Abstract
Newts are capable of regenerating several anatomical structures and organs, including their limbs. This remarkable regenerative capacity is thought to depend on cellular dedifferentiation. Terminally differentiated mammalian cells, by contrast, are normally incapable of reversing the differentiation process. Several factors could explain the absence of cellular dedifferentiation in mammals: (i) inadequate expression of genes that initiate dedifferentiation; (ii) insufficient intracellular signaling pathways; (iii) irreversible expression of differentiation factors; and (iv) structural characteristics that make dedifferentiation impossible. To investigate the causes underlying the lack of cellular plasticity in mammalian cells, we examined the effect of an extract derived from newt regenerating limbs on terminally differentiated mouse C2C12 myotubes. Approximately 18% of murine myotubes reentered the cell cycle when treated with regeneration extract, whereas 25% of newt myotubes exhibited cell cycle reentry. The muscle differentiation proteins MyoD, myogenin, and troponin T were reduced to undetectable levels in 15–30% of treated murine myotubes. We observed cellular cleavage in 11% of the treated murine myotubes and approximately 50% of these myotubes continued to cleave to produce proliferating mononucleated cells. These data indicate that mammalian myotubes can dedifferentiate when stimulated with the appropriate factors and suggest that one mechanism preventing dedifferentiation of mammalian cells is inadequate spatial or temporal expression of genes that initiate dedifferentiation.
Adult urodele amphibians and teleost fish can replace lost anatomical structures through a process known as epimorphic regeneration. Morgan (1) coined the term epimorphosis to refer to the regenerative process in which cellular proliferation precedes the development of a new anatomical structure. An adult urodele (e.g., a newt or axolotl) is capable of regenerating its limbs, spinal cord, heart ventricle, tail, retinas, eye lenses, and upper and lower jaws (2–4), whereas teleost fish can regenerate their fins and spinal cord (5, 6). These remarkable regenerative capabilities are lacking in mammals and other vertebrates.
The molecular and cellular mechanisms that govern epimorphic regeneration are incompletely defined. Following limb amputation, epithelial cells begin to migrate across the amputation site to form a wound epithelium (WE) a few layers thick within a single day of amputation. This wound epithelium thickens in response to continued epithelial cell migration and within days forms the mature apical epithelial cap (AEC) (7). The internal stump cells underlying the WE/AEC begin to dedifferentiate in response to undefined signals found in the early limb regenerate (8–15). These dedifferentiated cells then proliferate to form a mass of progenitor and pluripotent cells, known as the regeneration blastema, which harbors the cells that will later redifferentiate to form the regenerated limb. Although cellular dedifferentiation has been demonstrated in newts, terminally differentiated mammalian cells are normally thought to be incapable of reversing the differentiation process (16, 17). Several mechanisms could explain the lack of cellular plasticity in mammalian cells: (i) the extracellular factors that initiate dedifferentiation are not adequately expressed following amputation; (ii) the intrinsic cellular signaling pathways for dedifferentiation are absent; (iii) differentiation factors are irreversibly expressed in mammalian cells; and (iv) structural characteristics of mammalian cells make dedifferentiation impossible.
Here, we demonstrate that mouse myotubes can dedifferentiate when stimulated with an extract prepared from newt regenerating limb tissue. Mouse myotubes reenter the cell cycle, exhibit reduced levels of muscle differentiation proteins, and cleave to form smaller myotubes or proliferating, mononucleated cells. We demonstrate that proteins are an essential component of this dedifferentiation signal. These results indicate that mammalian cells can dedifferentiate when exposed to the appropriate factors.
Materials and Methods
Animals and Tissue Collection.
Adult newts, Notophthalmus viridescens, from Charles Sullivan & Co. (Nashville, TN), were maintained in a humidified room at 24°C and fed Tubifex worms two to three time each week. Operations were performed on animals anesthetized with 0.1% tricaine for ≈2–5 min. Regenerating limb tissue was collected as follows. Forelimbs were amputated by cutting just proximal to the elbow and soft tissue was pushed up the humerus to expose the bone. The bone and soft tissue were trimmed to produce a flat amputation surface. The newts were placed on ice for 1 h, transferred to a 0.5% sulfamerazine solution overnight, and then back into a normal water environment. Early regenerating tissue (days 1, 3, and 5 postamputation) was collected by reamputating the limb 0.5–1.0 mm proximal to the wound epithelium and removing any residual bone. Nonregenerating limb tissue was collected from limbs that had not been previously amputated. Tissue was extracted 2–3 mm proximal to the forelimb elbow and all bones were removed. Immediately after collection, all tissues were flash frozen in liquid nitrogen and stored at −80°C.
Preparation of Protein Extracts.
Tissues were thawed and all subsequent manipulations were performed at 4°C or on ice. Six grams of early regenerating tissue from days 1, 3, and 5 (2 g each) or 6 g of nonregenerating tissue were placed separately into 10 ml of DMEM containing three protease inhibitors (2 μg/ml leupeptin/2 μg/ml A-protinin/1 mM PMSF). The tissues were ground with an electronic tissue homogenizer for 1–2 min, hand homogenized for 10–15 min, and sonicated for 30 s. Cell debris was removed in two centrifugation steps. The homogenate was first spun at 2000 × g for 25 min and then the supernatant was spun again at 100,000 × g for 60 min. The insoluble lipid layer was aspirated and the remaining supernatant filter sterilized through a 0.45-μm filter. The protein content was assayed with a BCA protein assay kit (Pierce) and stored in 0.5 ml aliquots at −80°C.
Cell Culture.
Newt A1 limb cells were obtained as a gift from Jeremy Brockes (University College, London). Mouse C2C12 myoblast cell line was purchased from American Type Culture Collection. Newt A1 cells were passaged, myogenesis induced, and myotubes isolated and plated at low density as described by Ferretti and Brockes (18) and Lo et al. (14). Newt A1 cells were grown at 24°C in 2% CO2. The culture medium was adjusted to the axolotl plasma osmolality of 225 Osm (18), using an Osmette A Automated Osmometer (Precision Systems, Natick, MA). Culture medium contained MEM with Eagle's salt, 10% FBS, pen/strep (100 units/ml penicillin/100 μg/ml streptomycin), bovine pancreas insulin (0.28 units/ml), 2 mM glutamine, and distilled water. To induce myotube formation in newt A1 cells, a confluent culture of mononucleated cells was treated with medium containing 0.5% FBS [differentiation medium (DM)] for 4–6 days. The myotubes were isolated from the remaining mononucleated cells by gentle trypsinization (0.05% trypsin) and sequentially sieved through 100 μm and 35 μm nylon meshes. Larger debris and clumped cells were retained on the first sieve, most myotubes were retained on the second sieve, and most mononucleated cells were passed through both sieves. Myotubes were gently washed off the 35-μm sieve and plated at either 7–14 myotubes per mm2 or ≈2 myotubes per mm2 onto 35-mm plates precoated with 0.75% gelatin.
C2C12 cells were passaged and myogenesis induced as described by Guo et al. (19). C2C12 myotubes were isolated and plated at low density after trypsinization and sieving through a 100-μm mesh. Myotubes were retained on this sieve, whereas the mononucleated cells passed through the 100-μm mesh. Myotubes were washed off the sieve and plated at either 7–14 myotubes per mm2 or ≈2 myotubes per mm2 onto 35-mm plates precoated with 0.75% gelatin.
To induce myotube dedifferentiation, 0.025–2 mg/ml of regeneration extract was added to DM 24 h after plating at low density on 35-mm gelatin-coated plates. Medium and extract were changed daily. To identify cleavage events and subsequent cellular proliferation, individual myotubes (plated at ≈2 cells per mm2) were photographed on day 0, before the addition of extract, and every 12–24 h after the addition of extract for up to 10 days. Only isolated myotubes devoid of any surrounding mononucleated cells were chosen for study. To test for reduction in muscle-specific protein levels as well as reentry into the cell cycle, myotubes plated at 7–14 cells per mm2 were chosen. The cells were stained as described below on day four. Cells cultured in DM alone or in DM with nonregeneration extract were used as negative controls.
Immunofluorescence Assays.
Myotubes plated at low density in 35-mm plates were washed three times with PBS before fixation and immunostaining. Unless otherwise specified, all manipulations were at room temperature, all dilutions of antibodies were prepared in 2% normal goat serum (NGS)/0.1% Nonidet P-40 in PBS, and incubations were followed by washes with 0.1% Nonidet P-40 in PBS. Cells were fixed in cold methanol at −20°C for 10 min, rehydrated with PBS, and blocked with 10% NGS for 15 min. Primary antibodies were incubated with myotubes for 1 h at 37°C and included anti-troponin T (1:50 dilution, Sigma T6277), anti-myogenin (1:50 dilution, FSD clone, PharMingen 65121A), and anti-MyoD (1:10 dilution, Vector Laboratories). After three washes, cells were incubated 45 min at 37°C with secondary antibody. For troponin T, a goat anti-mouse IgG conjugated to Alexa 594 (1:100 dilution, Molecular Probes) was used, whereas myogenin and MyoD required incubation with a biotin-XX goat anti-mouse IgG (H + L) F(ab′)2 fragment conjugate (1:200 dilution, Molecular Probes), followed by a 45-min incubation with streptavidin Alexa 594 (1:100 dilution, Molecular Probes). To confirm that fluorescent signals depended on binding of primary antibodies, control experiments were performed in the absence of primary antibody.
Cells were labeled with BrdUrd for 12 h and staining was performed by using a 5-bromo-2′-deoxyuridine labeling and detection kit I according to the manufacturer's instructions (Boehringer Mannheim). Microscopic examination and photography were accomplished by using a Zeiss Axiovert 100 equipped with a mounted camera and fluorescent light source.
Characterization of the Newt Regeneration Extract.
C2C12 myotubes were plated at low density in DM as described above. Regeneration extract was treated in one of three ways: (i) boiled for 5 min; (ii) digested with 1% trypsin for 30 min at 37°C; or (iii) taken through several freeze–thaw cycles. In three separate experiments, the treated extracts were applied to cultured myotubes at a concentration of 0.3 mg/ml. DM and extract were changed daily. Immediately after the extract was digested with 1% trypsin, the appropriate amount of treated extract was added directly to the cell cultures. The serum in DM and the dilution of the extract served to inactivate the trypsin. In the freeze–thaw experiment, extract activity was tested after both two and three freeze–thaw cycles. The effect of the pretreated extracts on myotube S phase reentry was assessed after 4 days of treatment by performing the BrdUrd incorporation assay described above. The results were compared with BrdUrd incorporation in myotubes cultured in DM containing regeneration extract and myotubes cultured in DM alone or DM containing nonregeneration limb extract.
Results
Regeneration Extract Induces Newt Myotube Dedifferentiation.
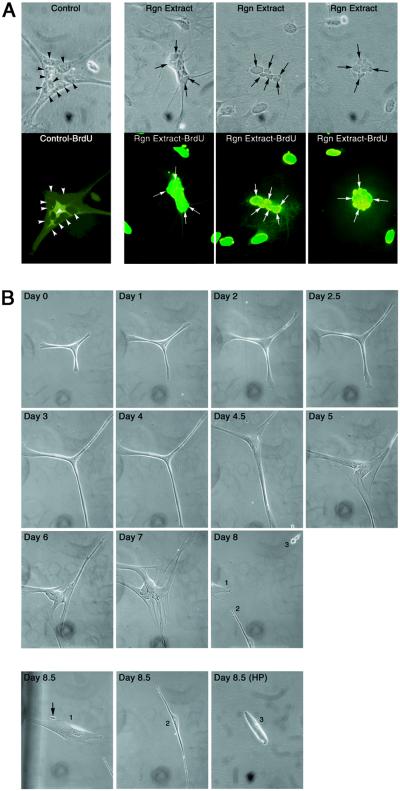
To determine the effect of newt regeneration extract on quiescent multinucleated newt myotubes, we applied extract to cultured myotubes and tested for BrdUrd incorporation. WE/AEC and adjacent proximal tissues from early newt limb regenerates (days 1–5 postamputation) were used to prepare regeneration extract as described in Materials and Methods. Newt A1 myotubes were plated at low density (7–14 cells per mm2) in DM and cultured with 0.025–2 mg/ml regeneration extract on day 0. Medium and extract were changed daily and myotubes were assayed for BrdUrd incorporation on day four. When quiescent newt A1 myotubes were cultured in DM with 0.3 mg/ml regeneration extract, 25% of the cells were stimulated to reenter the S phase of the cell cycle (Fig. 1A, Table 1). Whenever cell cycle reentry was observed in a newt myotube, all of the nuclei responded concordantly and incorporated BrdUrd. By contrast, only 2% of myotubes cultured in DM alone and 3% in DM with 0.3 mg/ml nonregenerating extract incorporated BrdUrd. This difference in DNA synthesis is significant at P = 6 × 10−8, using the Fisher–Irwin exact test. These data indicate that regenerating newt tissue contains factors that can induce newt myotubes to reenter the cell cycle.
Figure 1.
Limb regeneration extract induces newt myotube dedifferentiation. (A) Newt myotubes reentered the cell cycle when stimulated with limb regeneration extract. Top row shows phase contrast photomicrographs of newt myotubes. Bottom row shows fluorescent photomicrographs of the corresponding newt myotubes. Left panel shows a control myotube treated with nonregeneration extract. No nuclei reentered the cell cycle as evidenced by the lack of BrdUrd incorporation (arrowheads point to nuclei). The three panels on the right show myotubes that were treated with limb regeneration extract. All nuclei reentered the cell cycle as evidenced by BrdUrd incorporation (arrows point to representative nuclei that have reentered the cell cycle). (B) Limb regeneration extract induced newt myotube cleavage. Newt A1 myotubes were plated at very low density (≈2 cells per mm2) in DM on day 0 and treated daily with DM containing regeneration extract for 8.5 days. Multinucleated myotube is shown before treatment with limb regeneration extract (day 0). On day 1, the myotube began to exhibit altered morphology by elongating. This elongation process continued through day 4.5. By day 5, elongating myotube began to pull apart. Many centrally located nuclei are now readily visible. The cleavage process continued on days 6 and 7. By day 8, the myotube completely cleaved at the center to produce three smaller cells numbered 1, 2, and 3. The three products of the myotube cleavage are shown in separate photomicrographs at day 8.5. Cleavage product 3 is shown at high power (32× objective lens). All other photomicrographs in this figure are shown at low power (10× objective lens). Arrow points to a small cellular fragment protruding from the largest cleavage product, which suggests that additional cleavage is occurring (1). Control newt myotubes treated with nonregeneration limb extract or kept in DM alone showed no evidence of cleavage.
Table 1.
Effect of regeneration extract on newt and mouse myotubes
| Myotube treatment | % BrdUrd incorporation (n) | % of myotubes showing reduced levels (n)
|
% Cleavage (n) | ||
|---|---|---|---|---|---|
| MyoD | Myogenin | Troponin T | |||
| Newt | |||||
| Regeneration extract | 25 (102) | ND | ND | ND | 16 (56) |
| Nonregeneration extract | 3 (59) | ND | ND | ND | 0 (50) |
| Differentiation medium | 2 (96) | ND | ND | ND | 0 (43) |
| P value | 6 × 10−8 | 10−4 | |||
| Mouse | |||||
| Regeneration extract | 18 (76) | 18 (98) | 15 (82) | 30 (66) | 11 (92) |
| Nonregeneration extract | 0 (30) | 0 (46) | 0 (54) | 3 (32) | 0 (63) |
| Differentiation medium | 0 (32) | 0 (40) | 0 (48) | 6 (47) | 0 (61) |
| P value | 10−4 | 5 × 10−6 | 4 × 10−5 | 4 × 10−5 | 10−4 |
P values were determined by using the Fisher–Irwin exact test. Results from regeneration extract-treated myotubes were compared to results from the control myotubes (nonregeneration extract-treated plus differentiation medium alone). n, number of myotubes scored; ND, not done.
To determine whether factors contained in regenerating newt tissue can induce cellular cleavage, we applied the regeneration extract to cultured newt myotubes and followed cells with light microscopy. A1 myotubes were cultured at very low density (≈2 cells per mm2) in DM containing 0.3 mg/ml regeneration extract and each isolated, individual myotube was photographed every 24 h for 10 days. Only isolated myotubes with no surrounding mononucleated cells were studied. The first signs of cleavage were evident on days 3–5, when myotubes altered their shape and began to cleave into smaller myotubes (Fig. 1B). By day 10, 16% of the myotubes had cleaved to form smaller myotubes or mononucleated cells (Table 1). No cellular cleavage was seen in myotubes cultured in DM alone or in DM containing nonregeneration limb extract. This difference in the frequency of cellular cleavage is significant at P = 0.0001. These findings indicate that newt regeneration extract can induce cleavage of cultured newt myotubes.
Newt Regeneration Extract Induces Molecular and Cellular Dedifferentiation of Mouse Myotubes.
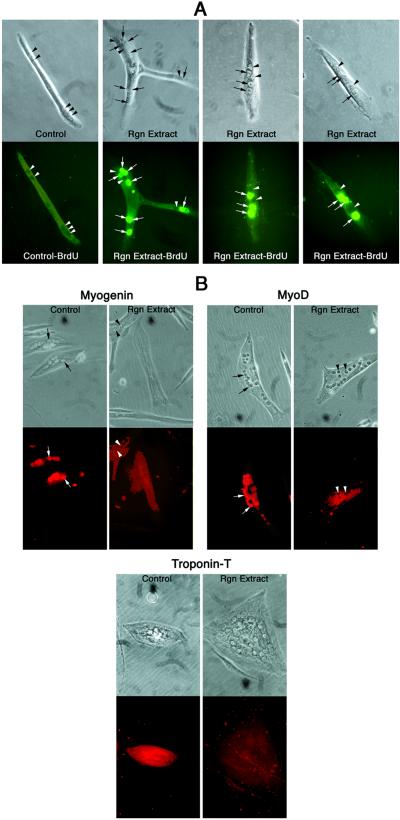
To determine whether mammalian myotubes could be induced to reenter the cell cycle, we applied regeneration extract to cultured C2C12 myotubes and tested for BrdUrd incorporation. C2C12 myotubes were plated at low density (7–14 cells per mm2) and cultured in DM containing 0.025–2 mg/ml of either regeneration or nonregeneration extract. Preliminary experiments indicated that both regeneration and nonregeneration extracts were toxic to mouse myotubes at concentrations ≥0.5 mg/ml and that cell cycle reentry could be induced when using regeneration extract at concentrations ≥0.125 mg/ml. The optimal regeneration extract concentration for cell cycle reentry was 0.3 mg/ml, so this concentration was chosen for all subsequent experiments. We also performed a preliminary time course experiment to determine when mouse myotube nuclei began to reenter the cell cycle in response to the regeneration extract. Within 2 days of the onset of extract treatment, individual nuclei within mouse myotubes already had reentered the S phase. Maximum cell cycle reentry was observed by day 3 and continued through day 4. These optimal conditions were used to assess both the percentage of myotubes containing DNA-synthesizing nuclei and the frequency with which nuclei would initiate DNA synthesis. Extract was added on day 0, medium and extract were changed daily, and cells were assayed for BrdUrd incorporation on day 4. Eighteen percent of regeneration extract-treated C2C12 myotubes showed S phase reentry (Fig. 2A, Table 1). By contrast, no BrdUrd incorporation was seen in C2C12 myotubes cultured in DM alone or in DM with nonregeneration extract (Fig. 2A, Table 1). This difference in DNA synthesis was significant at P = 0.0001. Thus, newt regeneration extract can induce cell cycle reentry in cultured mammalian myotubes. However, unlike their newt counterparts, only 19% of mouse myotubes that reentered the cell cycle exhibited concordant DNA synthesis, and overall, only 64% of the nuclei of responding myotubes reentered the cell cycle.
Figure 2.
Regeneration extract induces cell cycle reentry and reduction in myogenic protein levels in mouse myotubes. Mouse C2C12 myotubes were treated with regeneration or nonregeneration extract. (A) Myotubes were treated with BrdUrd for 12 h and then BrdUrd incorporation assays were performed. (Upper) Phase contrast photomicrographs; (Lower) corresponding fluorescent photomicrographs. Control-BrdU panel shows a myotube treated with nonregenerating limb extract. No nuclei reentered the cell cycle in this control myotube. Rgn Extract-BrdU panels show three mouse myotubes containing nuclei that reentered the cell cycle following treatment with regeneration extract. Arrowheads point to nuclei that did not incorporate BrdUrd. Arrows point to nuclei that incorporated BrdUrd in myotubes treated with regeneration extract. (B) Regeneration extract reduced myogenic protein levels in murine myotubes. (Upper) Phase contrast photomicrographs; (Lower) corresponding fluorescent photomicrographs. C2C12 myotubes cultured in DM containing regeneration extract (Rgn Extract) or nonregeneration limb extract (Control) are shown. The muscle-specific transcription factors myogenin and MyoD were reduced in myotubes treated with regeneration extract (arrowheads point to representative unstained nuclei), but present at normal levels in the controls (arrows point to representative fluorescent red nuclei). The muscle contractile protein troponin T was reduced in a treated myotube (unstained cytoplasm), but present in the control (fluorescent red cytoplasm).
To determine whether regeneration extract affects expression of muscle proteins, we applied the regeneration extract to C2C12 myotubes and performed indirect immunofluorescence assays to look for altered expression of myogenic regulatory proteins (myogenin and MyoD) and a muscle contractile protein (troponin T). The level of each of these myogenic factors was reduced in C2C12 myotubes when cultured with the regeneration extract for 4 days (Fig. 2B). Protein levels were judged to be reduced when the level of fluorescence was at or below background levels. Nuclear proteins, such as MyoD and myogenin, were often reduced to levels below the background staining of the cytoplasm, so that the nuclei appeared black, whereas the cytoplasm exhibited light background fluorescence. Reduced nuclear levels of MyoD and myogenin were observed, respectively, in 18% and 15% of the myotubes (Table 1). In addition, troponin T was reduced in the cytoplasm of 30% of the myotubes (Table 1). Reduced levels of all myogenic markers were most apparent by day 4. By contrast, MyoD and myogenin were consistently present in all control myotubes, whereas troponin T was identified in ≈94–97% of the controls (Table 1). All of these differences in protein levels were statistically significant (Table 1). These data indicate that newt regeneration extract can reduce the levels of muscle differentiation proteins in cultured mammalian myotubes.
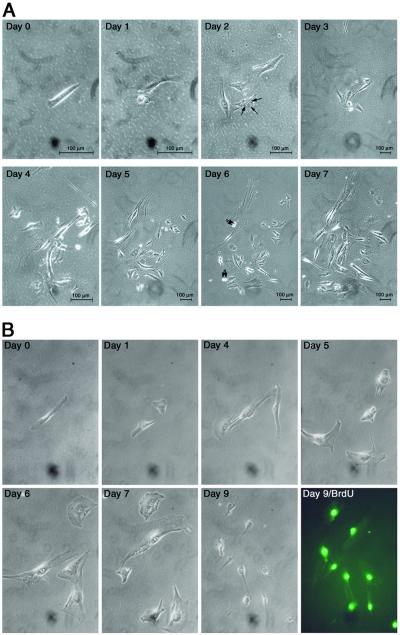
To determine whether newt regeneration extract contains factors that can induce cleavage of mammalian myotubes and subsequent cellular proliferation, we applied extract to C2C12 myotubes and followed isolated, individual myotubes by light microscopy. Only isolated myotubes in which no mononucleated cells were observed under low power were studied. The myotubes were plated at very low density (≈2 cells per mm2), cultured in DM containing 0.3 mg/ml regeneration extract on day 0, and individually photographed every 12–24 h to document cellular morphologic changes that occurred over a 10-day period. The medium and extract were changed daily. Cellular cleavage was noted by days 1–3 in 11% of myotubes (Fig. 3, Table 1), and cleavage was followed by cellular proliferation in half of these myotubes (Fig. 3). These cellular phenomena were not observed in any C2C12 myotubes cultured in DM alone or DM with 0.3 mg/ml nonregeneration limb extract (P = 0.0001). Thus, murine myotubes cultured with regeneration extract undergo cleavage to form smaller myotubes at nearly the same frequency as newt myotubes (11% vs. 16%). In addition, cleavage was often followed by cellular proliferation in the C2C12 myotubes. These data indicate that newt regeneration extract induces cleavage of mammalian myotubes and can promote proliferation of dedifferentiated mononucleated cells.
Figure 3.
Regeneration extract induces mouse myotube cleavage and cellular proliferation. (A) An isolated C2C12 myotube was treated daily for 7 days with DM containing regeneration extract. Day 0 shows the myotube cultured in DM before the addition of regeneration extract. Days 1–7 show the morphologic changes that occur as treatment continues for 7 days. On day 1, the myotube showed initial signs of fragmentation and cleavage. By day 2, the myotube cleaved to form four smaller fragments. Arrows point to three nuclei contained within a smaller myotube that resulted from the cleavage event. On days 3–5 the myotube continued to cleave to form numerous smaller cells. On day 6, there was evidence of cellular proliferation. Arrows point to cells progressing through cell division. On day 7, the cells continued to cleave and proliferate. (Scale bars, 100 μm.) (B) An example of a second isolated C2C12 myotube that was treated with newt regeneration extract for 9 days. Day 0, the myotube before treatment with the extract. Within 1 day of treatment, the myotube cleaved to form two smaller cellular products. Cleavage continued through day 9. On day 8 the cleaved cells were treated with BrdUrd, and on day 9 the mononucleated cells were assayed for BrdUrd incorporation (Day 9/BrdU).
Dedifferentiation Signal Is Comprised of Protein.
The dedifferentiation signal(s) found in the newt regeneration extract could belong to a number of different types of biomolecules, including proteins, lipids, nucleic acids, and polysaccharides. We performed several experiments to assign the dedifferentiation signal(s) to a particular biomolecular category. Our method of extraction reduced the probability that the dedifferentiation factor(s) contained lipids. During the extraction procedure, we removed the insoluble lipid layer following high-speed centrifugation; therefore, very little residual lipid remained in the extract. We also performed tests to determine whether the dedifferentiation factor was sensitive to freezing and thawing and whether it was heat labile. When the regeneration extract was subjected to two freeze–thaw cycles, its activity, as analyzed by BrdUrd incorporation, was reduced 3-fold when compared with extract that had been frozen and thawed only once. Three freeze–thaw cycles totally obliterated all activity. Boiling the extract for 5 min also eliminated all BrdUrd incorporation activity. When the regeneration extract was treated with 1% trypsin for 30 min at 37°C, the dedifferentiation signal was abolished. These results indicate that the dedifferentiation signal is comprised of protein. It is not known whether a single protein is responsible for the activity or whether conjugated proteins, such as glycoproteins, are involved.
Discussion
Cellular dedifferentiation, a phenomenon central to epimorphic regeneration in newts, is not normally observed in terminally differentiated mammalian myotubes. Mouse myotubes are incapable of reentering the cell cycle unless they have been genetically altered or treated with myoseverin, a microtubule-binding purine (20–27). We have previously demonstrated that ectopic expression of msx1, which encodes a homeobox-containing transcriptional repressor, can induce mouse myotubes to dedifferentiate to mononucleated cells that possess the properties of stem cells (28). Here, we show that unaltered mammalian myotubes can dedifferentiate when exposed to the same signals that induce dedifferentiation of newt cells during the early phases of limb regeneration. Mouse myotubes treated with a limb regeneration extract reenter the cell cycle, exhibit reduced levels of muscle differentiation proteins, and cleave to produce smaller myotubes or proliferating, mononucleated cells. We also demonstrate, for the first time, that cultured newt myotubes can respond to regeneration extract by reentering the cell cycle and by cleaving to form smaller myotubes or mononucleated cells. Thus, the cellular plasticity that was thought unique to urodeles can now be extended to mammals.
All of the nuclei within a given newt myotube responded concordantly to the newt limb regeneration extract by either reentering the cell cycle or by remaining in a quiescent state, whereas most often mouse myotube nuclei did not exhibit such concordance. In mouse myotubes that responded to the regeneration extract, ≈64% of the nuclei reentered the S phase, whereas the remaining nuclei retained their quiescence. Although we do not yet understand the reason for this S phase discordance within a single myotube, it must be noted that similar results were obtained following serum stimulation of newt/mouse hybrid myotubes (29). In the hybrid myotube study, the presence of the newt nuclei within the hybrid myotube made some, but not all, of the neighboring mouse nuclei competent to respond to serum and thrombin stimulation. This difference between newt and mouse myotubes may reflect the fact that newt myotubes appear to be primed for cellular dedifferentiation, as suggested by their response to serum or thrombin stimulation (30–32) and their minimal response to differentiation medium or nonregenerating limb extract in this present study.
We did not observe a size difference between myotubes that exhibited cell cycle reentry and those that did not. However, the procedure that we used to isolate myotubes selected for smaller multinucleated cells containing 4–23 nuclei (mean = 7.4 ± 4.5sd). Therefore, at this time, we do not know whether larger myotubes are capable of reentering the cell cycle in response to regeneration extract stimulation.
It has previously been demonstrated that cultured newt myotubes can dedifferentiate when transplanted back into the blastema of a regenerating newt limb (14, 15). Approximately 10–15% of the transplanted newt myotubes responded by cleaving to form mononucleated cells. In our study, newt regeneration extract induced cleavage of ≈16% of newt myotubes and 11% of the mouse myotubes, a finding that is remarkably consistent with the previous in vivo studies. The fact that the in vitro results recapitulate the in vivo studies suggest that the limb regeneration extract may have retained all of the dedifferentiation activity normally inherent in the regenerating limb and that the in vitro system may provide a reliable method for studying the dedifferentiation process.
Our findings are inconsistent with the interpretation that the proliferating cells arise from contaminating reserve cells in the myotube preparation. First, a careful photographic record was kept of the entire dedifferentiation process and no unexplained mononucleated cells suddenly appeared, as might be expected if hidden mononucleated cells were responsible for our observations. Second, we observed myotubes as they were beginning to pull apart and cleave (Fig. 3A, day 1). Third, cleavage events often lead to smaller multinucleated myotubes, which cannot be mistaken for contaminating mononucleated cells (Fig. 3A, day 2). Finally, if unobserved mononucleated cells were responsible for our observations, we would expect approximately the same number of apparent cleavage events in the controls. As noted above, no cleavage events of control myotubes were observed, whereas 11% of the treated myotubes cleaved. This difference in cleavage frequency is statistically significant at P = 0.0001. Therefore, we conclude that newt limb regeneration extract induces mouse myotubes to cleave to form smaller myotubes or proliferating, mononucleated cells.
These studies indicate that mammalian cells have retained the intracellular signaling pathways required for dedifferentiation. Therefore, it is likely that mammals fail to exhibit in vivo cellular dedifferentiation because they lack the signal that initiates the dedifferentiation process. These conclusions differ from those reached in a previous study (32). Based on the observation that newt myotubes reenter S phase when stimulated with serum or thrombin, whereas mammalian myotubes are refractory to these treatments (30–32), it was suggested that mammalian myotubes were intrinsically incapable of responding to dedifferentiation signals. Our results can be reconciled with these previous studies by recognizing that our work was performed by using crude extracts from the entire regenerating portion of the limb, whereas the previous studies (32) were performed by using either serum, crude thrombin, fractionated thrombin, or mixtures of thrombin and serum. The serum factor, which is thought to be a thrombin-activated ligand, induces newt myotubes to reenter the cell cycle but does not promote a continuation of the cell cycle through mitosis to produce mononucleated cells. Therefore, other factors found in the regenerating limb are required for complete dedifferentiation and may either cooperate with the thrombin-activated ligand or entirely substitute for it.
Our data suggest that proteins are required components of the dedifferentiation signal. This conclusion is based on the following observations: (i) the signal is contained in the soluble fraction of the extract; (ii) it is heat labile; (iii) it is sensitive to repeated freeze–thaw cycles; and (iv) the signal is abolished by treatment with the protease trypsin. We speculate that these factors are soluble extracellular proteins acting as ligands to activate receptors that transduce dedifferentiation signals through receptive cells, such as myotubes.
During newt limb regeneration, cellular dedifferentiation is followed by proliferation and redifferentiation of the blastemal cells. The patterning and differentiation signals involved in reforming the lost limb may be regulated by the regeneration blastema. If so, the primary obstacle preventing induced epimorphic regeneration in mammals may be the inability to form a mass of dedifferentiated cells from local tissues. If this hypothesis is correct, identifying essential dedifferentiation factors may lead to methods for enhancing the regenerative process in mammals.
Acknowledgments
We thank Jeremy Brockes for the newt A1 limb cells. We thank Diana Stafforini and Kirk Thomas for helpful suggestions. The Howard Hughes Medical Institute supported this work.
Abbreviation
- DM
differentiation medium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Morgan T H. Regeneration. New York: Columbia University; 1901. [Google Scholar]
- 2.Becker R O, Chapin S, Sherry R. Nature (London) 1974;248:145–147. doi: 10.1038/248145a0. [DOI] [PubMed] [Google Scholar]
- 3.Davis B M, Ayers J L, Koran L, Carlson J, Anderson M C, Simpson S B., Jr Exp Neurol. 1990;108:198–213. doi: 10.1016/0014-4886(90)90124-b. [DOI] [PubMed] [Google Scholar]
- 4.Brockes J P. Science. 1997;276:81–87. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- 5.Zottoli S J, Bentley A P, Feiner D G, Hering J R, Prendergast B J, Rieff H I. Prog Brain Res. 1994;103:219–228. doi: 10.1016/s0079-6123(08)61138-3. [DOI] [PubMed] [Google Scholar]
- 6.Johnson S L, Weston J A. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen R N, Tassava R A. Dev Dyn. 2000;217:216–224. doi: 10.1002/(SICI)1097-0177(200002)217:2<216::AID-DVDY8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Chalkley D T. J Morphol. 1954;94:21–70. [Google Scholar]
- 9.Thornton C S. J Exp Zool. 1957;134:357–382. doi: 10.1002/jez.1401340209. [DOI] [PubMed] [Google Scholar]
- 10.Bodemer C W, Everett N B. Dev Biol. 1959;1:327–342. [Google Scholar]
- 11.Hay E D, Fischman D A. Dev Biol. 1961;3:26–59. doi: 10.1016/0012-1606(61)90009-4. [DOI] [PubMed] [Google Scholar]
- 12.Thornton C S, Thornton M T. Experentia. 1965;21:146–151. [Google Scholar]
- 13.Steen T P. J Exp Zool. 1968;167:49–78. doi: 10.1002/jez.1401670105. [DOI] [PubMed] [Google Scholar]
- 14.Lo D C, Allen F, Brockes J P. Proc Natl Acad Sci USA. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Velloso C P, Imokawa Y, Brockes J P. Dev Biol. 2000;218:125–136. doi: 10.1006/dbio.1999.9569. [DOI] [PubMed] [Google Scholar]
- 16.Andres V, Walsh K. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh K, Perlman H. Curr Opin Genet Dev. 1997;7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 18.Ferretti P, Brockes J P. J Exp Zool. 1988;247:77–91. doi: 10.1002/jez.1402470111. [DOI] [PubMed] [Google Scholar]
- 19.Guo K, Wang J, Andres V, Smith R C, Walsh K. Mol Cell Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endo T, Nadal-Ginard B. In: Cellular and Molecular Biology of Muscle Development. Stockdale F, Kedes L, editors. New York: Liss; 1989. pp. 95–104. [Google Scholar]
- 21.Iujvidin S, Fuchs O, Nudel U, Yaffe D. Differentiation. 1990;43:192–203. doi: 10.1111/j.1432-0436.1990.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 22.Schneider J W, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 23.Tiainen M, Spitkovsky D, Jansen-Durr P, Sacchi A, Crescenzi M. Mol Cell Biol. 1996;16:5302–5312. doi: 10.1128/mcb.16.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novitch B G, Mulligan G J, Jacks T, Lassar A B. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo T, Nadal-Ginard B. J Cell Sci. 1998;111:1081–1093. doi: 10.1242/jcs.111.8.1081. [DOI] [PubMed] [Google Scholar]
- 26.Latella L, Sacco A, Pajalunga D, Tiainen M, Macera D, D'Angelo M, Felici A, Sacchi A, Crescenzi M. Mol Cell Biol. 2001;21:5631–5643. doi: 10.1128/MCB.21.16.5631-5643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosania G R, Chang Y T, Perez O, Sutherlin D, Dong H, Lockhart D J, Schultz P G. Nat Biotechnol. 2000;18:304–308. doi: 10.1038/73753. [DOI] [PubMed] [Google Scholar]
- 28.Odelberg S J, Kollhoff A, Keating M T. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 29.Velloso C P, Simon A, Brockes J P. Curr Biol. 2001;11:855–858. doi: 10.1016/s0960-9822(01)00234-2. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka E M, Gann A A, Gates P B, Brockes J P. J Cell Biol. 1997;136:155–165. doi: 10.1083/jcb.136.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka E M, Brockes J P. Wound Repair Regen. 1998;6:371–381. doi: 10.1046/j.1524-475x.1998.60413.x. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka E M, Drechsel D N, Brockes J P. Curr Biol. 1999;9:792–799. doi: 10.1016/s0960-9822(99)80362-5. [DOI] [PubMed] [Google Scholar]