ABSTRACT
The DNA damage response system (DDR) is crucial in addressing DNA double-strand breaks (DSBs), which pose a severe threat to genomic integrity. The SOG1 transcription factor is a master regulator of the Arabidopsis thaliana DDR. We previously showed that hyperphosphorylation of five Ser-Gln sites of SOG1 is the molecular switch to activate the DDR. In this study, we determined that SOG1 is hyperphosphorylated within 20 minutes following DSB-inducing treatment, followed by activation of several SOG1 target genes. Using SOG1 phosphorylation mutants, we demonstrated that although the hyperphosphorylation sites remain unchanged over time, the amount of hyperphosphorylation gradually increases. These observations suggest that rapid SOG1 hyperphosphorylation is limited by the amount of active kinases.
Abbreviations: SOG1, suppressor of gamma response; ATM, Ataxia telangiectasia mutated; ATR, ATM and Rad3-related
Keywords: DNA damage response, hyperphosphorylation, SOG1, A. thaliana
Introduction
A sophisticated network of DNA damage response (DDR) systems has evolved to address the fundamental problem of genomic erosion.1 The DDR of Arabidopsis thaliana involves DNA repair, cell-cycle arrest, endoreduplication, and programmed cell death.24 Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) kinases are the central regulators of this network.57ATM and ATR are activated by DNA double-strand breaks (DSBs) and DNA replication stress, respectively, resulting in the phosphorylation of hundreds of target proteins. These phosphorylation events are crucial for activating downstream pathways.
The SOG1 transcription factor is also critical for regulating an appropriate DDR.2 This transcription factor was originally isolated as a suppressor mutant of IR-induced cell cycle arrest of A. thaliana xpf-2 mutants, which lack functional XPF repair endonuclease.8 SOG1 is one of the NAC (NAM, ATAF1/2, and CUC2) proteins, which constitute one of the largest families of plant-specific transcription factors.2 SOG1 is the first identified plant-specific transcription factor involved in the DDR pathway. SOG1 protein is observed in meristematic tissues, such as the shoot and root apical meristem, lateral root primordium, root stele and young flowers.9,10 This observation is consistent with the fact that the DDR is important in actively dividing cells.
DNA damage in A. thaliana induces rapid and robust change in the transcriptional regulation of numerous genes.2,7 These changes in gene expression activate DNA repair, cell-cycle arrest, endoreduplication, and programmed cell death. Mutation of SOG1 causes various defects in the activation of these responses, indicating that SOG1 is a master regulator of the DDR. DSB-inducing treatment promotes hyperphosphorylation of SOG1, which is required to activate the DDR.
SOG1 contains three domains: the N-terminal extension, the central NAC domain, and the C-terminal transcription regulatory domain (Figure 1A). The C-terminus of SOG1 has five SQ (serine-glutamine) sites (350SQ, 356SQ, 372SQ, 430SQ, 436SQ), which are known as preferred sites for phosphorylation by ATM and ATR (Figure 1A). We previously showed that these five SQ sites are hyperphosphorylated in response to DSBs.9 Hyperphosphorylation of SOG1 induced by DSBs appears to be dependent on ATM, as it does not occur in ATM mutants.9 This suggests that SOG1 functions downstream of ATM and that ATM-dependent hyperphosphorylation of SQ motifs is essential for SOG1 functions.
Figure 1.

Time-dependent phosphorylation pattern of SOG1 and the activation of downstream genes.
We recently demonstrated that increased phosphorylation of SQ sites strengthens the DDR, as shown through mutation of SQ motifs (SQ to AQ) and study of SOG1 mutant lines with differing numbers of SQ phosphorylation sites.11 Our data also suggest that there is no relationship between the amount of DNA damage and the number of SOG1 hyperphosphorylation sites.11
Results
The function of SOG1 as a master regulator of DDR suggests that rapid SOG1 activation should occur in response to DNA damage. To examine this possibility, immunoblotting was used to observe SOG1 hyperphosphorylation over time after treatment with zeocin, a DSBs-inducing agent. We used A. thaliana sog1–1 transgenic plants expressing ProSOG1:SOG1(5SQ)-Myc (5SQ line), in which the promoter and coding regions of SOG1 are fused in-frame to a 10 × Myc tag. As we previously reported, independently of DNA damage, an anti-Myc antibody detected a band for nonphosphorylated SOG1-Myc (band a) and a slower migrating band for phosphorylated SOG1-Myc (band b), with both bands consistently observed (Figure 1B). We found that hyperphosphorylated SOG1-Myc (band c), which is DNA damage dependent, was visible at 20 min after zeocin treatment in SOG1-5SQ, with band intensity incrementally increasing in a time-dependent manner (20 min–60 min) (Figure 1B). This observation indicates that SOG1 hyperphosphorylation occurs rapidly and that the phosphorylation level is incrementally induced over time. This rapid phosphorylation is consistent with the function of SOG1, as an upstream regulator of DDR.
We then investigated whether downstream target genes of SOG1 are also rapidly activated following DNA damage. Therefore, we evaluated the expression of BRCA1 and RAD51, which are DNA repair genes and direct targets of SOG112. 5SQ seedlings were treated with 1 mM zeocin, and total RNA was extracted from root tips at several time points (0, 10, 20, 30, and 60 min). BRCA1 and RAD51 were faintly expressed at 0 min, the induction of both genes was observed at 30 min, and the induction became more pronounced at 60 min (Figure 1C). There was a time lag between SOG1 hyperphosphorylation and the activation of downstream genes. These results indicate that following DSB induction, SOG1 is hyperphosphorylated within 20 minutes and SOG1 target genes are induced within 30 minutes.
We next examined whether the number of SOG1 hyperphosphorylation sites changes over time following DNA damage exposure. We hypothesized that few SQ sites would be phosphorylated, immediately following DNA damage, with the number of phosphorylated SQ sites gradually increasing over time. The hyperphosphorylation patterns of various mutant strains [ProSOG1:SOG1(1SQ-4SQ)-Myc] (1SQ: 356SQ, 2SQ: 350SQ, 356SQ, 3SQ: 350AQ, 356AQ, 436AQ, 4SQ: 350SQ, 356SQ, 430SQ, 436SQ) were evaluated at different time points (Figure 2). Although hyperphosphorylation levels gradually increased over time for all mutant strains, there was no difference in the band pattern among phosphorylation mutants (Figure 2). These data suggest that there is no relationship between the time that has elapsed following DNA damage exposure and the number of SOG1 hyperphosphorylation sites.
Figure 2.
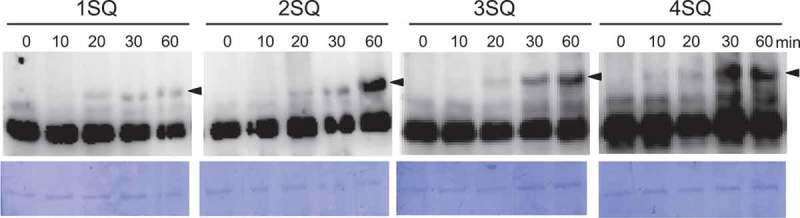
Time-dependent phosphorylation pattern of SOG1 phosphorylation mutants.
Discussion
The integrity of plant chromosomes is under constant assault from a variety of DNA damaging factors. DSBs are one of the most cytotoxic types of DNA damage, and prompt repair is essential. In this study, we demonstrated that SOG1 hyperphosphorylation is induced within 20 minutes following zeocin treatment, indicating that DDR mediated by SOG1 is rapidly activated by DSBs. Furthermore, we showed that there is a time lag of approximately 10 min after phosphorylation before the induction of several SOG1 target genes can be observed.
Our prior data show that SOG1 hyperphoshorylation (band c) increases with the amount of DNA damage9; here, we demonstrated that the intensity of hyperphosphorylated bands for SOG1 mutants gradually increases over time. Furthermore, it was shown that SQ sites seem to be equally phosphorylated, as the change in phosphorylation pattern is similar among mutants. These results indicate that hyperphosphorylation at SQ sites is not dependent on the amount of DNA damage or the time since this damage occurred; however, the hyperphosphorylation frequency at SQ sites seems to be dependent on these two factors. We previously showed that the phosphorylation of all five SQ sites is required for the full activation of SOG111. 356SQ is the first phosphorylation sites, and this phosphorylation triggers the phosphorylation of other SQ sites11. However, we were not able to identify a phosphorylation order in this study, perhaps because the phosphorylation events occur quite rapidly.
In mammalian cells, ATM has been shown to exist as inactive dimers in undamaged cells that rapidly undergo autophosphorylation after exposure to DNA damage-inducing agents and dissociate into active monomers.13 Because SQ motifs are target amino acids of ATM, active ATM may equally phosphorylate each SQ motif of SOG1. Therefore, the amount of hyperphosphorylation of SOG1 may be limited by the amount of active ATM, which is regulated by the amount of DNA damage and time. Further research is needed to determine how the 356SQ site undergoes phosphorylation first. As protein phosphorylation can directly affect distinct aspects of transcription factor function by regulating protein-protein interactions and DNA binding,14 it will be important to determine whether SOG1 hyperphosphorylation modifies target DNA directly or affects interacting factors. Future studies are needed to fully comprehend the role of SOG1 hyperphosphorylation in the DNA damage response.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana plants used in this study were grown on MS media [1 × MS salts including vitamins, 2% (w/v) sucrose, pH 6.0, 0.8% (w/v) gellan gum agar for solid medium] under continuous light conditions at 22°C. The accession Columbia (Col-0) was used as the wild-type strain, the sog1-1 line and transgenic SOG1 phosphorylation mutant lines have been described previously2,11.
Immunoblotting
Five-day-old seedlings were transferred to MS liquid medium containing 0 or 1 mM zeocin. After a 1 h incubation, a pool of root tips (approximately 100 seedlings) was excised and ground in the following buffer: 10 mM Tris (pH 7.6), 150 mM NaCl, 2 mM EDTA, 0.5% (v/v) Nonidet P-40 (Nacalai Tesque), 1 mM DTT, a protease inhibitor cocktail (Sigma-Aldrich), and a phosphatase inhibitor cocktail (0.1 mM Na3VO4, 1 mM NaF, 60 mM β-glycerophosphatase, and 20 mM p-nitrophenylphosphate). The slurry was centrifuged twice to remove debris, and the supernatant was recovered and used for subsequent analysis. Proteins (1 μg) were separated using an 8% SDS-PAGE gel containing 30 μM Phos-tag and 30 μM MnCl2. The Phos-tag reagent (NARD Institute) was used for identification of phosphorylated SOG1 protein. Phosphorylated SOG1 is visualized as bands that migrate slower than those of non-phosphorylated proteins. After electrophoresis, the proteins were electroblotted to a polyvinylidene difluoride (PVDF) membrane (Merck Millipore) in the following buffer: 6.3 mM NaHCO3, 4.3 mM Na2CO3, (pH 9.5), and 20% methanol. Because SOG1-Myc can be detected using an anti-Myc antibody, the membrane was incubated for 2 h at room temperature in the anti-Myc primary antibody A-14 (1:2000 dilution, Santa Cruz Biotechnology), rinsed 3 times with 1 × TBST, and incubated with an anti-rabbit immunoglobulin horseradish peroxidase-conjugated secondary antibody (1:4000, Promega) to detect SOG1-Myc. Next the membrane was washed 3 times with 1 × TBST and processed with a LAS-4000 luminescent image analyzer (Fujifilm) after incubation with the ECL Prime enhanced chemiluminescence kit (GE Healthcare).
Semi-quantitative RT-PCR analysis
Five-day-old seedlings were transferred to MS liquid medium containing 0 or 1000 μM zeocin. After 0, 10, 20, 30, and 60 min incubation, total RNA was extracted from a pool of root tips containing approximately 100 seedlings using an RNeasy Plant Mini Kit (Qiagen) following to the manufacturer’s instructions. All samples were treated with DNase I on a column using the Qiagen RNase-Free DNase Set (Qiagen) and quantified. To produce cDNA for qRT-PCR, 0.3 μg of total RNA was reverse-transcribed, using Prime Script RT reagent kit (TAKARA), according to the manufacturer’s protocol. The expression level relative to that measured at 0 min was calculated using the value of the relative RNA levels normalized to the internal control ELF4A-1 (eukaryotic translation initiation factor 4A-1, At3g13920). The following primer sets were used: ELF4A1 control primers (elf4A1 and elf4A5); BRCA1 primers (brca1F2 and brca1rtR2); and RAD51 primers (rad51AF1 and rad51ArtR1), which are found in Supplemental Table 1.
Structural features of SOG1 and phosphorylated mutant SOG1. The N-terminal extension, NAC domain, and transcription regulatory domain are shown. The five Ser-Gln (SQ) motifs are represented by dark gray boxes, and the mutated motifs (Ala-Gln) are represented by light gray boxes. (B) Detection of the phosphorylated form of SOG1. sog1-1 lines harboring ProSOG1:SOG1 [5SQ]-Myc were used. Five-day-old seedlings grown on MS plates were transferred to liquid medium containing 1 mM zeocin, and total protein from root tips was extracted 0, 10, 20, 30, and 60 min later. Phosphorylated forms of SOG1 were detected using an SDS-PAGE gel containing Phos-tag. Coomassie blue staining is shown below. Nonphosphorylated and phosphorylated SOG1-Myc (bands a and b) are indicated by white arrow heads, and hyperphosphorylated SOG1-Myc (bands c) is indicated by black arrowheads. (C) Semiquantative RT-PCR analysis of BRCA1 and RAD51. sog1-1 lines harboring ProSOG1:SOG1 [5SQ]-Myc were used. Five-day-old seedlings grown on MS plates were transferred to liquid medium containing 1 mM zeocin, and total RNA was extracted from root tips 0, 10, 20, 30, and 60 min later. Using the total RNA, cDNA was prepared and RT-PCR was performed. The RT-PCR product of eIF4A (eukaryotic initiation factor) was employed as a standard for RT-PCR amplification. The number below each band denotes its relative expression level (first normalized to elF4A) compared to the sample at 0 min. Average from two biological replicates was shown.
sog1-1 lines harboring ProSOG1:SOG1 [1SQ (356SQ), 2SQ, 3SQ, and 4SQ]-Myc were used. Hyperphosphorylated SOG1-Myc (bands c) is indicated by black arrowheads. Coomassie blue staining is shown below. This experiment was conducted similarly to the ones shown in Figure 1B.
Funding Statement
This work was supported by JSPS KAKENHI (13J40017 and 17K07455 to K.O.Y., 16H01472, 16K07408, 18H04787 and 18H04844 to S. K.) and by the MEXT Supported Program for the Strategic Research Foundation at Private Universities from the Ministry of Education, Culture, Sports, Science & Technology of Japan, Grant Number S1511023 to S.K.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Ciccia A, Elledge SJ.. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshiyama K, Conklin PA, Huefner ND, Britt AB.. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci U S A. 2009;106:12843–12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:10004–10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furukawa T, Curtis MJ, Tominey CM, Duong YH, Wilcox BW, Aggoune D, Hays JB, Britt AB. A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst). 2010;9:940–948. [DOI] [PubMed] [Google Scholar]
- 5.Culligan K, Tissier A, Britt A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell. 2004;16:1091–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell. 2003;15:119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. [DOI] [PubMed] [Google Scholar]
- 8.Preuss SB, Britt AB. A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics. 2003;164:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013;14:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshiyama KO. SOG1: a master regulator of the DNA damage response in plants. Genes Genet Syst. 2016;90:209–216. [DOI] [PubMed] [Google Scholar]
- 11.Yoshiyama KO, Kaminoyama K, Sakamoto T, Kimura S. Increased phosphorylation of Ser-Gln sites on SUPPRESSOR OF GAMMA RESPONSE1 strengthens the DNA damage response in arabidopsis thaliana. Plant Cell. 2017;29:3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogita N, Okushima Y, Tokizawa M, Yamamoto YY, Tanaka M, Seki M, Makita Y, Matsui M, Okamoto-Yoshiyama K, Sakamoto T, et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018;94:439–453 [DOI] [PubMed] [Google Scholar]
- 13.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. [DOI] [PubMed] [Google Scholar]
- 14.Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. [DOI] [PubMed] [Google Scholar]


