J., et al. (2013). Establishment of the Lotus japonicus gene expression atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 74 351–362. 10.1111/tpj.12119 [DOI] [PubMed] [Google Scholar]
- Vincent J. M. (1970). A Manual for the Practical Study of Root-Nodule Bacteria. Oxford: Blackwell Scientific Publications. [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26 136–138. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- Wang X., Yan Y., Li Y., Chu X., Wu C., Guo X. (2014). GhWRKY40, a multiple stress-responsive cotton WRKY gene, plays an important role in the wounding response and enhances susceptibility to Ralstonia solanacearum infection in transgenic Nicotiana benthamiana. PLoS One 9:e93577. 10.1371/journal.pone.0093577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dang F., Liu Z., Wang X., Eulgem T., Lai Y., et al. (2013). CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 14 131–144. 10.1111/j.1364-3703.2012.00836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingart H., Volksch B., Ullrich M. S. (1999). Comparison of ethylene production by Pseudomonas syringae and Ralstonia solanacearum. Phytopathology 89 360–365. 10.1094/PHYTO.1999.89.5.360 [DOI] [PubMed] [Google Scholar]
- Xie F., Murray J. D., Kim J., Heckmann A. B., Edwards A., Oldroyd G. E., et al. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. U.S.A. 109 633–638. 10.1073/pnas.1113992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X. F., He S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51 473–498. 10.1146/annurev-phyto-082712-102321 [DOI] [PubMed] [Google Scholar]
- Yamaya-Ito H., Shimoda Y., Hakoyama T., Sato S., Kaneko T., Hossain M. S., et al. (2017). Loss-of-function of ASPARTIC PEPTIDASE NODULE-INDUCED 1 (APN1) in Lotus japonicus restricts efficient nitrogen-fixing symbiosis with specific Mesorhizobium loti strains. Plant J. 93 5–16. 10.1111/tpj.13759 [DOI] [PubMed] [Google Scholar]
- Yamchi A., Ben C., Rossignol M., Zareie S. R., Mirlohi A., Sayed-Tabatabaei B. E., et al. (2018). Proteomics analysis of Medicago truncatula response to infection by the phytopathogenic bacterium Ralstonia solanacearum points to jasmonate and salicylate defence pathways. Cell Microbiol. 20:e12796. 10.1111/cmi.12796 [DOI] [PubMed] [Google Scholar]
- Yano K., Aoki S., Liu M., Umehara Y., Suganuma N., Iwasaki W., et al. (2017). Function and evolution of a Lotus japonicus AP2/ERF family transcription factor that is required for development of infection threads. DNA Res. 24 193–203. 10.1093/dnares/dsw052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang D., Chen J., Yang Y., Huang Z., Huang D., et al. (2004). Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol. Biol. 55 825–834. 10.1007/s11103-005-2140-3 [DOI] [PubMed] [Google Scholar]
- Zhao S., Guo Y., Sheng Q., Shyr Y. (2014). Advanced heat map and clustering analysis using heatmap3. Biomed. Res. Int. 2014:986048. 10.1155/2014/986048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.