Abstract
Purpose
On‐board MRI can provide superb soft tissue contrast for improving liver SBRT localization. However, the availability of on‐board MRI in clinics is extremely limited. On the contrary, on‐board kV imaging systems are widely available on radiotherapy machines, but its capability to localize tumors in soft tissue is limited due to its poor soft tissue contrast. This study aims to explore the feasibility of using an on‐board kV imaging system and patient prior knowledge to generate on‐board four‐dimensional (4D)‐MRI for target localization in liver SBRT.
Methods
Prior 4D MRI volumes were separated into end of expiration (EOE) phase (MRI prior) and all other phases. MRI prior was used to generate a synthetic CT at EOE phase (sCT prior). On‐board 4D MRI at each respiratory phase was considered a deformation of MRI prior. The deformation field map (DFM) was estimated by matching DRRs of the deformed sCT prior to on‐board kV projections using a motion modeling and free‐form deformation optimization algorithm. The on‐board 4D MRI method was evaluated using both XCAT simulation and real patient data. The accuracy of the estimated on‐board 4D MRI was quantitatively evaluated using Volume Percent Difference (VPD), Volume Dice Coefficient (VDC), and Center of Mass Shift (COMS). Effects of scan angle and number of projections were also evaluated.
Results
In the XCAT study, VPD/VDC/COMS among all XCAT scenarios were 10.16 ± 1.31%/0.95 ± 0.01/0.88 ± 0.15 mm using orthogonal‐view 30° scan angles with 102 projections. The on‐board 4D MRI method was robust against the various scan angles and projection numbers evaluated. In the patient study, estimated on‐board 4D MRI was generated successfully when compared to the “reference on‐board 4D MRI” for the liver patient case.
Conclusions
A method was developed to generate on‐board 4D MRI using prior 4D MRI and on‐board limited kV projections. Preliminary results demonstrated the potential for MRI‐based image guidance for liver SBRT using only a kV imaging system on a conventional LINAC.
Keywords: 4D MRI, deformable image registration, liver SBRT, MR guided radiotherapy, on‐board imaging, prior knowledge
1. Introduction
Reducing target localization errors for radiotherapy treatments can improve tumor control and reduce normal tissue toxicity.1, 2 This is especially important for liver stereotactic body radiation therapy (SBRT) treatments since (a) respiratory motion can cause treatment errors3 and (b) SBRT is more prone to significant dosimetric errors due to high dose per fraction delivery over a longer temporal duration. Therefore, on‐board 4D verification of the liver tumor before and during the SBRT treatment is critical.4, 5, 6, 7
Conventionally, cone‐beam CT (CBCT) has been used for on‐board target localization in liver SBRT. However, its localization accuracy is limited due to the poor soft tissue contrast and limited image quality when motion is involved.8, 9, 10 On‐board MRI has recently been introduced in radiation therapy for onboard target localization.11, 12, 13 Compared to CBCT, MRI has no ionizing radiation dose and much better soft tissue contrast, which can significantly improve the localization accuracy of liver SBRT. However, MRI‐radiotherapy machines are only available in a very limited number of radiation therapy facilities due to the high cost, which severely limits the usage of MR for radiotherapy guidance. On the contrary, conventional LINACs with on‐board kV imaging systems are widely available in most clinics. Therefore, it is highly desirable to use patient prior MR images and on‐board kV imaging systems to generate images with high soft tissue contrasts for target localization.
Previously, deformable image registration (DIR) has been used in the clinic for contouring and dose accumulation in adaptive radiation therapy.14, 15, 16, 17, 18, 19 Recently, we have developed DIR based optimization algorithms to estimate on‐board limited angle 4D CBCT and volumetric cine MRI, respectively, using prior images and on‐board data acquired within the same modality.5, 7, 20, 21 A previous method was also developed to use deformable motion reconstruction using x‐ray imaging for proton beam therapy.22, 23, 24 However, at present, no method has been developed to use DIR, prior images, and on‐board‐limited kV projections from a conventional LINAC to generate on‐board multimodality images to improve the soft tissue localization accuracy for SBRT treatment.
This study developed a novel approach to generate on‐board 4D MRI using a conventional LINAC with an on‐board kV imaging system and prior simulation MR images for liver SBRT localization. The method innovatively used patient prior 4D MRI images, on‐board, phase‐sorted, limited kV projections, and a deformation field map optimization algorithm to estimate on‐board, phase‐binned 4D MRI images. The feasibility of the method was evaluated through the digital XCAT phantom and a liver cancer patient.
2. Materials and methods
The overall workflow to generate the on‐board 4D MRI is shown in Fig. 1. Patient 4D MRI is acquired during the simulation stage, and MRIprior is defined as the end of expiration (EOE) phase of the 4D MRI. The on‐board 4D MRI at each respiratory phase is considered a deformation of the MRIprior, as shown in Eq. (1).
Figure 1.
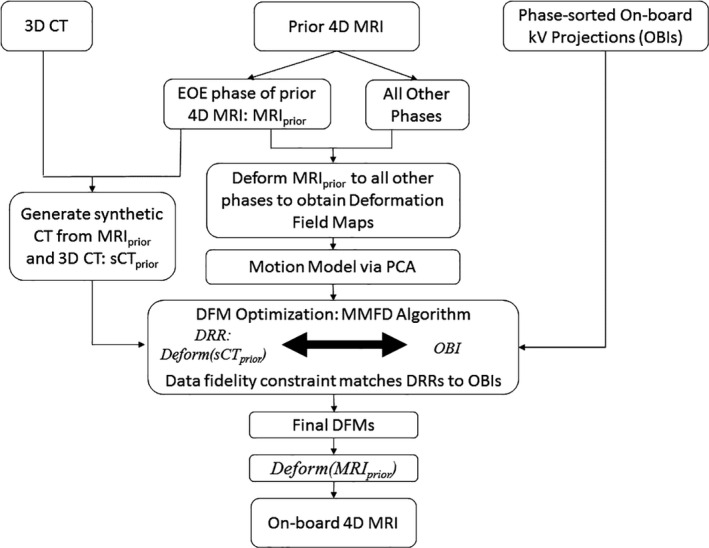
Overall workflow to generate on‐board 4D MRI from prior 4D MRI and on‐board kV projections.
| (1) |
DFM x , DFM y , and DFM z represent the deformation field maps (DFMs) along x, y, and z directions. The data fidelity constraint is used to solve the DFM based on phase‐sorted limited on‐board kV projections acquired. Specifically, MRIprior is used to generate a synthetic CT at EOE phase (sCTprior). The data fidelity constraint requires the digitally reconstructed radiographs (DRRs) calculated from the deformed sCTprior to match with the corresponding phase‐sorted on‐board kV projections acquired (OBI), as shown in Eq. (2).
| (2) |
A previously developed motion modeling and free‐form deformation (MMFD) method is used to solve the DFM in the ill conditioned problem defined in Eq. (2).4, 20 Specifically, MRIprior is deformably registered to all other phases of the prior 4D MRI to obtain a series of DFMs. Principal Component Analysis (PCA) is performed on the DFMs to extract the first three principal motion modes. The DFM to be solved is represented by a linear combination of the motion modes with a much fewer number of variables. A free‐form deformation model is used afterwards to fine‐tune the DFM obtained from motion modeling. Finally, the on‐board 4D MRI at each respiratory phase is generated by deforming the MRIprior based on the DFM solved for each phase, as shown in Eq. (1).
2.A. Generating synthetic CTprior from MRIprior
In order to perform the optimization of the DFMs, a synthetic CT prior, sCTprior, is generated so that DRRs can be calculated for matching with the on‐board kV projections in the data fidelity constraint defined in Eq. (2). Figure 2 shows a flowchart for generating sCTprior from MRIprior and a prior three‐dimensional (3D) CT (i.e., EOE phase from simulation 4D CT). DIR is performed using VelocityAI (Velocity Medical Solutions, Atlanta) to register the prior 3D CT with the MRIprior to generate a deformed CT. In this study, the EOE phase of a 4D CT was used for the 3D CT. Organ contours in both the MRIprior and deformed CT are extracted manually by a clinician. We consider the MRIprior contour as the ground‐truth, and override the deformed CT liver contour to be that of the MRIprior contour. The area of the MRIprior that does not overlap with the deformed CT contour is called the discrepancy region. In the discrepancy region, mean HU and noise values from a nearby region in the deformed CT are used to fill in realistic heterogeneous HU values. The internal textures of the liver including the tumor region were generated in the deformed CT directly based on the deformable registration, similar as the approach in other previous studies.2, 25, 26 The sCTprior is then used to optimize the deformation field map using the MMFD algorithm.
Figure 2.
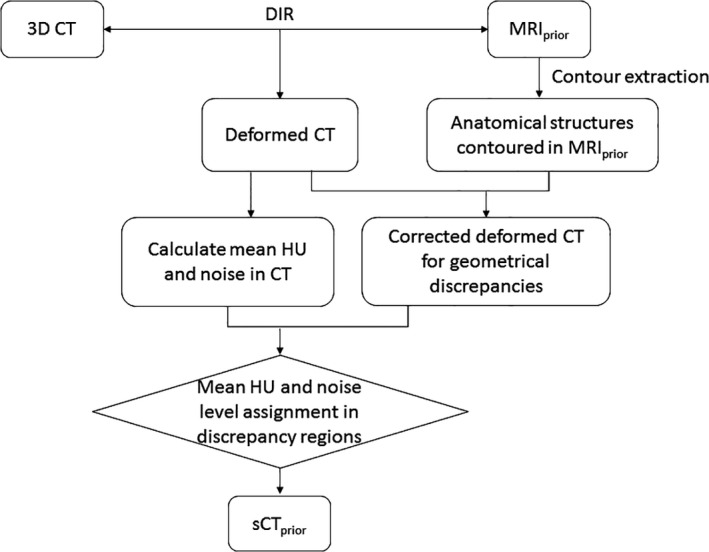
Workflow used to generate sCT prior from a 3D CT and MRI prior.
2.B. Initial estimation of deformation field maps from PCA‐based motion modeling
The MRIprior is deformed to all other phase volumes of the 4D MRI to obtain DFMs. The average of the DFMs is calculated and defined as DFM0,ave. PCA is used to extract patient motion patterns and the motion modeling (MM) optimization is used to get a rough estimation of DFM. The DFM is represented as a weighted linear combination of the motion patterns. The initial starting DFM into the MMFD optimization algorithm is DFM0,ave, and all weighting coefficients set to zero.
During the MM part of the MMFD algorithm, the weighting coefficients are optimized by using the data fidelity constraint shown in Eq. (2), which matches the DRRs from the deformed sCTprior to the onboard kV projections. The MM optimization generates a coarse DFM, which is used as the input to the free‐form deformation optimization. More details regarding the MM optimization can be found in previous work.4, 20
2.C. Fine‐tuning the deformation field maps using free‐form deformation
In the free‐form deformation (FD) optimization, the coarse DFM obtained from the MM is fine‐tuned by letting each voxel deform freely to correct any existing errors. In the FD optimization, the data fidelity constraint is met while minimizing the deformation energy of the DFM to preserve the smoothness. Equation (3) shows the equation for the deformation energy.
| (3) |
Details regarding the FD optimization can be found in previous work.4, 27 After the MMFD optimization, MRIprior is deformed based on the final DFM to obtain the on‐board MRI at a given respiratory phase.
2.D. XCAT simulation
A digital anthropomorphic phantom, XCAT, was used to simulate the prior 4D MRI, ground‐truth on‐board 4D MRI and ground‐truth on‐board 4D CT.28 In an XCAT simulation, the diaphragm curve sets the superior–inferior (SI) motion and the chest wall curve sets the anterior–posterior (AP) motion. Lateral motion was not simulated.
2.D.1. Prior 4D MRI simulation
A spherical lesion was generated in the middle of the liver in XCAT. Table 1 shows the image parameters for the prior 4D MRI simulation.
Table 1.
Simulation parameters for prior 4D MRI XCAT generation
| Lesion diameter size (mm) | 30 |
| Respiratory cycle (s) | 5 |
| Diaphragm curve amplitude (cm) | 3 |
| Chest wall curve amplitude (cm) | 2 |
| Image size | 256 × 256 × 150 |
| Resolution (mm3) | 1.67 × 1.67 × 1.67 |
The XCAT phantom was generated in activity mode in order to produce MRI‐like images. Texture was added using an in‐house XCAT‐MRI package to produce more realistic MRI images.
2.D.2. Ground‐truth on‐board 4D MRI and 4D CT simulation
Three on‐board scenarios were generated to reflect various respiratory changes with similar parameters used in Table 1, but with the following changes. (a) None. (b) Diaphragm curve and chest wall curve were set to 2 and 1.2 cm, respectively, to simulate amplitude decrease in both the SI and AP direction from simulation to treatment. (c) Diaphragm curve and chest wall curve were set to 3 and 2.5 cm, respectively, to simulate amplitude increase in the AP direction from simulation to treatment.
For all three scenarios, both 4D MRI and 4D CT XCAT phantoms were generated to represent the ground‐truth on‐board volumes. To generate the 4D CT, the XCAT was simulated in attenuation‐mode using an effective energy used in clinical CT scans. Both the ground‐truth on‐board 4D MRI and 4D CT had the same image size and resolution as the prior 4D MRI images.
2.D.3. On‐board kv projection simulation
Based on the simulated ground‐truth on‐board 4D CT, on‐board cone beam projections of different phases were simulated as full‐fan acquisition based on Siddon's ray‐tracing techniques.29, 30 The source to isocenter distance was 100 cm, and the isocenter to detector distance was 50 cm. Each projection contained 512 × 384 pixels, with each pixel being 0.78 × 0.78 mm in dimension.
Projections for each phase with the following acquisition schemes were generated based on previous publications.4, 20 (a) Orthogonal‐view 30° scan angle with 102 projections. (b) Orthogonal‐view 50° scan angle with 168 projections. (c) Single‐view 100° scan angle with 167 projections. (d) Single‐view 100° scan angle with 41 projections. (e) Single‐view 200° scan angle with 81 projections.
2.E. Patient study using liver cancer patient data
The on‐board 4D MRI method was retrospectively evaluated using one liver cancer patient. The liver cancer patient had both 4D MRI and 4D CT data. The 4D MRI was generated using a balanced steady‐state free precession (bSSFP) imaging acquisition technique to acquire two‐dimensional (2D) axial images continuously and then retrospectively sort the images based on respiratory phases.31, 32 The resolution of the 4D MRI was resized to match the resolution of the 4D CT, which was 1.27 × 1.27 × 2.5 mm3. The 4D MRI data were used as the “prior” 4D MRI and the 4D CT data were used as the “on‐board” volumes for the study. Orthogonal‐view 30° scan angle CBCT projections (102 total projections) were simulated from the 4D CT data and used as the on‐board kV projections. The same parameters were used to generate the projections as described in Section 2.D.3.
2.F. Evaluation methods
For the XCAT studies, the estimation accuracy for tumor location and volume in the on‐board 4D MRI was evaluated at the end of inspiration (EOI) phase since it has the largest deformation from MRIprior. An in‐house MATLAB (MathWorks, Natick, MA) code was used to automatically contour the tumors in both the estimated and ground‐truth volumes. Three metrics were defined to quantify the accuracy of the estimated lesion volume: volume percent difference (VPD), volume dice coefficient (VDC), and center‐of‐mass shift (COMS). Definitions for VPD and COMS can be found in previous work.5 VDC was defined as shown in Eq. (4).
| (4) |
where |V| and |V 0| are the numbers of elements in the estimated volume and ground‐truth volume, respectively.
For the patient data, a “reference” on‐board 4D MRI was generated by deforming the prior 4D MRI to the “on‐board” 4D CT phase by phase using VelocityAI. The estimated on‐board 4D MRI was then compared with the reference on‐board 4D MRI for evaluation.
3. Results
3.A. XCAT study
Table 2 shows the on‐board 4D MRI estimation accuracy for different XCAT scenarios with various scan angles and projections. All values are based on the end of inspiration (EOI) phase in the on‐board images, as it has the largest deformation from the prior data. Figure 3(a) shows MRIprior image, ground‐truth on‐board CT image in EOI phase, ground‐truth on‐board MRI image in EOI phase, and estimated on‐board MRI image in EOI phase. Figure 3(b) shows the subtraction images for the axial, coronal, and sagittal images shown in Fig. 1(a).
Table 2.
VPD (%), VDC and COMS (mm) values for the on‐board MRI images with the three XCAT scenarios. All results are shown for the EOI phase images
| Scenario 1 | Scenario 2 | Scenario 3 | ||
|---|---|---|---|---|
| VPD (%) | ||||
| Ortho‐view 30° | 102 projections | 9.89 | 9.00 | 11.58 |
| Ortho‐view 50° | 168 projections | 10.92 | 8.87 | 11.80 |
| Single‐view 100° | 167 projections | 9.93 | 8.61 | 14.35 |
| Single‐view 100° | 41 projections | 11.87 | 8.61 | 13.01 |
| Single‐view 200° | 81 projections | 11.40 | 7.21 | 11.15 |
| VDC | ||||
| Ortho‐view 30° | 102 projections | 0.95 | 0.96 | 0.94 |
| Ortho‐view 50° | 168 projections | 0.95 | 0.96 | 0.94 |
| Single‐view 100° | 167 projections | 0.95 | 0.96 | 0.93 |
| Single‐view 100° | 41 projections | 0.94 | 0.96 | 0.93 |
| Single‐view 200° | 81 projections | 0.94 | 0.96 | 0.94 |
| COMS (mm) | ||||
| Ortho‐view 30° | 102 projections | 0.79 | 0.80 | 1.05 |
| Ortho‐view 50° | 168 projections | 0.93 | 0.76 | 1.07 |
| Single‐view 100° | 167 projections | 0.82 | 0.67 | 1.25 |
| Single‐view 100° | 41 projections | 0.94 | 0.69 | 1.04 |
| Single‐view 200° | 81 projections | 0.92 | 0.57 | 0.95 |
Figure 3.
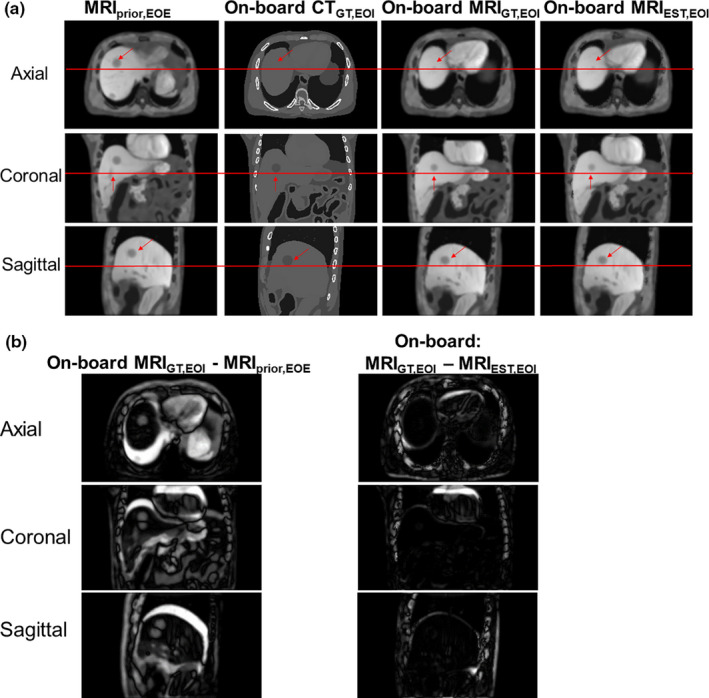
(a) The columns from left to right are the following. MRI prior at EOE phase, ground‐truth CT at EOI phase, ground‐truth on‐board MRI at EOI phase and estimated on‐board MRI at EOI phase for XCAT Scenario 2. The rows represent axial, coronal, and sagittal views, respectively. The horizontal line and arrows indicate areas for comparison. (b) Subtraction images for axial, coronal, and sagittal images shown in (a). [Color figure can be viewed at wileyonlinelibrary.com]
3.B. Patient study
Figure 4 shows prior MRI image at EOE phase, on‐board CT at EOI phase, reference on‐board MRI at EOI phase and estimated on‐board MRI at EOI phase for the patient data. The reference on‐board MRI was generated by deforming the EOI phase of the prior 4D MRI to the EOI phase of the on‐board 4D CT. The reference on‐board MRI visually match well with the estimated on‐board MRI.
Figure 4.
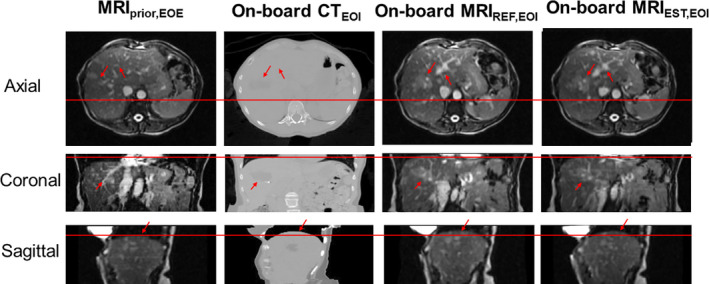
The columns from left to right show: Prior MRI (MRI prior) at EOE phase, on‐board CT at EOI phase, reference on‐board MRI at EOI phase and estimated on‐board MRI at EOI phase for the patient study. The rows show the axial, coronal, and sagittal views, respectively. The horizontal line and arrows indicate areas for comparison. [Color figure can be viewed at wileyonlinelibrary.com]
4. Discussion
To our knowledge, this is the first technique that uses prior knowledge and on‐board‐limited kV imaging to generate on‐board MR images for MR‐guided radiotherapy (MRgRT). Fundamentally, the method proposed is a 3D–2D multimodality deformable registration technique, which uses prior images to enhance the on‐board image contrast. Previously, deformable registration has been used for contouring and adaptive therapy, but it has not been fully explored to generate on‐board multimodality images for target localization. This is a novel concept that opens up a new avenue for the applications of deformable registration in image guidance.
Clinically, the method proposed can greatly improve localization accuracy of low‐contrast soft tissue tumors on x ray‐only radiotherapy machines. As we know, radiotherapy machines with x‐ray imaging systems are widely available in the clinics. However, the image quality of CBCT is rather limited especially for localizing low‐contrast tumors in soft tissue. MR‐Radiotherapy machines have been developed to address this issue. However, the availability of MR‐Radiotherapy machines is very limited in clinics partially due to its high cost. It is expected that x ray‐only radiotherapy machines will still remain dominant in clinical practice in the foreseeable future. This method provides a potential solution to address the main limitation and expand the capability of the x ray‐only radiotherapy units substantially to provide MRgRT to benefit the treatments of tumors in low‐contrast regions. Since the method is solely software based, it can be implemented on x ray‐only radiotherapy machines without requiring hardware modifications of the machine, which significantly limits the cost for implementing MRgRT to make it much more widely available for cancer patients.
The current study focused on using prior MRI images to enhance on‐board image contrast for liver patients treated on LINACS with an x‐ray imaging system. However, the idea and approach can be further broadened in three aspects: (a). Other prior images, such as contrast‐enhanced CT and functional images like PET/CT, can be used as well to generate on‐board multimodality images to provide multicontrast anatomical or functional guidance to further enhance the accuracy of target localization; (b). The technique can be applicable to other disease sites, besides liver, that have similar low‐contrast issues, such as breast patients; (c). The method can also be potentially applied for proton or heavy ion therapy machines with x‐ray imaging systems, or brachytherapy with in‐room CT/CBCT systems to generate on‐board or in‐room multimodality images to improve the low‐contrast tumor localization accuracy.
A previous technique was developed using prior images, PCA‐based motion modeling, and on‐board fiducial tracking based on x‐ray imaging to reconstruct respiratory motion for on‐board dose reconstruction or tracking for proton therapy.22, 23, 24 However, the method is potentially limited by the accuracy of the PCA modeling since patient breathing patterns can change from simulation to treatment, as shown in our previous studies.4, 6, 20 Furthermore, it required the invasive procedure of fiducial marker implantation, which may not always be applicable for patients. The marker migration would also affect the accuracy of the algorithm. In comparison, our method uses free‐form deformation to correct for inaccuracies in PCA modeling and does not require fiducial markers to be present. In addition, from a big picture, our study proposes a new concept of using prior images and deformable registration to generate on‐board multimodality images for target localization on conventional radiotherapy machines with only x‐ray imaging capability. The proposed method in this manuscript can greatly expand the prevalence and clinical impact of MRgRT or multimodality imaging in radiation therapy to enhance the precision of localizing low‐contrast tumors, which can lead to better tumor control and reduced toxicities. The improvement of precision also paves the road to further margin reduction and dose escalation.
In our studies, VPD, VDC, and COMS were used as metrics for evaluating the accuracy of the on‐board 4D MRI. Note that the accuracy that can be achieved is limited by the image resolution of the prior 4D MRI. In our studies, the image resolution was 1.67 × 1.67 × 1.67 mm3 for XCAT data. Therefore, we consider COMS within 2 mm as acceptable for the on‐board 4D MRI estimation. VPD is sensitive to target size, as it is calculated by dividing the target volume differences by the actual target volume size. Assuming the target volume is offset by 2 mm (the tolerance for COMS), the VPD for a 3‐cm‐diameter target is 20%, which was used as the tolerance for VPD in our studies.
Based on the XCAT data, on‐board 4D MRI was successfully estimated for all XCAT scenarios using orthogonal‐view 30° scan angles with 102 projections, orthogonal‐view 50° scan angles with 168 projections, single‐view 100° scan angles with 41 projections, single‐view 100° scan angles with 167 projection, and single‐view 200° scan angles with 81 projections. For the patient data, the estimated on‐board 4D MRI images visually matched well with the reference on‐board 4D MRI when using orthogonal‐view 30° scan angles. Please note that these projection numbers refer to only one phase (EOI) of the 4D image set, and the number of projections should be multiplied by 10 to get a better idea of the entire projection number across all 10 respiratory phase bins of the estimated on‐board 4D MRI. The results shown in Section III are values for the EOI phase, as this phase shows the greatest deformation from prior to on‐board. What may be more clinically relevant, however, may be to calculate the estimation of the ITV over the entire 10 phase on‐board 4D MRI. For this, we found that calculating the error in the ITV from the ground‐truth on‐board 4D MRI with the estimated on‐board 4D MRI using XCAT scenario 2 with orthogonal‐view 30° scan angles resulted in VPD/VDC/COMS = 6.64%/0.87/0.41 mm.
This study aimed to investigate the feasibility of generating on‐board 4D MRI using prior 4D MRI, limited projections from a conventional LINAC and deformation models. The estimation method utilized a synthetic CT in order to match DRRs to the on‐board projections in the data fidelity constraint. Different studies have been to done to investigate various ways of generating synthetic CT images, including use of bulk densities,33 voxel‐based intensity conversions,34 and deformable registration‐based methods.2, 25, 26 Each approach has its advantages and limitations. In our study, we used the deformable registration‐based approach, which is practical to implement and has been demonstrated to be an effective method to generate synthetic CT for regions affected by deformation, such as head–neck and prostate regions. The limitation of this approach is that the accuracy of the sCTprior generation is affected by the uncertainties in deformable registration. Other alternative approaches mentioned above can be explored in the future to minimize the effects of deformable registration in synthetic CT generation. Other DRR‐matching techniques can also be investigated to bypass synthetic CT generation by matching prior MRI to on‐board kV projections directly.
It is important to note that this study is the first to aim to investigate the feasibility of using kV projections and prior 4D MRI to generate on‐board 4D MRI. We have provided a proof of concept using XCAT simulation and one liver patient, and future studies are warranted to fully evaluate the innovative technique. To look at the effect of tumor contrast on the on‐board 4D MRI estimation, we did a minor XCAT simulation that reduced the tumor to liver contrast in the CT. The liver HU value was set to 62. The tumor HU value was reduced from −138 to 0. On‐board 4D MRI was estimated for XCAT scenario 2 using orthogonal‐view 30° scan angles with 102 projections. The lower tumor to liver contrast reduced the estimation accuracy with VPD/VDC/COMS from 9.00%/0.96/0.80 mm to 14.53%/0.93/1.13 mm. The lower contrast still resulted in accurate 4D MRI estimation based on the tolerance defined above. The accuracy of the current method may be limited for very low contrast regions. To address this, many other techniques can be implemented to further improve the robustness and accuracy of the method. For example, there may be other methods for synthetic CT generation with or without deformable registration, which can be further explored, as discussed above. For very low contrast tumors, the MMFD algorithm can be replaced by a finite element analysis (FEA)‐based biomechanical modeling to deform the low‐contrast regions within an organ based on surface matching of the organ.35 Future studies are warranted to fully evaluate the effect of tumor contrast.
More XCAT simulations will be conducted in the future, which can incorporate patient RPM breathing curves to generate XCAT simulations with more realistic breathing curves. The effect of irregular breathing pattern change, kV scan angle or number of projections can all be evaluated to optimize the scanning time and imaging dose of the kV acquisition for different patient scenarios in future studies. Additional patient studies need to be investigated, as well. In this study, we used one liver patient's data with both 4D MRI and 4D CT images. The patient study provides a further evaluation of the estimated on‐board 4D MRI using real liver patient images with real respiratory motions. It is important to note the challenges with the patient study. It is challenging to establish a ground‐truth on‐board 4D MRI image for the patient studies, since currently no places can acquire both on‐board 4D CBCT and on‐board 4D MRI in the same treatment room. One may deformably register prior 4D MRI to 4D CBCT to generate on‐board 4D MRI as an approximation of the ground‐truth for evaluation. However, the accuracy of the deformable registration will be very limited due to the very limited image quality of 4D CBCT caused by scattering, undersampling etc. In our study, we used 4D CT to simulate the on‐board 4D volume, and generated on‐board reference 4D MRI by deforming the prior 4D MRI to 4D CT, phase by phase, as the standard to compare with the estimated on‐board 4D MRI. DRRs were simulated from 4D CT and used as 4D CBCT projections for the estimation algorithm. In the future, Monte Carlo code may be used to simulate more realistic projections. Another possible approach for the patient study is to acquire two 4D MRI images, and use the first 4D MRI as prior and the second 4D MRI images as the on‐board ground‐truth. However, this would require the need to generate synthetic 4D CT images from the second 4D MRI and simulate 4D CBCT projections from the synthetic 4D CT, which has uncertainties associated as well.
Lastly, the current study used a single prior MRI set for generation of on‐board 4D MRI. In the future, multiple prior MRI sets with different contrasts may be used to generate on‐board multicontrast 4D MRI from on‐board kV imaging acquisition to further improve the localization accuracy.
5. Conclusion
A method has been developed to generate on‐board 4D MRI using prior 4D MRI and a limited number of on‐board kV projections. The preliminary results demonstrated the potential for MRI‐based image guidance for liver SBRT using only a kV imaging system on a conventional LINAC.
Acknowledgments
This work was supported by the National Institutes of Health Grant No. R01‐CA184173. We acknowledge the support of ARO Equipment Grant #W911NF‐13‐l‐0344 and an NVIDIA academic center equipment donation. Additionally, we also acknowledge Xiaoning Liu and Yawei Zhang for their help and contributions.
References
- 1. Zelefsky MJ, Kollmeier M, Cox B, et al. Improved clinical outcomes with high‐dose image guided radiotherapy compared with non‐IGRT for the treatment of clinically localized prostate cancer. Int J Radiat Oncol Biol Phys. 2012;84:125–129. [DOI] [PubMed] [Google Scholar]
- 2. Kraus KM, Jakel O, Niebuhr NI, Pfaffenberger A. Generation of synthetic CT data using patient specific daily MR image data and image registration. Phys Med Biol. 2017;62:1358–1377. [DOI] [PubMed] [Google Scholar]
- 3. Keall PJ, Mageras GS, Balter JM, et al. The management of respiratory motion in radiation oncology report of AAPM Task Group 76. Med Phys. 2006;33:3874–3900. [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y, Yin FF, Pan T, Vergalasova I, Ren L. Preliminary clinical evaluation of a 4D‐CBCT estimation technique using prior information and limited‐angle projections. Radiother Oncol. 2015;115:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harris W, Ren L, Cai J, Zhang Y, Chang Z, Yin FF. A technique for generating volumetric cine‐magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2016;95:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris W, Zhang Y, Yin FF, Ren L. Estimating 4D CBCT from prior information and extremely limited angle projections using structural PCA and weighted free‐form deformation for lung radiotherapy. Med Phys. 2017;44:1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren L, Zhang Y, Yin FF. A limited‐angle intrafraction verification (LIVE) system for radiation therapy. Med Phys. 2014;41:020701. [DOI] [PubMed] [Google Scholar]
- 8. Jaffray DA, Siewerdsen JH. Cone‐beam computed tomography with a flat‐panel imager: initial performance characterization. Med Phys. 2000;27:1311–1323. [DOI] [PubMed] [Google Scholar]
- 9. Siewerdsen JH, Jaffray DA. Cone‐beam computed tomography with a flat‐panel imager: magnitude and effects of x‐ray scatter. Med Phys. 2001;28:220–231. [DOI] [PubMed] [Google Scholar]
- 10. Siewerdsen JH, Jaffray DA. Cone‐beam computed tomography with a flat‐panel imager: effects of image lag. Med Phys. 1999;26:2635–2647. [DOI] [PubMed] [Google Scholar]
- 11. Fallone BG, Murray B, Rathee S, et al. First MR images obtained during megavoltage photon irradiation from a prototype integrated linac‐MR system. Med Phys. 2009;36:2084–2088. [DOI] [PubMed] [Google Scholar]
- 12. Raaymakers BW, Lagendijk JJ, Overweg J, et al. Integrating a 1.5 T MRI scanner with a 6 MV accelerator: proof of concept. Phys Med Biol. 2009;54:N229–N237. [DOI] [PubMed] [Google Scholar]
- 13. Mutic S, Dempsey JF. The ViewRay system: magnetic resonance‐guided and controlled radiotherapy. Semin Radiat Oncol. 2014;24:196–199. [DOI] [PubMed] [Google Scholar]
- 14. Rosenman JG, Miller EP, Tracton G, Cullip TJ. Image registration: an essential part of radiation therapy treatment planning. Int J Radiat Oncol Biol Phys. 1998;40:197–205. [DOI] [PubMed] [Google Scholar]
- 15. Mutic S, Dempsey JF, Bosch WR, et al. Multimodality image registration quality assurance for conformal three‐dimensional treatment planning. Int J Radiat Oncol Biol Phys. 2001;51:255–260. [DOI] [PubMed] [Google Scholar]
- 16. Nix MG, Prestwich RJD, Speight R. Automated, reference‐free local error assessment of multimodal deformable image registration for radiotherapy in the head and neck. Radiother Oncol. 2017;125:478–484. [DOI] [PubMed] [Google Scholar]
- 17. Brock KK, Mutic S, McNutt TR, Li H, Kessler ML. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. Med Phys. 2017;44:e43–e76. [DOI] [PubMed] [Google Scholar]
- 18. Brock KK; Deformable Registration Accuracy Consortium . Results of a multi‐institution deformable registration accuracy study (MIDRAS). Int J Radiat Oncol Biol Phys. 2010;76:583–596. [DOI] [PubMed] [Google Scholar]
- 19. Brock KK, Sharpe MB, Dawson LA, Kim SM, Jaffray DA. Accuracy of finite element model‐based multi‐organ deformable image registration. Med Phys. 2005;32:1647–1659. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Yin FF, Segars WP, Ren L. A technique for estimating 4D‐CBCT using prior knowledge and limited‐angle projections. Med Phys. 2013;40:121701. [DOI] [PubMed] [Google Scholar]
- 21. Harris W, Yin FF, Wang C, Zhang Y, Cai J, Ren L. Accelerating volumetric cine MRI (VC‐MRI) using undersampling for real‐time 3D target localization/tracking in radiation therapy: a feasibility study. Phys Med Biol. 2017;63:01NT01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Knopf A, Tanner C, Boye D, Lomax AJ. Deformable motion reconstruction for scanned proton beam therapy using on‐line x‐ray imaging. Phys Med Biol. 2013;58:8621–8645. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y, Knopf A, Tanner C, Lomax AJ. Online image guided tumour tracking with scanned proton beams: a comprehensive simulation study. Phys Med Biol. 2014;59:7793–7817. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y, Knopf AC, Weber DC, Lomax AJ. Improving 4D plan quality for PBS‐based liver tumour treatments by combining online image guided beam gating with rescanning. Phys Med Biol. 2015;60:8141–8159. [DOI] [PubMed] [Google Scholar]
- 25. Dowling JA, Lambert J, Parker J, et al. An atlas‐based electron density mapping method for magnetic resonance imaging (MRI)‐alone treatment planning and adaptive MRI‐based prostate radiation therapy. Int J Radiat Oncol Biol Phys. 2012;83:e5–e11. [DOI] [PubMed] [Google Scholar]
- 26. Guerreiro F, Burgos N, Dunlop A, et al. Evaluation of a multi‐atlas CT synthesis approach for MRI‐only radiotherapy treatment planning. Phys Med. 2017;35:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu X, Zhang D. Efficient parallel Levenberg‐Marquardt model fitting towards real‐time automated parametric imaging microscopy. PLoS ONE. 2013;8:e76665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Segars WP, Mahesh M, Beck TJ, Frey EC, Tsui BM. Realistic CT simulation using the 4D XCAT phantom. Med Phys. 2008;35:3800–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siddon RL. Fast calculation of the exact radiological path for a three‐dimensional CT array. Med Phys. 1985;12:252–255. [DOI] [PubMed] [Google Scholar]
- 30. Yan H, Ren L, Godfrey DJ, Yin FF. Accelerating reconstruction of reference digital tomosynthesis using graphics hardware. Med Phys. 2007;34:3768–3776. [DOI] [PubMed] [Google Scholar]
- 31. Cai J, Chang Z, Wang Z, Paul Segars W, Yin FF. Four‐dimensional magnetic resonance imaging (4D‐MRI) using image‐based respiratory surrogate: a feasibility study. Med Phys. 2011;38:6384–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang J, Cai J, Wang H, et al. Four‐dimensional magnetic resonance imaging using axial body area as respiratory surrogate: initial patient results. Int J Radiat Oncol Biol Phys. 2014;88:907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chin AL, Lin A, Anamalayil S, Teo BK. Feasibility and limitations of bulk density assignment in MRI for head and neck IMRT treatment planning. J Appl Clin Med Phys. 2014;15:4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koivula L, Kapanen M, Seppala T, et al. Intensity‐based dual model method for generation of synthetic CT images from standard T2‐weighted MR images ‐ generalized technique for four different MR scanners. Radiother Oncol. 2017;125:411–419. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y, Tehrani JN, Wang J. A biomechanical modeling guided CBCT estimation technique. IEEE Trans Med Imaging. 2017;36:641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]


