Abstract
Premise of the Study
Studies on the diversity of epiphyllous bryophytes have been limited because of minute and incomplete specimens and a lack of taxonomic expertise. The recent development of the DNA barcoding approach has allowed taxon identification and species discovery of many obscure groups of organisms.
Methods
With DNA extractions from 99 samples of 16 species, we compared the efficiencies of six DNA markers (rbcL, matK, trnL‐F, psbA, ITS1, and ITS2) in their ability to amplify, using a standard set of primers, as well as their discriminatory power, using distance‐based and tree‐based approaches with nucleotide data.
Results
The amplification success was relatively high (70–90%) with all of the markers, except for matK, which yielded no success. The barcoding gap, as calculated from the difference between inter‐ and intraspecific genetic distances, was the highest in ITS2, whereas the highest numbers of monophyletic groups were found with ITS2 and rbcL.
Discussion
rbcL should be used as a main barcoding marker with the addition of ITS2 for epiphyllous species. The development of DNA barcoding as a tool for quantifying species diversity will provide a rapid and reliable identification tool for epiphyllous bryophytes.
Keywords: biodiversity, ITS region, liverworts, molecular markers, rbcL region
Since its formal introduction, the concept of DNA barcoding as a tool for rapid taxon identification has continued to garner interest from the scientific community (Hebert et al., 2003; Hollingsworth et al., 2011). Although the use of molecular data to identify species is not new, remarkable successes using a single standardized region in taxon identification of animals (Hebert et al., 2003) and fungi (Schoch et al., 2014) have led to novel approaches in biodiversity inventories, as barcoding enhances the ability of taxonomists to gain more integrative insights into species delimitation (Hebert et al., 2004; Schindel and Miller, 2005; Pons et al., 2006). DNA barcoding has allowed a wide range of applications from authentication of traded plants and animals (Jiang et al., 2006; Phoolcharoen and Sukrong, 2012; Osathanunkul et al., 2015) to large‐scale ecological studies without obtaining the whole organisms, or even their tissue samples (Bohmann et al., 2014).
Leaf‐colonizing (epiphyllous) bryophytes offer an exciting system to test the utility of DNA barcoding. Over a thousand species of bryophytes from various taxonomic groups of mosses and liverworts can be epiphyllous. Ubiquitous in tropical ecosystems, epiphyllous bryophytes are often found on economically important plants, such as coffee and mangosteen (Roskoski, 1980; Zhu and So, 2001; Kraichak and Yaungthong, 2012). They also provide an excellent system for studying species assembly processes because each leaf represents a spatially and temporally discrete unit, and a large number of communities from host leaves can alleviate statistical power problems, which frequently hamper community assembly studies (Leibold et al., 2004; Zartman and Nascimento, 2006). Despite these features, studies on epiphyllous bryophytes have been somewhat limited. Aside from taxonomic challenges, specimens of these bryophytes are often minute and lack reproductive structures required for morphological identification. Although many bryologists have avoided working with this group, a few taxonomists who work on epiphyllous bryophytes have discovered a high level of undescribed genetic diversity (Gradstein et al., 2011; Yu et al., 2013a, 2013b) and a number of species new to science (Zhu and So, 1998; Pócs, 2011, 2012a, 2012b). The application of the DNA barcoding approach will facilitate diversity inventories of this fascinating but underappreciated group of epiphytes.
Unlike animals, bacteria, or fungi, a standardized barcoding region for land plants, including bryophytes, is far from settled. In 2009, the Consortium for the Barcode of Life (CBOL) Plant Working Group published a meta‐analysis of barcoding efficiency of individual major DNA markers and recommended two protein‐coding plastid regions, matK and rbcL, as standard markers for land plants. The potentially high discriminatory power of matK is hindered by the need for group‐specific primers, whereas rbcL demonstrates an impressively high success rate for amplifications across land plants, but is only mediocre in its ability to distinguish samples at the species level (CBOL Plant Working Group, 2009). Although it deviates from the original premise behind DNA barcoding, the multilocus approach with matK and rbcL was favored for the complementary potential of these two loci and eventually was approved as a standard set of barcoding regions for all land plants, with a provision that supplementary markers should also be studied. Among the proposed supplementary barcoding regions, the nuclear internal transcribed spacer (ITS hereafter; Li et al., 2011) and trnK‐psbA‐trnH (psbA hereafter) (Kress et al., 2005) are the most promising additions to the multilocus data set for DNA barcoding of land plants (Hollingsworth, 2011).
In bryological studies, rbcL and matK have rather limited success as barcoding regions, due to their low amplification rates and a lower variation among sequences below the rank of family (CBOL Plant Working Group, 2009). Two other regions have emerged as more promising candidates for the barcoding of bryophytes: trnL‐F and ITS (Stech and Quandt, 2010). These regions are consistently amplified and yield high‐quality sequences. The trnL‐F spacer, in particular, has been popular among molecular ecologists, as smaller parts of the region can be amplified from highly degraded DNA obtained from herbarium specimens and environmental sampling (Taberlet et al., 2006; Hollingsworth et al., 2011). Most trnL‐F and ITS sequences of bryophytes are the products of phylogenetic studies, so only a few studies have directly investigated their discriminatory power and reported relatively high resolution at the rank of species (Liu et al., 2010; Bell et al., 2011).
To find the best candidate loci for barcoding epiphyllous bryophytes, this study evaluated the efficiency of five candidate plant barcoding markers (rbcL, matK, trnL‐F, psbA, and ITS) in distinguishing a subset of epiphyllous bryophyte species from Thailand. These markers were amplified and sequenced, using a standard set of primers for bryophytes, to assess their amplification successes. Then, the nucleotide data were subjected to distance‐ and tree‐based analyses to determine their discriminatory power among the studied species.
METHODS
Taxon sampling and morphological identification
A total of 99 samples from 16 species of epiphyllous bryophytes from Thailand were selected for DNA sequencing (Appendix 1). Because an epiphyllous habit is typical of the Lejeuneaceae, 15 species belonged to that family, while Radula acuminata Stephani belongs to Radulaceae. The bryophyte tissue came from the dry preserved collection of leaves from previous studies in Ranong (Kraichak and Yaungthong, 2012) and Trat provinces (Kraichak, 2015), as well as from the current study in various locations in Thailand (Appendix 1). All bryophyte specimens were identified to species according to Zhu and So (2001), through examination under a dissecting and a compound microscope, based on morphological descriptions and keys (Jovet‐Ast, 1953, 1967; Tixier, 1985; Zhu and So, 2001). Voucher host plant specimens with additional bryophyte individuals were photographed for future reference. Vouchers were deposited in the herbarium at the Department of Botany, Faculty of Science, Kasetsart University, Bangkok, Thailand (Appendix 1). For each species, a minimum of two samples from different host leaves was acquired.
DNA isolation, amplification, and purification
Genomic DNA was isolated from dried plant material using the innuPREP Plant DNA Kit (Analytik Jena, Jena, Germany), following the manufacturer's protocol. We selected five DNA markers, including four chloroplast markers (trnL‐F, rbcL, matK, and trnH‐psbA) and one nuclear marker (ITS), based on their previous uses in barcoding and phylogenetic studies in bryophytes (reviewed in Stech and Quandt, 2010). Because of variable performance in past studies (Hartmann et al., 2006), we amplified and evaluated two regions of ITS: ITS1 (18S‐ITS1‐5.8S) and ITS2 (5.8S‐ITS‐26S). The chosen DNA markers were amplified with the following primers: (1) trnL‐F: trnL/trnF‐C and trnL‐trnF‐F (Taberlet et al., 1991); (2) rbcL: rbcL‐640‐F and rbcL‐1200‐R (Gradstein et al., 2006); (3) matK: RBGE‐LIV‐F1A and RBGE‐LIV‐R1A (Bell et al., 2011); (4) trnH‐psbA: trnK2F and psbA576R (Forrest et al., 2006); and (5) ITS with two sets of primers: Bryo18SF–Bryo5.8R for ITS1, and Bryo5.8SF–Bryo26SR for ITS2 (Hartmann et al., 2006).
Each 25‐μL reaction contained 9.5 μL of nuclease‐free water, 2.5 μL of OnePCR Plus mix (GeneDireX, Las Vegas, Nevada, USA), 2.5 μL of forward and reverse primers each, and 1 μL of genomic DNA. The PCR thermocycling conditions were specific to each primer pair. For trnL‐F, the cycle had initial denaturation at 92°C for 2 min; 30 cycles of denaturation at 92°C for 1 min, annealing at 51°C for 50 s, and elongation at 72°C for 90 s; with a final elongation at 72°C for 10 min. For rbcL, the cycle had initial denaturation at 94°C for 4 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 51°C for 50 s, and elongation at 72°C for 90 s; with a final elongation at 72°C for 10 min. For matK, the cycle had initial denaturation at 94°C for 4 min; 10 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 45 s, and elongation at 72°C for 1 min; 25 cycles of denaturation at 94°C for 45 s, annealing at 48°C for 45 s, and elongation at 72°C for 1 min; and a final elongation at 72°C for 10 min. For trnH‐psbA, the cycle had initial denaturation at 94°C for 1 min; 35 cycles of denaturation at 93°C for 1 min, annealing at 50°C for 1 min, and elongation at 72°C for 3 min; with a final elongation at 72°C for 7 min. For ITS1, the cycle had initial denaturation at 96°C for 3 min; 30 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 2 min, and elongation at 72°C for 3 min; with a final elongation at 72°C for 5 min. For ITS2, the cycle had initial denaturation at 94°C for 75 s; 35 cycles of denaturation at 95°C for 35 s, annealing at 55°C for 55 s, and elongation at 72°C for 42 s; with a final elongation at 72°C for 10 min. To assess the universality of these primers, we did not optimize the PCR conditions for individual taxa.
The PCR products were visualized on a 1% ethidium bromide–free agarose gel under UV light and then purified using USB ExoSAP‐IT PCR Product Cleanup (Applied Biosystems, Santa Clara, California, USA), following the manufacturer's instructions. The complementary strands were sequenced from the cleaned PCR products using the same primers as for amplifications. Sequencing reactions were performed with BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems) using the provided instructions. The samples were then run on an ABI 3730 automated sequencer at the Pritzker Laboratory for Molecular Systematics at the Field Museum (Chicago, Illinois, USA).
Sequence assembly and multiple sequence alignment
Resulting contigs and associated chromatograms were manually inspected, edited, and assembled using the program Geneious version 8.0.3 (Biomatters Ltd., Auckland, New Zealand). The identities of these sequences were examined using a “megaBLAST” search in the GenBank nucleotide database. For each marker, the verified sequences were aligned with the MUSCLE (Edgar, 2004) protocol through a Geneious plug‐in. The protocol was run for a maximum of 10 iterations with the first iteration using kmer4_6 distance and the CLUSTALW sequence weighting scheme, and the subsequent iterations were run with pctid_kimura distance and the CLUSTALW sequence weighting scheme. The resulting alignments were manually examined to remove ambiguous positions and gaps and were exported as FASTA files for further analyses. The sequences were submitted to the GenBank and BOLD (Barcode of Life Data Systems) (Ratnasingham and Hebert, 2007) databases (Appendix 1).
Evaluation of barcoding efficiency
To evaluate the efficiency of the markers as barcoding regions, the following criteria were used: (1) universality, (2) information content, and (3) discriminatory power (Hollingsworth, 2011). For universality, amplification successes were counted and divided by the total number of amplification attempts. For information content, the alignment length, number of variable positions, and GC content were compared. As for the discriminatory power, distance‐based and tree‐based approaches were employed to evaluate the markers' ability to distinguish the species with the sequence data. First, the distance‐based approach used genetic distance to determine whether the nearest neighbor was conspecific (the nearest neighbor test; Meier et al., 2006) and whether there was a sufficient gap between intraspecific and interspecific distances. The genetic distance among individual sequences was calculated using the Kimura 2‐parameter (K2P) model, a standard model in barcoding studies that has been shown to be appropriate for elucidating the barcoding gap with a standard barcode region (Hebert et al., 2003; Brown et al., 2012), with the function dist.dna in the R package “ape” (Paradis et al., 2004; R Core Development Team, 2013).
The nearest neighbor test calculates the genetic distances among all the studied sequences and identifies whether sequences with the shortest distance (“nearest neighbor”) are of the same species. The percentage of correct identification was calculated from the number of sequences with a conspecific nearest neighbor divided by the total number of sequences. The test was performed with the function “nearNeighbor” in the R package “spider” (Brown et al., 2012). The barcoding gap was calculated from the difference between the nearest non‐conspecific and the maximum conspecific distances. These distances were determined with the functions “nonConDist” and “maxInDist” in the R package “spider” (Brown et al., 2012). The Kruskal–Wallis test was also applied to determine whether the barcoding gap was significantly different among the chosen markers. A marker with high discriminatory power should have a high percent of correct identifications from the nearest neighbor test and a positive value for the barcoding gap.
Second, the tree‐based approach used the markers to reconstruct phylogenies of the studied species. In this study, neighbor‐joining (NJ) and maximum likelihood (ML) phylogenetic trees were reconstructed. The NJ trees were reconstructed using the K2P distance and the function “nj” in the R package “ape” (Paradis et al., 2004). A total of 1000 pseudo‐replicates was used to calculate bootstrap support for each node. A maximum likelihood phylogenetic reconstruction for each region was performed with the program RAxML‐HPC BlackBox version 8.1.11 (Stamatakis et al., 2008) on the online computing facility CIPRES (Miller et al., 2010). Following the model selection results for all of the loci from jModelTest 2 (Darriba et al., 2012), the “GTRGAMMA” model was used to perform likelihood searches to find the optimal tree and 1000 pseudo‐replicates were used to calculate bootstrap support for each node. The number of monophyletic groups was counted using the function “monophyly” in the R package “spider” (Brown et al., 2012). Phylogenetic reconstruction with a small data set can often result in poorly resolved relationships among the species and is often avoided in a systematic study. However, the main focus of a tree‐based test for barcoding efficiency is to determine the ability of a marker to recover monophyly among sequences of the same species, and the relationships among the studied taxa are not used as a criterion for the discriminating power of a barcoding marker (Hebert et al., 2003; Brown et al., 2012).
RESULTS
DNA extraction and amplification success
All of the studied markers, except for matK, were successfully amplified for 76.84% to 90.53% of the samples (Table 1). The psbA spacer had the highest success (90.53%), whereas the amplification of matK yielded no products, despite repeated attempts. From the PCR products, 304 high‐quality sequences were obtained and used for the subsequent analyses. The alignment length ranged between 492 and 632 bp with 30.22% to 89.23% of positions variable and the GC content between 31.59% and 59.29% (Table 1).
Table 1.
PCR success and characteristics of the studied markers in epiphyllous bryophytes from Thailand
| Markers | PCR success (%) | No. of sequencesa | No. of speciesb | Alignment length (bp) | Variable site (%) | GC content (%) |
|---|---|---|---|---|---|---|
| matK | 0 | — | — | — | — | — |
| ITS1 | 85.26 | 46 | 9 | 526 | 65.21 | 59.29 |
| ITS2 | 76.84 | 49 | 11 | 492 | 89.23 | 59.12 |
| trnH‐psbA | 90.53 | 79 | 13 | 632 | 30.22 | 35.79 |
| rbcL | 87.37 | 56 | 9 | 522 | 49.62 | 38.82 |
| trnL‐F | 86.32 | 74 | 12 | 430 | 58.84 | 31.59 |
Number of high‐quality sequences used in the analysis. The total number of samples included in the study was 99.
The total number of species included in the study was 16.
Distance‐based evaluation
The distribution of barcoding gaps showed that the interspecific distances were mostly greater than intraspecific distances in ITS2 and rbcL (positive barcoding gap), while the intraspecific distances were mostly greater than interspecific distances in trnL‐F and psbA (negative barcoding gap; Fig. 1A). For ITS1, roughly half of the interspecific distances were greater than intraspecific distances. For ITS2 and rbcL, most differences between inter‐ and intraspecific distance were close to zero, whereas differences were more widely distributed in ITS2 (Fig. 1A). The barcoding gaps varied significantly among the studied markers (Kruskal–Wallis test, P < 0.01). The nearest neighbor test showed the highest percentage of conspecific nearest neighbors in ITS2 (100%) and the lowest percentage in psbA (72.15%) (Fig. 1B).
Figure 1.
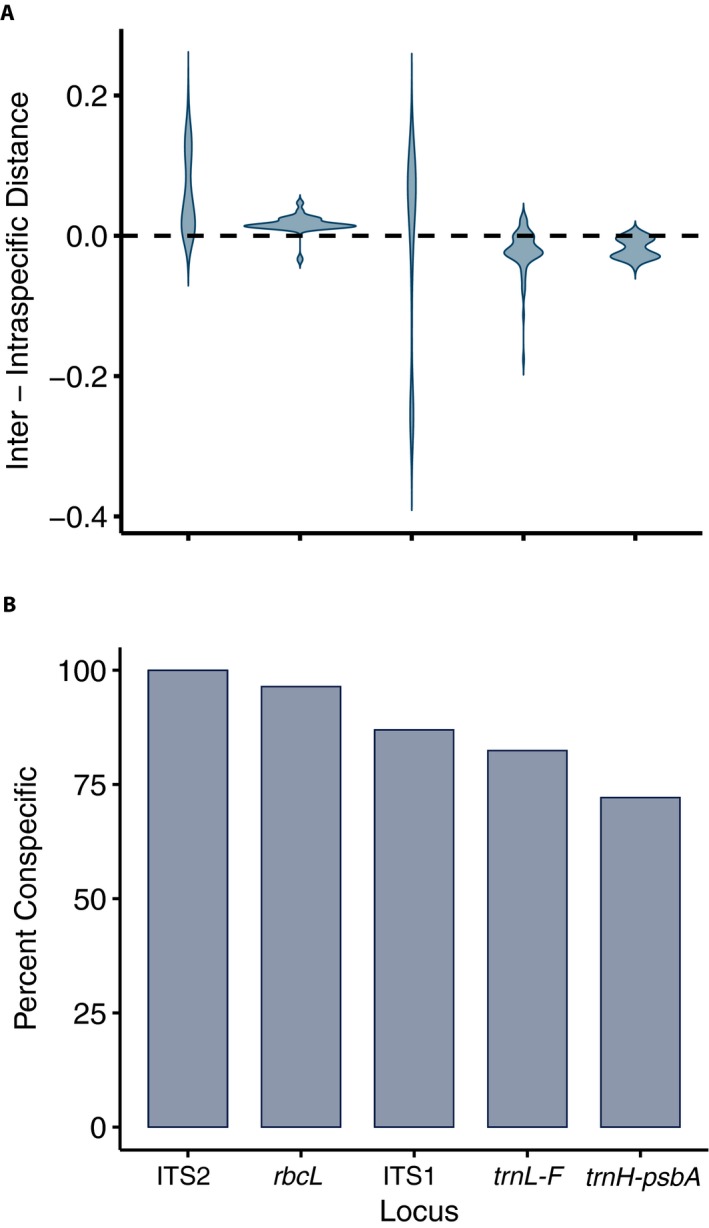
Distance‐based comparison of efficiency among the studied barcoding markers for epiphyllous liverworts from Thailand. (A) Distribution of barcoding gap, as defined by the difference between the minimum non‐conspecific distance and the maximum conspecific distance. (B) The percentage of correct identifications from the nearest neighbor test.
Tree‐based evaluation
ITS2 recovered the highest percentage of monophyletic groups in both NJ and ML reconstructions at 100% and 90%, respectively. The rbcL phylogeny recovered 77.78% and 88.89% of the monophyletic groups in NJ and ML reconstructions, respectively. The rest of the markers recovered less than half of the monophyletic groups. The psbA spacer yielded the lowest number of monophyletic groups at 7.69% in the ML reconstruction, whereas trnL‐F recovered the lowest number of monophyletic groups at 16.67% in the NJ reconstruction (Fig. 2; Appendices S1, S2).
Figure 2.
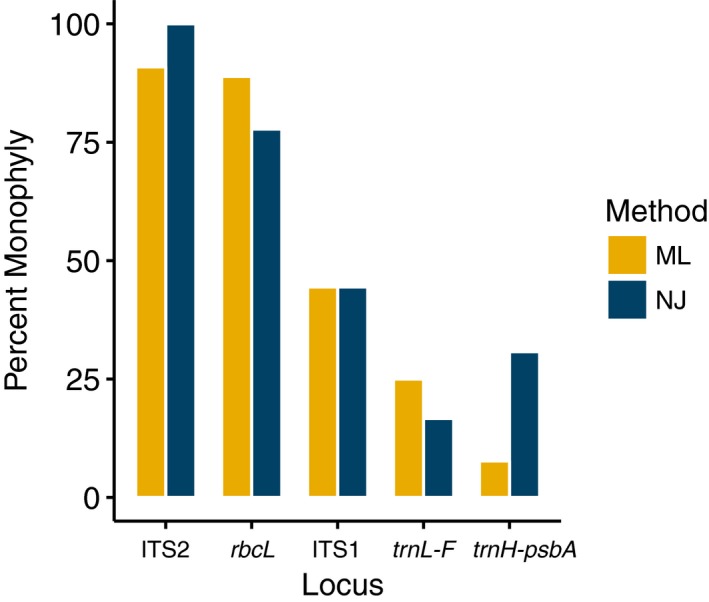
Tree‐based comparison of efficiency among the studied barcoding markers for epiphyllous liverworts from Thailand, using the percentage of monophyletic groups recovered from the neighbor‐joining (NJ = dark blue) and the maximum likelihood (ML = yellow) phylogenetic reconstructions.
DISCUSSION
The current study evaluated the efficiencies of six barcoding markers in distinguishing epiphyllous bryophyte species from Thailand. The amplification success was similar among most of the markers, with the notable exception of matK, suggesting that these markers can be successfully amplified equally well with a proper protocol and set of primers. However, the discriminatory power varied substantially, with rbcL and ITS2 showing the highest discriminatory power. These markers have also been proposed as part of the standard barcoding set for land plants. However, for epiphyllous bryophytes, rbcL and ITS2 displayed different strengths and weaknesses in their barcoding applications.
In the proposal for the standardized barcoding regions for land plants, rbcL was proposed, along with matK, as a core barcoding region (Kress and Erickson, 2007; CBOL Plant Working Group, 2009). Some of the key attributes for rbcL are its universality across the plant kingdom and the ease of alignment. Nevertheless, it suffers from moderate discriminatory power in many groups and has somewhat lower universality in bryophytes (CBOL Plant Working Group, 2009; Hollingsworth et al., 2009). In our study, rbcL was similar to ITS2 in its high discriminatory power among the epiphyllous species, even though the barcoding gap was close to zero. Such a small gap is the direct result of the conserved nature of protein‐coding genes, such as rbcL, which makes them easy to align and simultaneously less suitable for providing resolution at the species rank (Stech and Quandt, 2010; Hassel et al., 2013). In our case, this small barcoding gap reduced the number of successes in the distance‐based approaches. Moreover, we obtained rbcL sequences from fewer species, suggesting a potential issue of universality of primers for this region. Existing rbcL data for bryophytes in databases are uneven across the group because it is not a typical marker for systematic studies and has only been thoroughly sampled in specific groups (Stech and Quandt, 2010). This uneven distribution of data has made it difficult to test and develop universal primers for bryophytes to date, but the gradually increasing amount of data for rbcL, both from single‐locus and genomic studies, will allow us to see the full potential of rbcL as a barcoding marker for bryophytes in the future (Forrest et al., 2006; Hassel et al., 2013; Myszczyński et al., 2017).
ITS has been widely used in plant systematics and has only recently begun to gain traction as a barcoding region for land plants. In an early attempt to standardize barcoding regions, ITS was proposed as the most promising marker from a relatively small data set from flowering plants (Kress et al., 2005). However, issues of multiple copies and fungal contamination led to a decline in the use of ITS as a barcoding region (Hollingsworth, 2011; Cheng et al., 2016). However, ITS has since reemerged as a barcoding region with the separate consideration of two regions (ITS1 and ITS2), along with studies with a broader taxon sampling (Hollingsworth et al., 2009; Liu et al., 2010; Li et al., 2011). Many recent studies have included ITS2 in plant barcoding and even support ITS2 as the best candidate for plant barcoding (e.g., Yao et al., 2010; Li et al., 2011; Feng et al., 2015). Our study similarly demonstrated that ITS2 had the highest discriminatory power among the tested regions for epiphyllous bryophytes, although the sequences were difficult to align and often of low quality due to the low specificity of primers. Although we observed large barcoding gaps, we also observed a large variation in the inter‐ and intraspecific distances, a problem that can potentially be worsened with broader taxon sampling. Despite these difficulties, the use of ITS2 can be beneficial for advancing barcoding studies in bryophytes, as a relatively large amount of data already exist in global databases from phylogenetic studies (Stech and Quandt, 2010). The recent development of universal plant‐specific markers for ITS (Cheng et al., 2016) will most likely enhance our ability to produce data from ITS2 and increase its use as a barcoding region.
For the other markers, their subpar performance in bryophytes was not entirely surprising. For matK, data have been extremely difficult to obtain in bryophytes and ferns (CBOL Plant Working Group, 2009) due to secondary structures at the priming sites of this marker (Wicke and Quandt, 2009). Even with attempts to design specific matK primers for bryophytes, success has been limited to only a few groups (Wicke and Quandt, 2009; Bell et al., 2011), as reflected in a small amount of existing data of matK for bryophytes in global nucleotide databases. In our study, the amplification success for matK was zero with every tested primer set. Combinations of PCR conditions were also attempted for matK on our genomic DNA that could be amplified for other markers, suggesting the ongoing problem with priming sites for this region. Therefore, at this point, it is not clear whether the discriminatory power of matK in flowering plants will extend to bryophytes. The other two markers, psbA and trnL‐F, amplified well for epiphyllous bryophytes but showed only limited success in distinguishing species. Although psbA has been used in phylogenetic studies of various groups of bryophytes (Shaw et al., 2003; Forrest et al., 2006) and has a high discriminatory power within flowering plants, it is considered unsuitable as a standalone barcoding region, especially for pleurocarpous mosses (Stech and Quandt, 2010). We provided yet another example of how the conserved nature of psbA sequences among bryophyte species renders this marker less optimal for barcoding purposes. Finally, trnL‐F is one of the most popular phylogenetic markers and was expected to be a prime candidate for barcoding in bryophytes (Taberlet et al., 2006; Stech and Quandt, 2010). However, owing to its short length (the shortest alignment in our study), it can only offer a limited amount of information for species identification (Liu et al., 2010; Stech and Quandt, 2010; Bell et al., 2011).
From our results, ITS2 and rcbL exhibited the greatest potential for discriminating epiphyllous liverwort species with a DNA barcoding approach, owing to their primer universality, sequencing success, and high discriminatory power. However, these two markers still had some limitations: rbcL showed small differences between intra‐ and interspecific genetic distances, whereas the ITS2 sequences showed problems with low sequence quality and resulted in numerous gaps in the alignment, making it difficult to unambiguously use the data. In future work, a broader selection of species will validate the efficiency of these markers as barcoding regions for bryophytes in Thailand and elsewhere.
DATA ACCESSIBILITY
The sequence data are deposited in and accessible from the U.S. National Center for Biotechnology Information's GenBank database (https://www.ncbi.nlm.nih.gov; accession no. MH579787–MH580157).
Supporting information
APPENDIX S1. Single‐locus neighbor‐joining trees of five studied markers for studied epiphyllous bryophyte species: ITS1 (A), ITS2 (B), psbA (C), rbcL (D), and trnL‐F (E). Black circles at the nodes indicate nodes with bootstrap support greater than 70.
APPENDIX S2. Single‐locus maximum likelihood trees of five studied markers for studied epiphyllous bryophyte species: ITS1 (A), ITS2 (B), psbA (C), rbcL (D), and trnL‐F (E). Black circles at the nodes indicate nodes with bootstrap support greater than 70.
ACKNOWLEDGMENTS
The funding for this project came from the Thailand Research Fund (MRG5580012), Kasetsart University's Faculty of Science Preproposal Fund, and a Visiting Researcher Grant from the Field Museum of Natural History. This research is supported in part by the Graduate Program Scholarship from the Graduate School, Kasetsart University. The authors also thank Thomas Gleason for his help with the laboratory portion of this project at the Pritzker Laboratory for Molecular Systematics and Evolution at the Field Museum, Chicago, Illinois, USA.
APPENDIX 1. List of specimens, their locality, and GenBank accession numbers for the sequences used in the study. Vouchers were deposited in the herbarium at the Department of Botany, Faculty of Science, Kasetsart University, Bangkok, Thailand.
| Specimen | GenBank accession no. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Province | Country | Latitude | Longitude | DNA no. | ITS1 | ITS2 | psbA | rbcL | trnL‐F | |
| EK1720 | Leptolejeunea elliptica | Chumpon | Thailand | 10°45′30″N | 99°3′34″E | 1 | MH579856 | MH579787 | — | — | MH579920 |
| EK1720A | Cololejeunea tenella | Chumpon | Thailand | 10°45′30″N | 99°3′34″E | 2 | MH579857 | MH579788 | — | — | MH579921 |
| EK1707 | Colura ornata | Chumpon | Thailand | 10°45′30″N | 99°3′34″E | 3 | — | MH579789 | — | — | MH579922 |
| EK1712 | Leptolejeunea epiphylla | Chumpon | Thailand | 10°45′30″N | 99°3′34″E | 4 | MH579858 | MH579790 | — | — | MH579923 |
| EK1719 | Caudalejeunea reniloba | Chumpon | Thailand | 10°45′30″N | 99°3′34″E | 5 | MH579859 | MH579791 | MH580002 | MH580131 | MH579924 |
| EK1723 | Cololejeunea lanciloba | Uthaithani | Thailand | 15°36′31″N | 99°19′15″E | 11 | MH579860 | — | — | MH580083 | MH579925 |
| EK893_1 | Radula acuminata | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 13 | — | — | — | — | MH579926 |
| EK883_2 | Caudalejeunea reniloba | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 15 | — | MH579792 | — | — | MH579927 |
| EK1726 | Leptolejeunea elliptica | Uthaithani | Thailand | 15°36′31″N | 99°19′15″E | 16 | — | — | — | MH580084 | MH579928 |
| EK1724 | Leptolejeunea elliptica | Uthaithani | Thailand | 15°36′31″N | 99°19′15″E | 17 | — | — | — | — | MH579929 |
| EKEP001 | Radula acuminata | Pang‐Nga | Thailand | 9°2′32″N | 98°26′55″E | 29 | — | MH579793 | — | — | — |
| EKEP003 | Cololejeunea lanciloba | Pang‐Nga | Thailand | 9°2′32″N | 98°26′55″E | 30 | — | — | — | — | MH579930 |
| EKEP002 | Radula acuminata | Pang‐Nga | Thailand | 9°2′32″N | 98°26′55″E | 31 | — | MH579794 | — | — | MH579931 |
| SRS071 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 32 | MH579861 | — | MH580003 | MH580085 | MH579932 |
| SRS078 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 33 | — | — | MH580004 | MH580087 | MH579933 |
| SRS067 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 34 | MH579862 | — | MH580005 | MH580088 | MH579934 |
| SRS070 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 36 | — | — | MH580006 | MH580089 | — |
| SRS038 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 37 | MH579863 | — | MH580007 | MH580093 | MH579935 |
| SRS037 | Colura inflata | Trat | Thailand | 12°22′55″N | 102°39′21″E | 38 | MH579864 | — | MH580008 | MH580094 | MH579936 |
| SRS046 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 39 | MH579865 | MH579795 | MH580009 | MH580090 | MH579937 |
| SRS034 | Colura inflata | Trat | Thailand | 12°22′55″N | 102°39′21″E | 40 | — | MH579796 | MH580010 | MH580132 | MH579938 |
| SRS083 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 41 | — | — | MH580011 | MH580095 | MH579939 |
| SRS035 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 42 | MH579866 | MH579797 | MH580012 | MH580096 | MH579940 |
| SRS045 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 44 | MH579867 | MH579798 | MH580013 | MH580097 | MH579941 |
| SRS043 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 46 | MH579868 | — | MH580014 | MH580133 | MH579942 |
| SRS095 | Colura ornata | Trat | Thailand | 12°22′55″N | 102°39′21″E | 47 | MH579869 | — | MH580015 | MH580134 | MH579943 |
| JW002 | Diplasiolejeunea cavifolia | Ranong | Thailand | 10°30′49″N | 98°54′26″E | 48 | — | MH579799 | — | — | MH579944 |
| SRS088 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 49 | — | — | — | MH580098 | — |
| EKE002 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 50 | MH579870 | — | MH580016 | MH580099 | MH579945 |
| EKE001 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 51 | — | MH579800 | MH580017 | MH580135 | MH579946 |
| SRS039 | Cololejeunea denticulata | Trat | Thailand | 12°22′55″N | 102°39′21″E | 52 | MH579871 | MH579801 | MH580018 | MH580091 | MH579947 |
| SRS087 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 53 | MH579872 | MH579802 | MH580019 | MH580100 | MH579948 |
| SRS027 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 55 | — | — | MH580020 | — | MH579949 |
| SRS044 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 56 | MH579873 | MH579803 | MH580021 | MH580103 | MH579950 |
| SRS051 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 57 | MH579874 | MH579804 | MH580022 | MH580101 | MH579951 |
| JW001 | Diplasiolejeunea cavifolia | Ranong | Thailand | 10°30′49″N | 98°54′26″E | 58 | MH579875 | MH579805 | MH580023 | MH580136 | MH579952 |
| SRS011 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 60 | MH579876 | MH579806 | MH580024 | MH580086 | MH579953 |
| SRS012 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 61 | — | MH579807 | MH580025 | MH580104 | MH579954 |
| SRS042 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 62 | — | MH579808 | MH580026 | MH580105 | MH579955 |
| SRS026 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 63 | — | MH579809 | MH580027 | MH580106 | MH579956 |
| SRS021 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 65 | MH579877 | MH579810 | MH580028 | MH580137 | MH579957 |
| SRS040 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 66 | MH579878 | — | MH580029 | MH580138 | MH579958 |
| SRS016 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 67 | — | MH579811 | MH580030 | MH580092 | MH579959 |
| SRS018 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 68 | MH579879 | MH579812 | MH580031 | MH580139 | MH579960 |
| SRS108 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 69 | MH579880 | — | MH580032 | MH580140 | MH579961 |
| SRS079 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 70 | — | — | MH580033 | MH580107 | MH579962 |
| SRS097 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 71 | — | MH579813 | MH580034 | MH580108 | MH579963 |
| SRS009A | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 73 | MH579881 | — | MH580035 | MH580142 | MH579964 |
| SRS102 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 75 | MH579882 | MH579814 | MH580036 | MH580109 | MH579965 |
| SRS104 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 76 | MH579883 | MH579815 | MH580037 | MH580143 | MH579966 |
| SRS106 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 77 | MH579884 | MH579816 | MH580038 | MH580144 | MH579967 |
| SRS006 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 78 | MH579885 | MH579817 | MH580039 | MH580145 | MH579968 |
| SRS081 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 79 | — | — | MH580040 | MH580111 | MH579969 |
| SRS105 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 80 | MH579886 | MH579818 | MH580041 | MH580146 | MH579970 |
| SRS107 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 81 | MH579887 | MH579819 | MH580042 | MH580147 | MH579971 |
| SRS004 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 82 | MH579888 | — | MH580043 | MH580148 | MH579972 |
| SRS007 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 83 | MH579889 | MH579820 | MH580044 | MH580150 | MH579973 |
| SRS025 | Cololejeunea gottschei | Trat | Thailand | 12°22′55″N | 102°39′21″E | 84 | MH579890 | MH579821 | MH580045 | MH580151 | MH579974 |
| SRS025 | Cololejeunea tenella | Trat | Thailand | 12°22′55″N | 102°39′21″E | 85 | — | — | MH580046 | MH580112 | MH579975 |
| SRS002 | Cololejeunea denticulata | Trat | Thailand | 12°22′55″N | 102°39′21″E | 86 | MH579891 | — | MH580047 | MH580152 | — |
| SRS003 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 87 | MH579892 | — | MH580048 | MH580113 | MH579976 |
| SRS033 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 88 | MH579893 | — | MH580049 | MH580153 | MH579977 |
| SRS010 | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 89 | MH579894 | — | MH580050 | MH580149 | MH579978 |
| SRS013 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 90 | MH579895 | MH579822 | MH580051 | MH580154 | MH579979 |
| SRS009B | Cololejeunea lanciloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 91 | MH579896 | — | MH580052 | MH580141 | MH579980 |
| SRS014 | Caudalejeunea reniloba | Trat | Thailand | 12°22′55″N | 102°39′21″E | 92 | MH579897 | MH579823 | MH580053 | MH580155 | MH579981 |
| SRS023 | Cololejeunea indosinica | Trat | Thailand | 12°22′55″N | 102°39′21″E | 93 | MH579898 | MH579824 | MH580054 | MH580114 | MH579982 |
| SRS024 | Cololejeunea indosinica | Trat | Thailand | 12°22′55″N | 102°39′21″E | 94 | — | — | MH580055 | — | MH579983 |
| EKE005 | Leptolejeunea epiphylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 95 | MH579899 | MH579825 | MH580056 | MH580156 | — |
| EKE006 | Leptolejeunea epiphylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 96 | MH579900 | MH579826 | MH580057 | MH580115 | MH579984 |
| EKE007 | Leptolejeunea epiphylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 97 | MH579901 | MH579827 | MH580058 | MH580116 | MH579985 |
| EKE008 | Cololejeunea tenella | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 98 | — | MH579828 | MH580059 | — | MH579986 |
| EKE009 | Cololejeunea tenella | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 99 | MH579902 | MH579829 | MH580060 | MH580117 | MH579987 |
| EKE010 | Cololejeunea tenella | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 100 | — | MH579830 | MH580061 | — | — |
| EKE011 | Leptolejeunea epiphylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 101 | MH579903 | MH579831 | MH580062 | MH580118 | MH579988 |
| EKE012 | Cololejeunea tenella | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 102 | — | MH579832 | MH580063 | — | MH579989 |
| EKE013 | Cololejeunea goebelii | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 103 | MH579904 | MH579833 | MH580064 | MH580119 | MH579990 |
| EKE014 | Cololejeunea goebelii | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 104 | MH579905 | MH579834 | MH580065 | MH580120 | MH579991 |
| EKE015 | Cololejeunea goebelii | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 105 | — | MH579835 | MH580066 | MH580121 | MH579992 |
| EKE016 | Cololejeunea goebelii | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 106 | MH579906 | MH579836 | MH580067 | MH580122 | — |
| EKE017 | Leptolejeunea maculata | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 107 | MH579907 | MH579837 | MH580068 | MH580123 | MH579993 |
| EKE018 | Leptolejeunea maculata | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 108 | MH579908 | MH579838 | MH580069 | MH580124 | MH579994 |
| EKE019 | Leptolejeunea maculata | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 109 | MH579909 | MH579839 | MH580070 | MH580125 | MH579995 |
| EKE020 | Leptolejeunea maculata | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 110 | — | MH579840 | MH580071 | MH580126 | — |
| EKE021 | Lejeunea anisophylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 111 | — | MH579841 | MH580072 | — | — |
| EKE022 | Lejeunea anisophylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 112 | MH579910 | MH579842 | MH580073 | MH580127 | — |
| EKE023 | Lejeunea anisophylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 113 | MH579911 | MH579843 | MH580074 | MH580128 | MH579996 |
| EKE024 | Lejeunea anisophylla | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 114 | MH579912 | MH579844 | — | — | — |
| EKE025 | Cololejeunea lanciloba | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 115 | MH579913 | MH579845 | MH580075 | MH580110 | MH579997 |
| EKE026 | Cololejeunea lanciloba | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 116 | MH579914 | MH579846 | MH580076 | — | MH579998 |
| EKE028 | Cololejeunea lanciloba | Ranong | Thailand | 9°22′31″N | 98°23′53″E | 118 | MH579915 | MH579847 | MH580077 | MH580102 | MH579999 |
| JW003 | Diplasiolejeunea cavifolia | Ranong | Thailand | 10°30′49″N | 98°54′26″E | 119 | — | MH579848 | — | — | — |
| JW002 | Diplasiolejeunea cavifolia | Ranong | Thailand | 10°30′49″N | 98°54′26″E | 120 | MH579916 | MH579849 | MH580078 | — | MH580000 |
| JW004 | Diplasiolejeunea cavifolia | Ranong | Thailand | 10°30′49″N | 98°54′26″E | 121 | MH579917 | MH579850 | MH580079 | MH580129 | — |
| JW005 | Diplasiolejeunea cavifolia | Ranong | Thailand | 10°30′49″N | 98°54′26″E | 122 | — | MH579851 | — | — | — |
| SRS091A | Cololejeunea sp. | Trat | Thailand | 12°22′55″N | 102°39′21″E | 123 | MH579918 | MH579852 | MH580080 | MH580130 | MH580001 |
| SRS091B | Cololejeunea sp. | Trat | Thailand | 12°22′55″N | 102°39′21″E | 124 | — | MH579853 | MH580081 | MH580157 | — |
| SRS091C | Cololejeunea sp. | Trat | Thailand | 12°22′55″N | 102°39′21″E | 125 | — | MH579854 | — | — | — |
| SRS091D | Cololejeunea sp. | Trat | Thailand | 12°22′55″N | 102°39′21″E | 126 | MH579919 | MH579855 | MH580082 | — | — |
Yodphaka, S. , Boonpragob K., Lumbsch H. T., and E. Kraichak . 2018. Evaluation of six regions for their potential as DNA barcodes in epiphyllous liverworts from Thailand. Applications in Plant Sciences 6(8): e1174.
LITERATURE CITED
- Bell, D. , Long D. G., Forrest A. D., Hollingsworth M. L., Blom H. H., and Hollingsworth P. M.. 2011. DNA barcoding of European Herbertus (Marchantiopsida, Herbertaceae) and the discovery and description of a new species. Molecular Ecology Resources 12: 36–47. [DOI] [PubMed] [Google Scholar]
- Bohmann, K. , Evans A., Gilbert M. T. P., Carvalho G. R., Creer S., Knapp M., Yu D. W., and de Bruyn M.. 2014. Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology & Evolution 29: 358–367. [DOI] [PubMed] [Google Scholar]
- Brown, S. D. J. , Collins R. A., Boyer S., Lefort M.‐C., Malumbres‐Olarte J., Vink C. J., and Cruickshank R. H.. 2012. Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Molecular Ecology Resources 12: 562–565. [DOI] [PubMed] [Google Scholar]
- CBOL Plant Working Group . 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences USA 106: 12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, T. , Xu C., Lei L., Li C., Zhang Y., and Zhou S.. 2016. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Molecular Ecology Resources 16: 138–149. [DOI] [PubMed] [Google Scholar]
- Darriba, D. , Taboada G. L., Doallo R., and Posada D.. 2012. jModelTest 2: More models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. 2004. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S. , Jiang Y., Wang S., Jiang M., Chen Z., Ying Q., and Wang H.. 2015. Molecular identification of Dendrobium species (Orchidaceae) based on the DNA barcode ITS2 region and its application for phylogenetic study. International Journal of Molecular Sciences 16: 21975–21988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest, L. L. , Davis E. C., Long D. G., Crandall‐Stotler B. J., Clark A., and Hollingsworth M. L.. 2006. Unraveling the evolutionary history of the liverworts (Marchantiophyta): Multiple taxa, genomes and analyses. Bryologist 109: 303–334. [Google Scholar]
- Gradstein, S. R. , Wilson R., Ilkiu‐Borges A. L., and Heinrichs J.. 2006. Phylogenetic relationships and neotenic evolution of Metzgeriopsis (Lejeuneaceae) based on chloroplast DNA sequences and morphology. Botanical Journal of the Linnean Society 151: 293–308. [Google Scholar]
- Gradstein, S. R. , Iilkiu‐Borges A. L., and Vanderpoorten A.. 2011. Habitat specialization triggers the evolution of unusual morphologies: The case of Cololejeunea stotleriana sp. nov. from Ecuador. Bryologist 114: 9–22. [Google Scholar]
- Hartmann, F. A. , Wilson R., Gradstein S. R., Schneider H., and Heinrichs J.. 2006. Testing hypotheses on species delimitations and disjunctions in the liverwort Bryopteris (Jungermanniopsida: Lejeuneaceae). International Journal of Plant Sciences 167: 1205–1214. [Google Scholar]
- Hassel, K. , Segreto R., and Ekrem T.. 2013. Restricted variation in plant barcoding markers limits identification in closely related bryophyte species. Molecular Ecology Resources 13: 1047–1057. [DOI] [PubMed] [Google Scholar]
- Hebert, P. D. N. , Ratnasingham S., and de Waard J. R.. 2003. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society B. Biological Sciences 270 Suppl: S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, P. D. N. , Penton E. H., Burns J. M., Janzen D. H., and Hallwachs W.. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator . Proceedings of the National Academy of Sciences USA 101: 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, M. L. , Andra Clark A., Forrest L. L., Richardson J., Pennington R. T., Long D. G., Cowan R., et al. 2009. Selecting barcoding loci for plants: Evaluation of seven candidate loci with species‐level sampling in three divergent groups of land plants. Molecular Ecology Resources 9: 439–457. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, P. M. 2011. Refining the DNA barcode for land plants. Proceedings of the National Academy of Sciences USA 108: 19451–19452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, P. M. , Graham S. W., and Little D. P.. 2011. Choosing and using a plant DNA barcode. PLoS ONE 6: e19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H. , Xie Z., Koo H. J., McLaughlin S. P., Timmermann B. N., and Gang D. R.. 2006. Metabolic profiling and phylogenetic analysis of medicinal Zingiber species: Tools for authentication of ginger (Zingiber officinale Rosc.). Phytochemistry 67: 1673–1685. [DOI] [PubMed] [Google Scholar]
- Jovet‐Ast, S. 1953. Le genre Colura: Hépatiques: Lejeuneaceae, Diplasiae. Revue Bryologique et Lichénologique 22: 206–312. [Google Scholar]
- Jovet‐Ast, S. 1967. Colura récoltes du Pakistan aux Philippines par Pierre Tixier. Revue Bryologique et Lichénologique 35: 138–142. [Google Scholar]
- Kraichak, E. 2015. Relationships between moisture and community structure of epiphyllous bryophytes. Ninth Botanical Conference of Thailand BCT–Stepping toward Global Forum, Bangkok, Thailand.
- Kraichak, E. , and Yaungthong K.. 2012. Ecology and diversity of epiphyllous bryphytes in the Naka Wildlife Sanctuary, Ranong Province: A preliminary study. Report to Department of National Parks, Wildlife, and Plant Conservation No. 0907.1/13812, Bangkok, Thailand.
- Kress, W. J. , and Erickson D. L.. 2007. A two‐locus global DNA barcode for land plants: The coding rbcL gene complements the non‐coding trnH‐psbA spacer region. PLoS ONE 2: e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress, W. J. , Wurdack K. J., Zimmer E. A., Weigt L. A., and Janzen D. H.. 2005. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences USA 102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold, M. A. , Holyoak M., Mouquet N., Amarasekare P., Chase J. M., Hoopes M. F., Holt R. D., et al. 2004. The metacommunity concept: A framework for multi‐scale community ecology. Ecology Letters 7: 601–613. [Google Scholar]
- Li, D.‐Z. , Gao L.‐M., Li H.‐T., Wang H., Ge X. J., Liu J.‐Q., Chen Z.‐D., et al. 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences USA 108: 19641–19646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Yan H. F., Cao T., and Ge X. J.. 2010. Evaluation of 10 plant barcodes in Bryophyta (Mosses). Journal of Systematics and Evolution 48: 36–46. [Google Scholar]
- Meier, R. , Shiyang K., Vaidya G., and Ng P. K. L.. 2006. DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Systematic Biology 55: 715–728. [DOI] [PubMed] [Google Scholar]
- Miller, M. A. , Pfeiffer W., and Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, Louisiana, USA.
- Myszczyński, K. , Bączkiewicz A., Buczkowska K., Ślipiko M., Szczecińska M., and Sawicki J.. 2017. The extraordinary variation of the organellar genomes of the Aneura pinguis revealed advanced cryptic speciation of the early land plants. Scientific Reports 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osathanunkul, M. , Madesis P., and de Boer H.. 2015. Bar‐HRM for authentication of plant‐based medicines: Evaluation of three medicinal products derived from Acanthaceae species. PLoS ONE 10: e0128476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis, E. , Claude J., and Strimmer K.. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics (Oxford, England) 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Phoolcharoen, W. , and Sukrong S.. 2012. Molecular analysis of Vitex species using candidate DNA barcoding and PCR‐RFLP of the matK gene for authentication of Vitex glabrata . Natural Product Communications 8: 125–128. [PubMed] [Google Scholar]
- Pócs, T. 2011. New or little known epiphyllous liverworts, XIV. The genus Colura (Lejeuneaceae) in São Tomé Island, with the description of Colura thomeensis sp. nov. Bryologist 114: 362–366. [Google Scholar]
- Pócs, T. 2012a. Bryophytes from the Fiji Islands, VI. The genus Cololejeunea Raddi (Jungermanniopsida), with the description of seven new species. Acta Botanica Hungarica 54: 145–188. [Google Scholar]
- Pócs, T. 2012b. New or little known epiphyllous liverworts, XVI. A small collection from Laos. Acta Biologica Plantarum Agriensis 2: 5–10. [Google Scholar]
- Pons, J. , Barraclough T., Gomez‐Zurita J., Cardoso A., Duran D., Hazell S., Kamoun S., et al. 2006. Sequence‐based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology 55: 595–609. [DOI] [PubMed] [Google Scholar]
- R Core Development Team . 2013. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Website http://www.R-project.org [accessed 30 July 2018].
- Ratnasingham, S. , and Hebert P. D. N.. 2007. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Molecular Ecology Notes 7: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski, J. P. 1980. N2 fixation (C2H2 reduction) by epiphylls on coffee, Coffea arabica . Microbial Ecology 6: 349–355. [DOI] [PubMed] [Google Scholar]
- Schindel, D. E. , and Miller S. E.. 2005. DNA barcoding a useful tool for taxonomists. Nature 435: 17. [DOI] [PubMed] [Google Scholar]
- Schoch, C. L. , Robbertse B., Robert V., Vu D., Cardinali G., Irinyi L., Meyer W., et al. 2014. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database 2014: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, A. J. , Cox C. J., and Boles S. B.. 2003. Polarity of peatmoss (Sphagnum) evolution: Who says bryophytes have no roots? American Journal of Botany 90: 1777–1787. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. , Hoover P., and Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stech, M. , and Quandt D.. 2010. 20,000 species and five key markers: The status of molecular bryophyte phylogenetics. Phytotaxa 9: 196. [Google Scholar]
- Taberlet, P. , Gielly L., Pautou G., and Bouvet J.. 1991. Universal primers for amplification of three non‐coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Taberlet, P. , Coissac E., Pompanon F., Gielly L., Miquel C., Valentini A., Vermat T., et al. 2006. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Research 35: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixier, P. 1985. Contribution à la connaissance des Cololejeunoideae. Bryophytorum Bibliotheca, vol. 27. J. Cramer, Vaduz, Liechtenstein.
- Wicke, S. , and Quandt D.. 2009. Universal primers for the amplification of the plastid trnK/matK region in land plants. Anales del Jardín Botánico de Madrid 66: 285–288. [Google Scholar]
- Yao, H. , Song J., Liu C., Luo K., Han J., Li Y., Pang X., et al. 2010. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE 5: e13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Heinrichs J., Zhu R.‐L., and Schneider H.. 2013a. Empirical evidence supporting frequent cryptic speciation in epiphyllous liverworts: A case study of the Cololejeunea lanciloba complex. PLoS ONE 8: e84124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Pócs T., Schäfer‐Verwimp A., Heinrichs J., Zhu R.‐L., and Schneider H.. 2013b. Evidence for rampant homoplasy in the phylogeny of the epiphyllous liverwort genus Cololejeunea (Lejeuneaceae). Systematic Botany 38: 553–563. [Google Scholar]
- Zartman, C. E. , and Nascimento H. E. M.. 2006. Are habitat‐tracking metacommunities dispersal limited? Inferences from abundance‐occupancy patterns of epiphylls in Amazonian forest fragments. Biological Conservation 127: 46–54. [Google Scholar]
- Zhu, R. L. , and So M. L.. 1998. Two epiphyllous liverworts, Cololejeunea dozyana (Sande Lac.) Schiffn. and Cololejeunea sigmoidea Jovet‐Ast & Tixier (Hepaticae, Lejeuneaceae), new to Taiwan. Botanical Bulletin of Academia Sinica 39: 125–130. [Google Scholar]
- Zhu, R.‐L. , and So M. L.. 2001. Epiphyllous liverworts of China. Nova Hedwigia Beiheft, vol. 121. J. Cramer, Vaduz, Liechtenstein.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Single‐locus neighbor‐joining trees of five studied markers for studied epiphyllous bryophyte species: ITS1 (A), ITS2 (B), psbA (C), rbcL (D), and trnL‐F (E). Black circles at the nodes indicate nodes with bootstrap support greater than 70.
APPENDIX S2. Single‐locus maximum likelihood trees of five studied markers for studied epiphyllous bryophyte species: ITS1 (A), ITS2 (B), psbA (C), rbcL (D), and trnL‐F (E). Black circles at the nodes indicate nodes with bootstrap support greater than 70.
Data Availability Statement
The sequence data are deposited in and accessible from the U.S. National Center for Biotechnology Information's GenBank database (https://www.ncbi.nlm.nih.gov; accession no. MH579787–MH580157).


