ABSTRACT
Production and isolation of recombinant proteins are key steps in modern Molecular Biology. Expression vectors and platforms for various hosts, including both prokaryotic and eukaryotic systems, have been used. In basic plant research, Arabidopsis thaliana is the central model for which a wealth of genetic and genomic resources is available, and enormous knowledge has been accumulated over the past years – especially since elucidation of its genome in 2000. However, until recently an Arabidopsis platform had been lacking for preparative-scale production of homologous recombinant proteins. We recently established an Arabidopsis-based super-expression system, and used it for a structural pilot study of a multi-subunit integral membrane protein complex. This review summarizes the benefits and further potential of the model plant system for protein productions.
Abbreviations: Nb, Nicotiana benthamiana; OT, oligosaccharyltransferase
Keywords: Arabidopsis, recombinant proteins, protein N-glycosylation, complex-type N-glycans, Endoplasmic Reticulum, Golgi apparatus, plasma membrane, trans-Golgi network
Recombinant proteins enable the study of structure and function of proteins from virtually any origin in a common platform. Typically, a cDNA of the gene of interest is expressed in a heterologous host for overexpression, and the purified recombinant protein is used for a series of biochemical characterizations. For decades, this routine has been successfully used for identifying peptides responsible for the enzymatic activities in (often partially) purified fractions from the native host. This also allowed for in-depth analysis of enzymes/proteins identified by reverse genetics, whose abundance could be extremely low in the native host.
A range of host organisms are available for recombinant protein production. Most popular is the gram-negative bacterium Escherichia coli (E. coli), but other bacteria such as Bacillus subtilis and eukaryotic hosts are also used. The latter includes yeasts, like Saccharomyces cerevisiae and Pichia pastoris, insect cells (Spodoptera frugiperda, Sf), mammalian cells (chinese hamster ovary, CHO), and plant-cell systems (tobacco bright yellow 2, BY2).1–12 While results vary according to the nature of proteins and expression hosts, the yield of recombinant proteins can reach as much as 50 mg/L of E. coli culture under standard laboratory conditions.
E. coli is the preferred host for production of recombinant proteins of plant origin as well. However, expression of plant proteins in E. coli still poses a fair challenge with respect to expression level, protein folding, and post-translational modifications. In particular, if a recombinant protein needs to be glycosylated or otherwise post-translationally modified, eukaryotic hosts have been used.8 In plant research, transient and stable expression in the wild tobacco species Nicotiana benthamiana (Nb) has proven successful for functional analyses of recombinant proteins in many studies.13,14 However, use of Nb in basic research is mostly limited to rather small analytical experiments, such as in situ-observation of fluorescent proteins or co-precipitation assays to prove protein-protein interaction. By contrast, the biopharmaceutical industry adapted the Nb system for large-scale production of clinical proteins, such as vaccines and antibodies.15 Possible drawbacks of the Nb system for routine preparative-scale applications are the requirement for sizable growth facilities15 and a special set-up for mass-infiltration of Agrobacteria into leaf tissues.16
Arabidopsis thaliana is a widely used model for developmental and molecular plant biology.17 Availability of a collection of indexed/characterized mutants and sequenced genomes of different Arabidopsis accessions promoted the generation of a large body of knowledge on the molecular mechanisms governing growth, development and stress responses. Well-studied are also the processes governing gene expression, post-translational modifications, and protein targeting in this organism. On the other hand, our current understanding of Arabidopsis biology is rather genetics-centered, and understanding the biochemical basis of cellular machineries can become a challenge. This is partially due to the lack of reliable protein production in the homologous host, where post-translational modifications and complex formation with endogenous interaction partners is possible. We recently reported an Arabidopsis-based recombinant protein production platform (super-expression system) suitable for biochemical and structural studies (Figure 1).18 This platform yielded as much as 0.4 mg of purified protein per gram fresh weight (FW) using a heterologous model protein (mCherry) for expression and purification. The benefit as homologous overexpression system was demonstrated for oligosaccharyltransferase (OT), an integral-membrane protein complex consisting of seven or moresubunits that could be assembled on a single overexpressed central STT3a subunit. The OT-ribosome super-complex was then purified and its three-dimensional structure determined by transmission electron-microscopy at 30Å resolution.18 Combined with our pilot study applying this technology to a soluble enzyme (CTD phosphatase-like 4),19 which was difficult to express in E. coli as an active form, we propose the Arabidopsis super-expression system as a versatile recombinant protein production platform.
Figure 1.
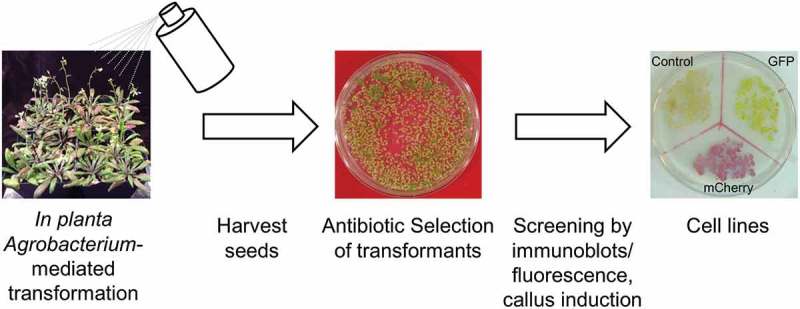
Workflow of establishing Arabidopsis cell lines for recombinant protein expression. The Arabidopsis thaliana rdr6-11 host is transformed by Agrobacterium-mediated floral transformation. Seeds are harvested and primary transformants screened under proper selection. Resistant plants will be further tested, e.g. by immunoblotting using a specific antibody raised against the transgene product (typically using a single fully-expanded cotyledon), or by fluorescence if the target protein is fused to a fluorescent protein. Ten to twenty seedlings with high transgene expression are sliced into small pieces and placed on callus induction medium. Induced calli are transferred to fresh medium every week (established cell lines).
Although unexplored, Arabidopsis provides various advantages as protein expression platform, particularly for Arabidopsis research. First of all, Agrobacterium-mediated floral transformation is very easy and widely used in standard laboratory set-ups.20 Each transformed seed produced with this protocol represents an independent transformation event.21 Hence, individual transformants are physically separated from each other, and those with high level expression do not suffer competitive growth disadvantages compared with tissue-culture-based transformation protocols. Once identified, high expressors can be maintained in a petri dish-based cell culture system at ambient temperature (25°C) in darkness, and do not require large growth space or sophisticated incubators. Established cell lines typically double their mass in one week, and 20–30 g can be harvested for laboratory-scale experiments. Second, unlike in heterologous systems, Arabidopsis proteins produced in Arabidopsis cells will undergo proper post-translational modifications and associate with their native partner(s) to form active complexes. It should be noted, that the above advantages could be a drawback in certain cases. Proteins involved in the Arabidopsis programmed cell death (PCD) pathway may induce PCD in Arabidopsis, and proteins whose expression levels are tightly regulated may not accumulate to sufficiently high levels. However, availability of many genotypes is another merit of Arabidopsis. Our current system uses rdr6-11 as the standard host to avoid gene silencing (similar to P19 co-expression in the Nb system),18,22 but one can use various genotypes or mutant backgrounds to improve yield and activity of recombinant proteins, or study the role of a particular post-translational modification.
Beyond production of plant proteins, there is also potential for the production of enzymes/proteins for pharmaceutical use. More than 60% of them are secreted glycoproteins, including monoclonal antibodies (mAbs, about half of the targets), clotting factors and cytokines, like erythropoietin (EPO).23 Moreover, recent epidemics of the flu or Ebola, and efforts to prepare vaccines to fight them, further highlight the potential of plant-based production systems for pharmaceutical proteins.24-29 While the Arabidopsis system is not yet suitable for fast, large-scale production in response to a new epidemic, it will be suitable for known targets by low-cost, stable production of proteins, especially for patients who require long-term treatments.30,31 Unlike the Nb leaf-infiltration system, established Arabidopsis cultures are bacteria-free, and the purified protein products will contain no endotoxins from bacterial contaminations, reducing requirements for purification and testing. For this application to be successful, the well-known issue of plant-specific N-glycan modifications should be addressed by humanization of host plant N-glycans.32 We and others have prepared lines containing various mutations in the plant N-glycan modification pathway that eliminate/reduce immunogenic potential.33-38 We have introgressed selected mutant alleles/combinations into the rdr6-11 background, of which rdr6-11 cgl1-3 and rdr6-11 fucTa fucTb xylT, will serve as models for future efforts in this direction.
A further advantage of Arabidopsis as expression host is the potential for customizing the host for the production of a protein of interest. While the rdr6-11 background may be satisfactory to overcome transgene-silencing in many cases,22 plant cells harbor multiple mechanisms that can limit recombinant protein yields at the level of translation, post-translational processing/targeting, and turnover.39 Since these mechanisms monitor endogenous protein levels, they will also do so upon over-accumulation of foreign proteins (e.g. trigger the unfolded protein response/ER-associated degradation).40–44 When a protein of interest should not accumulate to satisfactorily high levels, one could apply various genetic manipulations, including chemical mutagenesis, activation tagging,45 and targeted/untargeted overexpression (for example, FOX hunting)46 to the transgenic line of interest and screen for individuals with high expression levels, like used in bacterial engineering.47 The factors identified by such approaches can also be incorporated into other large-scale plant expression systems, via CRISPR/Cas9-targeted mutagenesis or overexpression of endogenous factors.48,49
Overall, the new super-expression system complements Arabidopsis as excellent genetic model, by combining the benefit of homologous protein production with that of well-established genetic resources/manipulations, for improved production of recombinant proteins.
Funding Statement
This work was in part financially supported by Deutsche Forschungs-gemeinschaft (DFG) grant SCHA 541/11 (to A.v.S.) and by the National Science Foundation (IOS1547551) (to H.K.).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Wurm F, Bernard A.. Large-scale transient expression in mammalian cells for recombinant protein production. Curr Opin Biotechnol. 1999;10156–159. [DOI] [PubMed] [Google Scholar]
- 2.Hacker DL, Balasubramanian S.. Recombinant protein production from stable mammalian cell lines and pools. Curr Opin Struct Biol. 2016;38129–136. doi: 10.1016/j.sbi.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Overton TW. Recombinant protein production in bacterial hosts. Drug Discov Today. 2014;19590–601. doi: 10.1016/j.drudis.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Trowitzsch S, Bieniossek C, Nie Y, Garzoni F, Berger I. New baculovirus expression tools for recombinant protein complex production. J Struct Biol. 2010;17245–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Mattanovich D, Branduardi P, Dato L, Gasser B, Sauer M, Porro D. Recombinant protein production in yeasts. Methods Mol Biol. 2012;824329–358. doi: 10.1007/978-1-61779-433-9_17. [DOI] [PubMed] [Google Scholar]
- 6.Niimi T. Recombinant protein production in the eukaryotic protozoan parasite leishmania tarentolae: a review. Methods Mol Biol. 2012;824307–315.doi: 10.1007/978-1-61779-433-9_15. [DOI] [PubMed] [Google Scholar]
- 7.Cregg JM, Cereghino JL, Shi J, Higgins DR. Recombinant protein expression in pichia pastoris. Mol Biotechnol. 2000;1623–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- 8.Reitz C, Li J, Neubauer P. Recombinant protein production: a comparative view on host physiology (Laupheim, Germany, March 2013). N Biotechnol. 2013;30405–409. doi: 10.1016/j.nbt.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Assenberg R, Wan PT, Geisse S, Mayr LM. Advances in recombinant protein expression for use in pharmaceutical research. Curr Opin Struct Biol. 2013;23393–402. doi: 10.1016/j.sbi.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Cox MM. Recombinant protein vaccines produced in insect cells. Vaccine. 2012;301759–1766. doi: 10.1016/j.vaccine.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarroll L, La K. Stable insect cell cultures for recombinant protein production. Curr Opin Biotechnol. 1997;8590–594. [DOI] [PubMed] [Google Scholar]
- 12.Reuter LJ, Bailey MJ, Joensuu JJ, Ritala A. Scale-up of hydrophobin-assisted recombinant protein production in tobacco BY-2 suspension cells. Plant Biotechnol J. 2014;12402–410. doi: 10.1111/pbi.12147. [DOI] [PubMed] [Google Scholar]
- 13.Wydro M, Kozubek E, Lehmann P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of nicotiana benthamiana. Acta Biochim Pol. 2006;53289–298. [PubMed] [Google Scholar]
- 14.Wagner B, Fuchs H, Adhami F, Ma Y, Scheiner O, Breiteneder H. Plant virus expression systems for transient production of recombinant allergens in Nicotiana benthamiana. Methods. 2004;32227–234. doi: 10.1016/j.ymeth.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Holtz BR, Berquist BR, Bennett LD, Kommineni VJM, Munigunti RK, White EL, Wilkerson DC, Wong KYI, Ly LH, Marcel S. Commercial-scale biotherapeutics manufacturing facility for plant-made pharmaceuticals. Plant Biotechnol J. 2015;131180–1190. doi: 10.1111/pbi.12469. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q, Lai H. Gene delivery into plant cells for recombinant protein production. Biomed Res Int. 2015;2015932161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282(662):79–82. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- 18.Jeong IS, Lee S, Bonkhofer F, Tolley J, Fukudome A, Nagashima Y, May K, Rips S, Lee SY, Gallois P, et al. Purification and characterization of Arabidopsis thaliana oligosaccharyltransferase complexes from the native host: a protein super-expression system for structural studies. Plant J. 2018; doi: 10.1111/tpj.13847. [DOI] [PubMed] [Google Scholar]
- 19.Fukudome A, Aksoy E, Wu X, Kumar K, Jeong IS, May K, Russell WK, Koiwa H. Arabidopsis CPL4 is an essential C-terminal domain phosphatase that suppresses xenobiotic stress responses. Plant J. 2014;8027–39. doi: 10.1111/tpj.12612. [DOI] [PubMed] [Google Scholar]
- 20.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16735–743. [DOI] [PubMed] [Google Scholar]
- 21.Defeauz C, Clough SJ, Bent AF. Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral dip. Plant Physiol. 2000;123895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butaye KM, Goderis IJ, Wouters PF, Pues JM, Delaure SL, Broekaert WF, Depicker A, Cammue BP, De Bolle MF. Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 2004;39440–449. doi: 10.1111/j.1365-313X.2004.02144.x. [DOI] [PubMed] [Google Scholar]
- 23.Planinc A, Bones J, Dejaegher B, Van Antwerpen P, Delporte C. Glycan characterization of biopharmaceuticals: updates and perspectives. Anal Chim Acta. 2016;92113–27. doi: 10.1016/j.aca.2016.03.049. [DOI] [PubMed] [Google Scholar]
- 24.Olinger GG Jr., Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci U S A. 2012;10918030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeitlin L, Pettitt J, Scully C, Bohorova N, Kim D, Pauly M, Hiatt A, Ngo L, Steinkellner H, Whaley KJ, et al. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc Natl Acad Sci U S A. 2011;10820690–20694. doi: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castilho A, Bohorova N, Grass J, Bohorov O, Zeitlin L, Whaley K, Altmann F, Steinkellner H. Rapid high yield production of different glycoforms of Ebola virus monoclonal antibody. PLoS One. 2011;6e26040. doi: 10.1371/journal.pone.0026040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai H, He J, Engle M, Diamond MS, Chen Q. Robust production of virus-like particles and monoclonal antibodies with geminiviral replicon vectors in lettuce. Plant Biotechnol J. 2012;1095–104. doi: 10.1111/j.1467-7652.2011.00649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phoolcharoen W, Bhoo SH, Lai H, Ma J, Arntzen CJ, Chen Q, Mason HS. Expression of an immunogenic Ebola immune complex in Nicotiana benthamiana. Plant Biotechnol J. 2011;9807–816. doi: 10.1111/j.1467-7652.2011.00593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Z, Phoolcharoen W, Lai H, Piensook K, Cardineau G, Zeitlin L, Whaley KJ, Arntzen CJ, Mason HS, Chen Q. High-level rapid production of full-size monoclonal antibodies in plants by a single-vector DNA replicon system. Biotechnol Bioeng. 2010;1069–17. doi: 10.1002/bit.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosales-Mendoza S, Tello-Olea MA. Carrot cells: a pioneering platform for biopharmaceuticals production. Mol Biotechnol. 2015;57219–232. doi: 10.1007/s12033-014-9837-y. [DOI] [PubMed] [Google Scholar]
- 31.Shaaltiel Y, Bartfeld D, Hashmueli S, Baum G, Brill-Almon E, Galili G, Dym O, Boldin-Adamsky SA, Silman I, Sussman JL, et al. Production of glucocerebrosidase with terminal mannose glycans for enzyme replacement therapy of Gaucher’s disease using a plant cell system. Plant Biotechnol J. 2007;5579–590. doi: 10.1111/j.1467-7652.2007.00263.x. [DOI] [PubMed] [Google Scholar]
- 32.Strasser R. Biological significance of complex N-glycans in plants and their impact on plant physiology. Front Plant Sci. 2014;5363. doi: 10.3389/fpls.2014.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank J, Kaulfurst-Soboll H, Rips S, Koiwa H, von Schaewen A. Comparative analyses of Arabidopsis cgl1 (complex glycan 1) mutants and genetic interaction with stt3a (staurosporin & temperature-sensitive 3a). Plant Physiol. 2008;1481354–1367. doi: 10.1104/pp.108.127027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kajiura H, Koiwa H, Nakazawa Y, Okazawa A, Kobayashi A, Seki T, Fujiyama K. Two Arabidopsis thaliana Golgi alpha-mannosidase I enzymes are responsible for plant N-glycan maturation. Glycobiology. 2010;20235–247. doi: 10.1093/glycob/cwp170. [DOI] [PubMed] [Google Scholar]
- 35.Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA. 2008;1055933–5938. doi: 10.1073/pnas.0800237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rips S, Bentley N, Jeong IS, Welch JL, von Schaewen A, Multiple KH. N-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell. 2014;263792–3808. doi: 10.1105/tpc.114.129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strasser R, Bondili JS, Vavra U, Schoberer J, Svoboda B, Glossl J, Leonard R, Stadlmann J, Altmann F, Steinkellner H, et al. A unique {beta}1,3-galactosyltransferase is indispensable for the biosynthesis of n-glycans containing lewis a structures in Arabidopsis thaliana. Plant Cell. 2007;192278–2292. doi: 10.1105/tpc.107.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strasser R, Altmann F, Mach L, Glossl J, Steinkellner H. Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett. 2004;561132–136. doi: 10.1016/S0014-5793(04)00150-4. [DOI] [PubMed] [Google Scholar]
- 39.Nelson CJ, Millar AH. Protein turnover in plant biology. Nat Plants. 2015;115017. doi: 10.1038/nplants.2015.176. [DOI] [PubMed] [Google Scholar]
- 40.De Ruijter JC, Frey AD. Analysis of antibody production in Saccharomyces cerevisiae: effects of ER protein quality control disruption. Appl Microbiol Biotechnol. 2015;999061–9071. doi: 10.1007/s00253-015-6807-7. [DOI] [PubMed] [Google Scholar]
- 41.Heimel K. Unfolded protein response in filamentous fungi-implications in biotechnology. Appl Microbiol Biotechnol. 2015;99121–132. doi: 10.1007/s00253-014-6192-7. [DOI] [PubMed] [Google Scholar]
- 42.Prashad K, Mehra S. Dynamics of unfolded protein response in recombinant CHO cells. Cytotechnology. 2015;67237–254. doi: 10.1007/s10616-013-9678-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hesketh AR, Castrillo JI, Sawyer T, Archer DB, Oliver SG. Investigating the physiological response of Pichia (Komagataella) pastoris GS115 to the heterologous expression of misfolded proteins using chemostat cultures. Appl Microbiol Biotechnol. 2013;979747–9762. doi: 10.1007/s00253-013-5186-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lajoie P, Snapp EL. Changes in BiP availability reveal hypersensitivity to acute endoplasmic reticulum stress in cells expressing mutant huntingtin. J Cell Sci. 2011;1243332–3343. doi: 10.1242/jcs.087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigel D, Ahn J-H. Activation tagging in Arabidopsis. Plant Physiol. 2000;1221003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, Tsuhara Y, Suzuki K, Ishikawa A, Seki M, et al. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 2006;48974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- 47.Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact. 2011;1032. doi: 10.1186/1475-2859-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grav LM, La Cour Karottki KJ, Lee JS, Kildegaard HF. Application of CRISPR/Cas9 genome editing to improve recombinant protein production in cho cells. Methods Mol Biol 2017; 1603:101–18. [DOI] [PubMed] [Google Scholar]
- 49.Ma X, Zhu Q, Chen Y, Liu Y-G. CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant. 2016;9961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]


