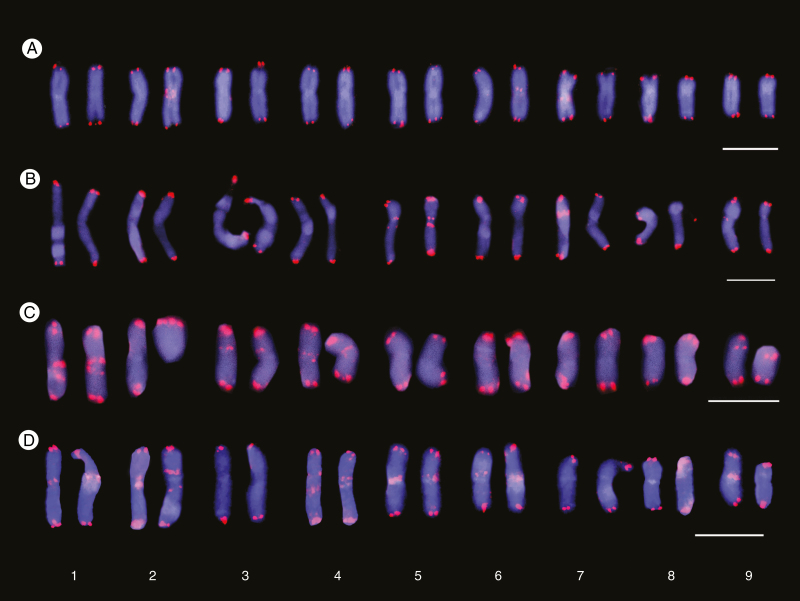Abstract
Background and Aims
Interstitial telomeric repeat (ITR) sites, consisting of tandem repeats of telomeric motifs localized at intrachromosomal sites, have been reported in a few unrelated organisms including plants. However, the causes for the occurrence of ITRs outside of the chromosomal termini are not fully understood. One possible explanation are the chromosomal rearrangements involving telomeric sites, which could also affect the location of other structural genome elements, such as the 45S rDNA. Taking advantage of the high dynamism in 45S rDNA loci previously found in Anacyclus (Asteraceae, Anthemideae), the occurrence and patterns of variation of ITRs were explored in this genus with the aim of finding common underlying causes.
Methods
In total, 132 individuals from 44 populations of nine species were analysed by fluorescence in situ hybridization using an Arabidopsis-type telomeric sequence as a probe.
Key results
Variable presence of ITR sites was detected in six out of nine species of Anacyclus, ranging from two to 45 sites and showing contrasting chromosomal locations and a differential presence of the ITR site on homologous chromosome pairs. At the intraspecific level, the ranges were as large as 0–12 ITR sites. Although only 26 % of the total observed ITR sites were localized in chromosomes bearing 45S rDNA loci, all cases of interstitial 45S rDNA reported in a previous work co-occurred with ITRs in close proximity in the same chromosome arms.
Conclusions
High levels of ITR polymorphism within a single species have not been previously reported in plants and suggest that this pattern might have been overlooked due to insufficient sampling. Although ancient Robertsonian translocations or the amplification of terminal 45S rDNA sites cannot, on their own, explain all of the levels of variability in ITRs reported here, there are suggestions that they may have been involved in the evolutionary history of this genus or its ancestors in Anthemideae.
Keywords: Interstitial Arabidopsis-type telomere repeats, FISH, Asteraceae, cytogenetic evolution, rDNA, Anthemideae, Anacyclus
INTRODUCTION
Telomeres are universal components of linear eukaryotic chromosomes and are essential for chromosome stability. They protect the chromosome caps against end-to-end fusions or from the elicitation of a cellular response, such as apoptosis (Blackburn, 2001). Telomeres are formed by nucleoprotein/DNA complexes that cap the ends of chromosomes, consist of simple G-rich motifs that are tandemly repeated and constitute long repetitive stretches, up to 160 kb in some plants (e.g. Nicotiana tabacum; Fajkus et al., 1995). In plants, the dominant consensus telomere repeat is composed of seven nucleotides of the Arabidopsis-type (TTTAGGG)n (Richards and Ausubel, 1988) following a sequence that is highly similar to those of other organisms from distant lineages of flowering plants (for the presence of unusual repeats in plants, see Weiss and Scherthan, 2002; Sýkorová et al., 2003; Shibata and Hizume, 2011; Peška et al., 2015; Tran et al., 2015; Fajkus et al., 2016). Conservation of the terminal telomeric loci and concomitant sequences in eukaryotes probably reflects the strong selective pressures associated with their essential function, which facilitates the complete replication of the chromosome termini and protects them from being covalently joined (Nelson et al., 2014).
An interstitial telomeric repeat (ITR) site consists of tandem repeats of telomeric motifs that are localized at intrachromosomal sites (Lin and Yan, 2008). They have been reported in unrelated organisms, such as vertebrates (Meyne et al., 1990) and plants (Cox et al., 1993; Fuchs et al., 1995). Telomeric-like sequences have been reported to be associated with terminal nucleolus organizer regions (NORs) in some organisms (Copenhaver and Pikaard, 1996; Liu and Fredga, 1999; Armstrong et al., 2001; Dvořáčková et al., 2015). However, whether such associations between rRNA genes and telomeric sequences occur in organisms showing interstitial rRNA genes has not been investigated. Addressing this question may provide clues about the mechanisms of chromosomal and genomic rearrangements involved in the origin of ITRs.
Recently, we reported exceptionally high levels of 45S rDNA locus diversity in the plant genus Anacyclus (Asteraceae, Anthemideae), a diploid (2n = 18) Mediterranean lineage including nine species. Such diversity affects site number, chromosomal location, presence in accessory chromosomes, and differential presence of the 45S rDNA site on the homologous chromosome pairs (i.e. homozygosis and heterozygosis; Rosato et al., 2017). There is an uneven distribution of this diversity, particularly at the inter- and intrapopulation levels, which is particularly pronounced in sympatric populations of Anacyclus clavatus and A. valentinus and in areas where species ranges overlap even if no hybridization has been reported. One of the possible mechanisms involved in such dynamism is chromosomal rearrangements. Therefore, we examined if other structural elements of the genome that could have been affected by such rearrangements show comparable levels of polymorphism to 45S rDNA sites. We focused on telomeric elements and the possibility of ITR sites.
To this end, we used an Arabidopsis-type telomeric sequence as a probe to search for the presence of ITRs in mitotic chromosomes using fluorescence in situ hybridization (FISH), to assess the degree of stability of these elements across Anacyclus. Specifically, our objectives were: (1) to determine whether ITR loci are present within the karyotype of Anacyclus species and, if so, to assess the patterns of variation at the population and species levels in wild accessions; (2) to ascertain whether variation in 45S ribosomal loci is associated with the presence of ITR; and (3) to analyse the results from an evolutionary perspective. For this purpose, we used a large sampling size covering the whole genus, which also allowed us to document the frequency of ITRs in natural populations, and performed ancestral character-state reconstruction for the number of ITR sites.
MATERIALS AND METHODS
Plant materials
A subset of the accessions included in Rosato et al. (2017) to analyse 45S rDNA site variation in Anacyclus was used. Specifically, 132 individuals from 44 populations of nine Anacyclus species were analysed in this study (Supplementary Data Table S1). This sampling covers all species of the genus according to the most recent molecular phylogenetic analysis (Vitales et al., 2018), which has revised the circumscription of Anacyclus by Humphries (1979). Seeds were collected in the field and those with the fastest rate of germination (Torices et al., 2013) were selected. After germination, seedlings were grown individually in a mix of COMPO SANA Universal Potting Soil and siliceous sand (3: 1) until roots were developed. Root tips were collected following Rosato et al. (2008).
Cytogenetic analysis
Fluorescence in situ hybridization
The telomeric sequence TTTAGGG was localized using the pAtT4 clone isolated from Arabidopsis thaliana (Richards and Ausubel, 1988). The probe was labelled with biotin-16-dUTP through a nick translation procedure according to the manufacturer’s protocols (Roche, Germany). We followed the in situ hybridization protocols of Rosato et al. (2008). Probe detection was conducted using the method of Zhong et al. (1996) with modifications according to Galián et al. (2014). Because we were interested in the possible association between 45S rDNA sites and ITRs as well as the potential underlying causes for their co-occurrence, the same slides analysed by Rosato et al. (2017) to detect the 45S rDNA sites using the pTa71 clone (Gerlach and Bedbrook, 1979) were sequentially hybridized with the telomeric DNA probe pAtT4 clone (Fig. S1).
Karyotype analysis
Chromosome measurements were made on digital images using the computer application MicroMeasure version 3.2 (Reeves, 2001). Cariograms were obtained from chromosome measurements of at least five well-spread metaphase plates. Each chromosome arm was divided into four regions of equal size according to Roa and Guerra (2012): proximal (p), interstitial–proximal (ip), interstitial–terminal (it) and terminal (t). Each ITR (and 45S rDNA) site was positioned in one of the four regions. Neither the ITR nor the 45S rDNA signals coincided with the position of the centromere.
Evolutionary trends in ITR site change
To explore evolutionary trends in ITR site change, we chose two characters (the occurrence of ITRs and the modal number of ITR sites per species) to be mapped onto the available phylogenetic tree of Anacyclus based on sequences from the nrDNA internal transcribed spacer (ITS) region (Oberprieler, 2004). The presence of ITRs was coded as binary (with three species being polymorphic: A. clavatus, A. valentinus and A. monanthos), whereas the modal number of ITR sites was coded as multistate discrete. Intervals were established so that no species were polymorphic for this character (0, 1–10, 11–20, >20). Reconstruction of ancestral character states was carried out using the maximum likelihood (ML) approach in Mesquite, version 3.04 (Maddison and Maddison, 2015), which assigns to each internal node the character state that maximizes the probability of obtaining the observed character states in the terminal taxa under the specified model of evolution. The ML reconstructions were conducted using the Mk1 model of evolution (Schluter et al., 1997; Pagel, 1999). The Mk1 (Markov k-state 1 parameter model) is a k-state generalization of the Jukes–Cantor model, corresponding to Lewis’ (2001) Mk model, which assigns equal probability to changes between any two character states. In addition, we mapped the geographical distribution of the number of ITRs per species and population to search for possible patterns.
RESULTS
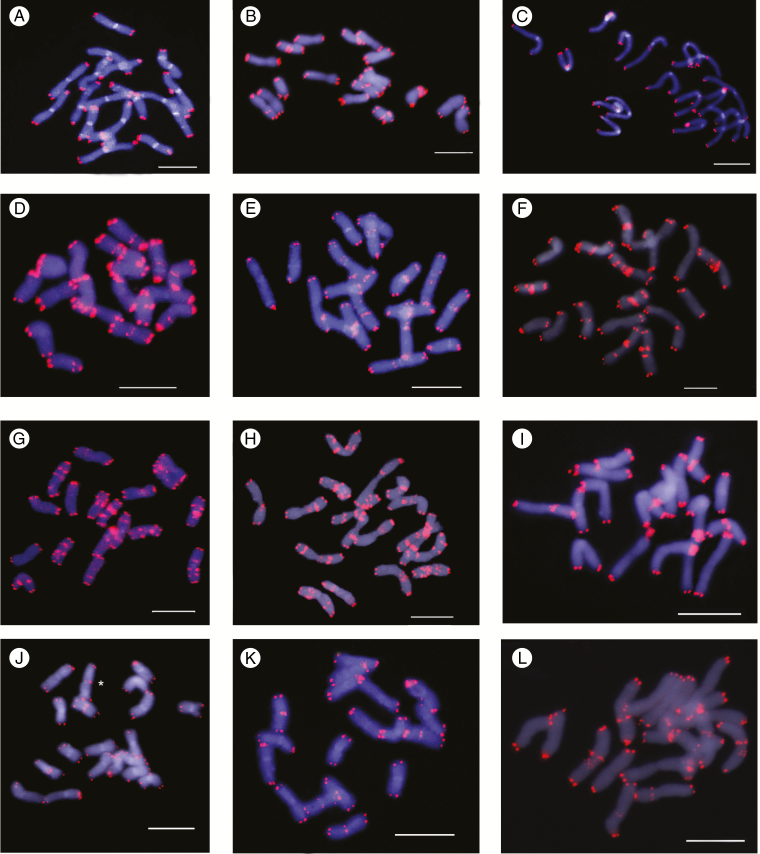
FISH experiments using the Arabidopsis-type telomeric probe (TTTAGGG)n revealed, as expected, strong and consistent signals in the terminal ends of both chromosome arms (Fig. 1). In addition, a variable number of interstitial telomeric signals were detected in six out of nine species of Anacyclus (Table 1, Figs 1 and 2). Interstitial telomeric sites were absent in A. homogamos, A. maroccanus and A. radiatus (both subspecies). In contrast, all analysed accessions of A. atlanticus, A. linearilobus and A. pyrethrum showed ITRs. The remaining three species (A. monanthos, A. clavatus and A. valentinus) were polymorphic for the presence and distribution of ITRs, showing a contrasting pattern of variation at the inter- and intrapopulation levels (Fig. 3). For instance, one population of A. monanthos showed no ITRs (GAB), another was fixed for the presence of ITRs (MAT) and the ZER population showed individuals both with and without ITRs. This overall pattern was also found in A. clavatus. In contrast, A. valentinus showed populations fixed for the presence of ITRs, or with intrapopulation polymorphisms but none fixed for the absence of ITRs.
Fig. 1.
Telomeric-like sequence (TTTAGGG)n sites in Anacyclus assessed by FISH analysis. Telomeric sites and interstitial telomeric repeat (ITR) sites are shown as red fluorescent signals and the chromosomes are counterstained with 4, 6-diamidino-2-phenylindole (DAPI) (blue colour). (A) Anacyclus homogamos (no ITR sites); (B) A. atlanticus (six ITR sites); (C–E) A. clavatus showing three (C), nine (D) and 14 (E) ITR sites; (F) A. linearilobus (19 ITR sites); (G-H) A. pyrethrum showing 26 (G) and 45 (H) ITR sites; (I–L) A. valentinus showing three (I, J), four (K) and ten (L) ITR sites. The occurrence of an extra chromosome in A. valentinus (J) is indicated by an asterisk. Scale bars: 10 µm.
Table 1.
Distribution of ITR sites in the studied populations of Anacyclus
| Species | Number of analysed populations (sample size) | ITR site number (number of individuals) |
|---|---|---|
| A. atlanticus | 3 (10*) | 6 (2), 8 (2), 17 (1) |
| A. clavatus | 14 (38) | 0 (8), 2 (5), 3 (2), 4 (5), 6 (3), 8 (9), 9 (3), 12 (2), 14 (1) |
| A. homogamos | 2 (8) | 0 (8) |
| A. linearilobus | 3 (5*) | 19 (1) |
| A. maroccanus | 3 (9) | 0 (9) |
| A. monanthos | 3 (10) | 0 (5), 4 (5) |
| A. radiatus | ||
| subsp. coronatus | 2 (7) | 0 (7) |
| subsp. radiatus | 3 (8) | 0 (8) |
| A. pyrethrum | 2 (6*) | 26 (1), 30 (1), 45(2) |
| A. valentinus | 9 (31) | 0 (4), 2 (11), 3 (3), 4 (6), 5 (2), 6 (2), 10 (3) |
*Five individuals of A. atlanticus, four of A. linearilobus and two of A. pyrethrum unambiguously showed the presence of ITR sites, but their number could not be accurately determined.
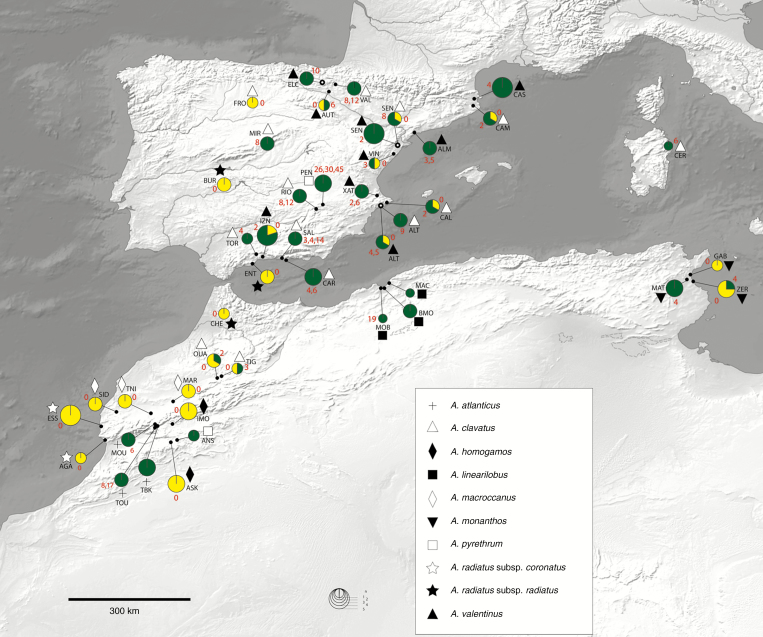
Fig. 2.
Geographical distribution of the occurrence of interstitial telomeric repeats (ITRs) in Anacyclus. Green indicates the occurrence of ITRs, with the number of ITR sites found in individual samples shown in red. Eleven individuals were unequivocally found to contain ITRs but the exact number could not be determined (populations ANS, BMO, MAC, MOB, TBK; see Table 1). Yellow indicates no ITRs detected. Circle size is proportional to the sampling in each population. Sympatric populations of A. clavatus and A. valentinus are indicated by an empty dot. The map is made with Natural Earth. Free vector and raster map data @naturalearthdata.com.
Fig. 3.
Cariograms showing inter- and intrapopulation site number variation of interstitial telomeric repeats (ITRs) in Anacyclus clavatus. Telomeric sites and ITRs are shown as red fluorescent signals and the chromosomes are counterstained with 4, 6-diamidino-2-phenylindole (DAPI) (blue colour). Representative individuals from Salobreña (A, D), Carchuna (B) and Altea (C) populations are shown. (A) Four ITR sites; (B) six sites; (C) nine sites; (D) 14 sites. Scale bars: 10 µm.
Overall, when present, the number of ITRs in the genome ranged from two (A. clavatus, A. valentinus) to 45 sites (A. pyrethrum). The ranges were also outstanding at the intra- and interpopulation levels (Table 1, Fig. 2). The most striking results were found in A. pyrethrum where ITRs ranged from 26 to 45 signals within a single population. Interestingly, a significant number of those individuals showing ITRs were heterozygous for at least one locus (approx. 31 %). Uneven numbers of ITR signals occurred in all species, except in A. monanthos, which was homozygous for all ITR sites.
The number of chromosome pairs showing ITR signals ranged from one (A. clavatus and A. valentinus) to all nine pairs of the complement (A. pyrethrum). The modal value of chromosomal pairs showing ITRs was three. The range of variation for this feature was remarkable in some species, such as A. clavatus, where one to six chromosome pairs showed ITRs.
The lowest average number of ITR signals observed per genome (mean ± SD) was 3.45 ± 0.48 in A. valentinus followed by 5.03 ± 0.63 in A. clavatus. The highest average value was 36.5 ± 4.30 recorded in A. pyrethrum.
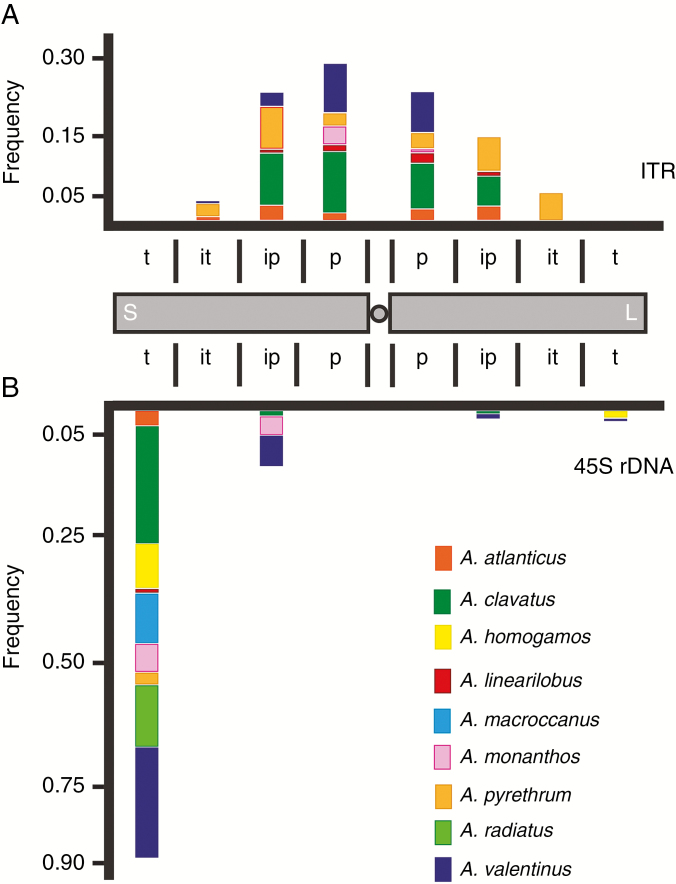
The distribution of ITR sites in the chromosome arms was slightly but significantly (χ2 = 11.34, P = 0.0034) biased towards the short arms (containing 56 % of the overall signals) compared to the long arms (44 %). Overall, the presence of ITR signals along the chromosome arms was uneven. No ITR signals were localized at the centromeric or subtelomeric regions and their presence was mostly recorded in the p and ip regions of the short (95 %) and long arms (89 %) (Fig. 4A). When comparing Anacyclus species there were differences in the ITR distribution along the chromosomal regions, but no fixed species-specific sites could be detected.
Fig. 4.
Frequency of ITR sites (A) and 45S rDNA sites (B) along the chromosome arms in Anacyclus species. Each chromosome arm was divided into four regions of equal size according to Roa and Guerra (2012): proximal (p), interstitial–proximal (ip), interstitial–terminal (it) and terminal (t). Short arm (S), long arm (L).
Only 26 % of the total observed ITR sites were localized in chromosomes bearing 45S rDNA loci, suggesting that these chromosomes were not a hotspot of ITR amplification. However, in those chromosomes with both, the ITRs and interstitial 45S rDNA signals were found in close proximity (Fig. 4, Fig. S2).
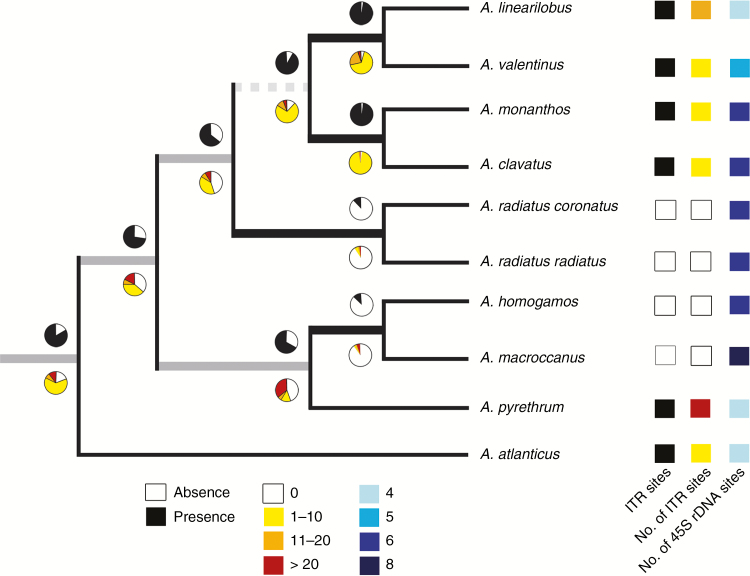
The ML reconstruction of the ancestral states of the two ITR characters suggests that the presence of ITRs was the most likely condition in the basalmost node leading to Anacyclus diversification (Fig. 5). It is inferred that ITR losses occurred at least twice during the evolutionary history of this genus. The hypothesis that the ancestral state in Anacyclus was the presence of a moderate number of ITR sites (1–10) showed the highest likelihood scores (Fig. 5). No clear geographical pattern was found for the distribution of the number of ITRs across populations and species (Fig. 2).
Fig. 5.
Maximum likelihood reconstruction of ancestral character states for two characters, presence/absence of ITR sites and number of ITR sites in Anacyclus species, using the phylogenetic tree in Oberprieler (2004) with a slight modification (Heliocauta atlantica, the sister group to Anacyclus, is labelled as A. atlanticus). Pie charts illustrate the relative likelihood for each character state at each node under the specified model of evolution (Mk1 model, in this study). Bootstrap support for the nodes obtained in the original study of Oberprieler are indicated as follows: black (90–100 %), grey (71–89 %), dashed (<70 %). The presence of ITR sites, the ranges of modal numbers of ITR sites and the number of 45S rDNA loci (Rosato et al., 2017) for each species are indicated on the right margin.
DISCUSSION
Is Anacyclus an isolated case of site number ITR hypervariability in plants?
Following the early compilations of Cox et al. (1993) and Fuchs et al. (1995), there have been several new reports of interstitial telomere-like sequences in plants (e.g. Tek and Jiang, 2004; Gaspin et al., 2010; Mandáková et al., 2016). In particular, a broad survey using FISH detected Arabidopsis-type or human-type (TTAGGG)n repeats at interstitial or proximal chromosome regions in eight of 87 species (Shibata and Hizume, 2011). Under this perspective, our reports of ITR in Anacyclus, belonging to a plant family (Asteraceae) in which ITR sites have been previously recorded (Hanmoto et al., 2007; Shibata and Hizume, 2011), are of limited novelty.
However, in contrast to previous findings, the situation in Anacyclus is unique due to the unexpected variation in site number and chromosomal distribution of the ITRs at the intra- and interpopulation, and interspecific levels. Our experimental approach, covering all known species from a single genus and sampling a significant number of populations and individuals, contrasts with all previous reports dealing with the presence (or absence) of ITRs in plants on the basis of FISH analysis.
Our results in Anacyclus suggest a hypervariability and dynamism previously unrecorded in plants, although the possibility that other studies have partly underestimated this due to inadequate sampling cannot be discarded. All analysed Anacyclus individuals showing ITRs may be distinguished by their cytogenetic pattern. However, a closer characterization beyond the cytogenetic level, based on extensive sequencing data, could provide additional information about sequence homogeneity and copy number within and between ITR sites. This may open new perspectives on our understanding of the evolutionary turnover of ITR sites in plants.
ITR occurrence is ancestral in Anacyclus
The ML character-state reconstruction of ITR presence and site number in Anacyclus suggests that ITRs were probably present in the ancestor of the genus. Shibata and Hizume (2011) reported the presence of ITRs in two other genera of Anthemideae (Matricaria and Tanacetum) that are closely related to Anacyclus (Sonboli et al., 2012; Vitales et al., 2018). However, the distribution of ITRs within Anthemideae could not be simply inferred from phylogenetic relatedness because we detected the presence of a high number of ITR sites (52) in Cota [C. nigellifolia (Boiss.) Alv.Fern. & Vitales; M. Rosato et al., unpubl. res.], which is distantly related to Anacyclus (Vitales et al., 2018). In contrast, no ITRs were detected in individuals from four species of Argyranthemum (Borgen et al., 2003), a genus also belonging in the tribe Anthemideae, but not closely related to Anacyclus (Oberprieler, 2004). These scattered results are compatible with the possibility that ITRs originated early in the diversification of the Eurasian grade of Anthemideae (Oberprieler et al., 2009) and were followed by complex and recurrent cycles of amplification, genomic spread and contraction.
Trends in ITR change in Anacyclus
Our results have consistently shown that, contrary to what was assumed previously in other plant species, the occurrence of ITRs is a labile genomic feature in Anacyclus. The ranges of ITR presence within and between populations of several species are similar or even higher than those reported for interspecific comparisons in other genera. Such a high level of polymorphism suggests caution when inferring the evolutionary dynamics of ITR change because the low sample size analysed for some populations and species might be masking ITR polymorphisms. For instance, the fixed absence of ITR sites in A. homogamos, A. maroccanus and A. radiatus and the fixed presence of ITRs in A. pyrethrum and A. linearilobus may turn out to contain some variation if sampling is increased. These cautions, however, do not alter the conclusion that ITR losses and gains have occurred independently many times during the evolutionary history of Anacyclus.
Patterns in A. valentinus, A. monanthos and A. clavatus, where the presence of ITRs is either fixed, polymorphic or absent at the population level, are particularly noticeable. However, the relatively large number of populations and individuals analysed suggests that our results mirror the overall pattern of ITR polymorphisms present in these species. With the exception of the geographically restricted A. atlanticus and A. linearilobus, the other annual species are weedy, grow in unstable anthropogenic environments and show contrasting changes in population sizes (I. Álvarez, pers. observ.), leading to severe demographic bottlenecks. Thus, it could be hypothesized that recurrent cycles of breeding isolation between populations, together with stochastic population bottlenecks and founder effects, are associated with balancing selection and genetic drift leading to the fixation of ITR genotypes. Additional biological factors may counteract genetic drift and be associated with the generation and maintenance of ITR diversity. Hybridization may be one of those factors given that all tested species in Anacyclus are obligate outcrossers and interfertile (Humphries, 1981), and there is evidence for its occurrence in natural populations (Rosato et al., 2017; I. Álvarez, unpubl. res.). The heterozygosity for several ITR loci found in several individuals (Table 1, Fig. 3) is consistent with the structural heterozygosity reported in highly repeated DNA families in Anacyclus (Schweizer and Ehrendorfer, 1976; Ehrendorfer et al., 1977), and both groups of observations are likely to be contributed to by outcrossing and eventual hybridization.
The origin of ITRs in Anacyclus
The first observations of ITRs in centromeric regions of many animal and plant species suggested that ITRs were cytological hallmarks of chromosome rearrangements (Fuchs et al., 1995). Specifically, it was first hypothesized that ITRs derived from centric fusions (Robertsonian translocation) between two telocentric (or acrocentric) chromosomes to form metacentric ones (Slijepcevic, 1998), causing a change in chromosome number (Guerra, 2008).
The fact that no ITR sites are present in centromeric locations in Anacyclus apparently does not support such an origin, although the possibility that ITRs were present in ancient fusions and later spread to other chromosome regions through chromosome rearrangements (inversions, breakage–fusion–bridge cycle, translocation as single events or combined) cannot be totally discarded. Recently, Huang et al. (2016) postulated that two whole-genome duplication events occurred during the early evolutionary history of Asteraceae (Late Cretaceous–Palaeocene). In addition, they suggested that most of the major clades are palaeopolyploids that may have undergone drastic chromosome rearrangements after polyploidization, including descending dysploidies, leading to low chromosomal base numbers (e.g. x = 9 in tribe Anthemideae, to which Anacyclus belongs). Interestingly, the occurrence of ITRs associated with translocation-based descending dysploidy was recently reported in Cardamine cordifolia A. Gray (Mandáková et al., 2016), suggesting that similar cytogenetic processes may have occurred in Asteraceae and Brassicaceae (Lysak et al., 2016). The fact that the three species presenting interstitial 45S rDNA loci (i.e. A. clavatus, A. monanthos and A. valentinus) also show closely situated ITRs in the chromosomes lends some support for a role of chromosomal rearrangements in the occurrence of ITRs in Anacyclus.
Alternatively, interstitial telomeric-like sequences have been reported to be associated with dynamic changes in terminal NORs and could be linked to transposition events (Copenhaver and Pikaard, 1996; Liu and Fredga, 1999; Armstrong et al., 2001). However, because a preferential association between ITRs and terminal 45S rDNA sites was not observed in any of the nine species analysed, our results suggest instead that the transposition of a 45S rDNA multigene family has not played a significant role in the origin of ITRs in Anacyclus.
Another possible causal mechanism based on the heterochomatin distribution model (Schweizer and Loidl, 1987) is equilocal dispersion of telomeric DNA to interstitial regions of the chromosome via transposition or heterologous recombination. This was recently suggested in Nothoscordum (Souza et al., 2016).
Finally, telomeric sequences can be considered to be a particular kind of microsatellite sequence, due to their short length, high copy number and tandem arrangement in the genome. Thus, it may be hypothesized that the generation of telomeric-like repeats is caused by similar mechanisms to those driving the evolutionary dynamics of the repetitive satellite DNA sequences (Charlesworth et al., 1994). In fact, the repair mechanism of DNA double-strand breaks could also lead to short stretches of ITR sequences (Lin and Yan, 2008). Instead, the mechanism of rolling-circle replication of extra-chromosomal circular DNA has been proposed as a mechanism for the amplification of longer repetitive sequences, such as the megabase-sized ITR formation detected in Solanum (He et al., 2013). The fact that we detected signals of long stretches of telomeric-like sequences, some of them as strong or even stronger than those in the telomeric regions, suggests the rolling-circle replication of extra-chromosomal circular DNA as a possible mechanism to explain the hypervariable ITR pattern in Anacyclus. This requires that the ITR had already jumped out of the chromosome termini. Therefore, it could be hypothesized that repeats of the TTTAGGG telomeric motif were already present in interstitial locations of Anacyclus ancestors and their amplification generated long stretches of telomeric-like sequences discernible as ITR signals on a reduced evolutionary timescale. This hypothesis is consistent with some published works and our own preliminary results from related genera in which ITR signals have been detected (see above under the section on ancestral occurrence of ITRs). However, short stretches of telomeric-like regions are undetectable by FISH (Majerová et al., 2011) and yet these have been detected in interstitial chromosomal locations using DNA sequencing (Majerová et al., 2014). Therefore, the possibility that there are short stretches of ITRs, as reported by Lin and Yan (2008), in addition to the long ones cannot be excluded. Next-generation sequencing approaches can be used not only to test for the presence of short stretches of ITRs but also to shed light on the mechanisms responsible for ITRs in Anacyclus and on whether more than one of them underlie the patterns reported here.
CONCLUSION
Telomeres are one of the best conserved landmarks in eukaryotic genomes because the natural ends of linear chromosomes require structural adaptations that are essential for chromosome stability. Tandem repeats of telomeric motifs have been reported at intrachromosomal sites in unrelated organisms. The level of variation of ITRs in wild plants had not been assessed to date. However, it is assumed that the occurrence of ITRs in plants is stable and generated by unusual genomic events. Here, we report for the first time in plants high levels of ITR polymorphism in site number, chromosome distribution and differential presence of the ITR site on homologous chromosome pairs within and between populations in Anacyclus. Such hypervariability shows that, contrary to expectations, the occurrence of ITRs is a labile genomic feature in Anacyclus, and suggests that this phenomenon has been possibly overlooked in plants due to the low sample sizes used in previous studies. The high levels of polymorphism found opens new perspectives on our understanding of the evolutionary turnover of ITR sites in plants and, possibly, also on the dynamics and maintenance of the telomeric regions themselves. Finally, our results suggest caution for those studies using ITRs as markers of species’ phylogenetic relationships without a thorough sampling.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: Accessions of Anacyclus species analysed by FISH with their origins. Fig. S1: Localization of 45S rDNA sites, telomeric sites and interstitial telomeric repeat (ITR) sites. Fig. S2: Association between 45S rDNA and interstitial telomeric repeat (ITR) sites.
ACKNOWLEDGEMENTS
We thank A. Herrero for technical support, and R. Gonzalo, L. Medina, A. Quintanar and M. Santos for help in seed collection. We acknowledge the constructive criticisms and valuable comments of the two anonymous reviewers and the Handling Editor, Dr Martin Lysak, who provided insightful comments that improved the manuscript. This work was supported by funds from the Spanish Ministry of Economy and Competitiveness [Project CGL2013-49097-C2-1-P].
LITERATURE CITED
- Armstrong SJ, Franklin FCH, Jones GH. 2001. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. Journal of Cell Science 114: 4207–4217. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. 2001. Switching and signaling at the telomere. Cell 106: 661–673. [DOI] [PubMed] [Google Scholar]
- Borgen L, Leitch L, Santos‐Guerra A. 2003. Genome organization in diploid hybrid species of Argyranthemum (Asteraceae) in the Canary Islands. Botanical Journal of the Linnean Society 141: 491–501. [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. 1994. The evolutionary dynamics of the repetitive DNA of eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS. 1996. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. The Plant Journal 9: 259–272. [DOI] [PubMed] [Google Scholar]
- Cox AV, Bennett ST, Parokonny AS, Kenton A, Callimassia MA, Bennett MD. 1993. Comparison of plant telomere locations using a PCR-generated synthetic probe. Annals of Botany 72: 239–247. [Google Scholar]
- Dvořáčková M, Fojtová M, Fajkus J. 2015. Chromatin dynamics of plant telomere and ribosomal genes. The Plant Journal 83: 18–37. [DOI] [PubMed] [Google Scholar]
- Ehrendorfer F, Schweizer D, Greger H, Humphries C. 1977. Chromosome banding and synthetic systematics in Anacyclus (Asteraceae Anthemideae). Taxon 26: 387–394. [Google Scholar]
- Fajkus J, Kovařík A, Královics R, Bezděk M. 1995. Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Molecular and General Genetics 247: 633–638. [DOI] [PubMed] [Google Scholar]
- Fajkus P, Peška V, Sitová Z, et al. 2016. Allium telomeres unmasked: the unusual telomeric sequence (CTCGGTTATGGG)n is synthesized by telomerase. The Plant Journal 85: 337–347. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Brandes A, Schubert I. 1995. Telomere sequence localization and karyotype evolution in higher plants. Plant Systematics and Evolution 196: 227–241. [Google Scholar]
- Galián JA, Rosato M, Rosselló JA. 2014. Incomplete sequence homogenisation in 45S rDNA multigene families: intermixed IGS heterogeneity within the single NOR locus of the polyploid species Medicago arborea (Fabaceae). Annals of Botany 114: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspin C, Rami JF, Lescure B. 2010. Distribution of short interstitial telomere motifs in two plant genomes: putative origin and function. BMC Plant Biology 10: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Research 7: 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra M. 2008. Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenetic and Genome Research 120: 339–350. [DOI] [PubMed] [Google Scholar]
- Hanmoto H, Kataoka R, Ohmido N, Yonezawa Y. 2007. Interstitial telomere-like repeats in the Haplopappus gracilis (Asteraceae) genome revealed by fluorescence in situ hybridization. Cytologia 72: 483–488. [Google Scholar]
- He L, Liu J, Torres GA, Zhang H, Jiang J, Xie C. 2013. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Research 21: 5–13. [DOI] [PubMed] [Google Scholar]
- Huang CH, Zhang C, Liu M, et al. 2016. Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Molecular Biology and Evolution 33: 2820–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries CJ. 1979. A revision of the genus Anacyclus L. (Compositae: Anthemideae). Bulletin of the British Museum (Natural History) 7: 83–142. [Google Scholar]
- Humphries CJ. 1981. Cytogenetic and cladistic studies in Anacyclus (Compositae: Anthemidae). Nordic Journal of Botany 1: 83–96. [Google Scholar]
- Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Lin KW, Yan J. 2008. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutation Research/Reviews in Mutation Research 658: 95–110. [DOI] [PubMed] [Google Scholar]
- Liu WS, Fredga K. 1999. Telomeric (TTAGGG)n sequences are associated with nucleolus organizer regions (NORs) in the wood lemming. Chromosome Research 7: 235–240. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Mandáková T, Schranz ME. 2016. Comparative paleogenomics of crucifers: ancestral genomic blocks revisited. Current Opinion in Plant Biology 30: 108–115. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 3.04. 2015. http://mesquiteproject.org [Google Scholar]
- Majerová E, Fojtová M, Mandáková T, Fajkus J. 2011. Methylation of plant telomeric DNA: what do the results say?Plant Molecular Biology 77: 533–536. [DOI] [PubMed] [Google Scholar]
- Majerová E, Mandáková T, Vu GT, Fajkus J, Lysak MA, Fojtová M. 2014. Chromatin features of plant telomeric sequences at terminal vs. internal positions. Frontiers in Plant Science 5: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Gloss AD, Whiteman NK, Lysak MA. 2016. How diploidization turned a tetraploid into a pseudotriploid. American Journal of Botany 103: 1187–1196. [DOI] [PubMed] [Google Scholar]
- Meyne J, Baker RJ, Hobart HH, et al. 1990. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 99: 3–10. [DOI] [PubMed] [Google Scholar]
- Nelson AD, Beilstein MA, Shippen DE. 2014. Plant telomeres and telomerase; In Howell SH, ed. Molecular Biology. New York: Springer, 25–49. [Google Scholar]
- Oberprieler C. 2004. On the taxonomic status and the phylogenetic relationships of some unispecific Mediterranean genera of Compositae-Anthemideae I. Brocchia, Endopappus and Heliocauta. Willdenowia 34: 39–57. [Google Scholar]
- Oberprieler C, Himmelreich S, Källersjö M, Vallès J, Watson LE, Vogt R. 2009. Anthemideae. In: Funk V, Stuessy T, Bayer R, eds. Systematics, Evolution and Biogeography of the Compositae. Washington: IAPT, 631–666. [Google Scholar]
- Pagel M. 1999. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Systematic Biology 48: 612–622. [Google Scholar]
- Peška V, Fajkus P, Fojtová M, et al. 2015. Characterisation of an unusual telomere motif (TTTTTTAGGG)n in the plant Cestrum elegans (Solanaceae), a species with a large genome. The Plant Journal 82: 644–654. [DOI] [PubMed] [Google Scholar]
- Reeves A. 2001. MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome 44: 439–443. [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. 1988. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127–136. [DOI] [PubMed] [Google Scholar]
- Roa F, Guerra M. 2012. Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evolutionary Biology 12: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato M, Castro M, Rosselló JA. 2008. Relationships of the woody Medicago species (Section Dendrotelis) assessed by molecular cytogenetic analyses. Annals of Botany 102: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato M, Álvarez I, Nieto Feliner G, Rosselló JA. 2017. High and uneven levels of 45S rDNA site-number variation across wild populations of a diploid plant genus (Anacyclus, Asteraceae). PloS One 12: e0187131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers AO, Ludwig D. 1997. Likelihood of ancestor states in adaptive radiation. Evolution 51: 1699–1711. [DOI] [PubMed] [Google Scholar]
- Schweizer D, Ehrendorfer F. 1976. Giemsa banded karyotypes, systematics, and evolution in Anacyclus (Asteraceae-Anthemideae). Plant Systematics and Evolution 126: 107–148. [Google Scholar]
- Schweizer D, Loidl J. 1987. A model for heterochromatin dispersion and the evolution of C-band patterns. Chromosome Today 9: 61–74. [Google Scholar]
- Shibata F, Hizume M. 2011. Survey of Arabidopsis- and human-type telomere repeats in plants using fluorescence in situ hybridisation. Cytologia 76: 353–360. [Google Scholar]
- Slijepcevic P. 1998. Telomeres and mechanisms of Robertsonian fusion. Chromosoma 107: 136–140. [DOI] [PubMed] [Google Scholar]
- Sonboli A, Stroka K, Osaloo SK, Oberprieler C. 2012. Molecular phylogeny and taxonomy of Tanacetum L. (Compositae, Anthemideae) inferred from nrDNA ITS and cpDNA trnH–psbA sequence variation. Plant Systematics and Evolution 298: 431–444. [Google Scholar]
- Souza G, Vanzela AL, Crosa O, Guerra M. 2016. Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae). Genetica 144: 157–166. [DOI] [PubMed] [Google Scholar]
- Sýkorová E, Lim KY, Kunická Z, et al. 2003. Telomere variability in the monocotyledonous plant order Asparagales. Proceedings of the Royal Society of London B 270: 1893–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tek A, Jiang J. 2004. The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similar sequence. Chromosoma 113: 77–83. [DOI] [PubMed] [Google Scholar]
- Torices R, Agudo A, Álvarez I. 2013. Not only size matters: achene morphology affects time of seedling emergence in three heterocarpic species of Anacyclus (Anthemideae, Asteraceae). Anales del Jardín Botánico de Madrid 70: 48–55. [Google Scholar]
- Tran TD, Cao HX, Jovtchev G, et al. 2015. Centromere and telomere sequence alterations reflect the rapid genome evolution within the carnivorous plant genus Genlisea. The Plant Journal 84: 1087–1099. [DOI] [PubMed] [Google Scholar]
- Vitales D, Nieto Feliner G, Vallès J, Garnatje T, Firat M, Álvarez I. 2018. A new circumscription of the Mediterranean genus Anacyclus (Anthemideae, Asteraceae) based on chloroplast and nuclear DNA markers. Phytotaxa 349: 1–17. [Google Scholar]
- Weiss H., Scherthan H. 2002. Aloe spp. – plants with vertebrate-like telomeric sequences. Chromosome Research 10: 155–164. [DOI] [PubMed] [Google Scholar]
- Zhong X, Fransz PF, Wennekes-van Eden J, Zabel P, van Kammen A, de Jong HJ. 1996. High-resolution mapping on pachytene chromosomes extended DNA fibres by fluorescence in situ hybridisation. Plant Molecular Biology Reporter 14: 232–242. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.