Abstract
Objective
Traditional prognostic factors have proved insufficient to account for heterogeneity in the clinical behavior of chronic lymphocytic leukemia (CLL). Cryptochrome-1 (CRY-1) is a circadian clock gene essential in maintaining the circadian rhythm and regulating cell proliferation. We evaluated CRY-1 gene expression in CLL and addressed its putative role as a prognostic indicator for the clinical course of CLL.
Materials and Methods
A total of 100 CLL patients at diagnosis were studied for CRY-1 gene expression by real-time reverse-transcription polymerase chain reaction and were followed for assessment of time to first treatment (TFT).
Results
CRY-1 was expressed in 94% of the CLL patients at diagnosis. The median CRY-1 relative gene expression level (0.006) stratified patients into high and low expression groups. Forty of 100 (40%) CLL patients showed high CRY-1, 54/100 (54%) showed low CRY-1, and 6/100 (6%) had undetectable CRY-1 gene expression. High CRY-1 gene expression was concordant with CD38+, Zap-70+, and double CD38+Zap-70+ expression; unfavorable/intermediate cytogenetics; unmutated immunoglobulin heavy-chain variable-region gene; and diffuse marrow infiltration. The high CRY-1 gene expression patient group exhibited shorter TFT than the patients with low CRY-1 gene expression. A Cox proportional hazard regression model identified CRY-1 gene expression to be independently predictive for TFT.
Conclusion
CRY-1 is differentially expressed among CLL patients, stratifying them into low-risk and high-risk groups. CRY-1 gene expression could constitute a reliable prognostic indicator for CLL progression, complementing the role of standard well-established prognostic factors. CRY-1 gene expression could be employed as a prognostic indicator for disease progression during the initial prognostic work-up and follow-up for CLL patients.
Keywords: Chronic lymphocytic leukemia, Cryptochrome-1, Circadian genes, Time to first treatment, Prognosis, Real-time polymerase chain reaction, CD38, Zap-70
Abstract
Amaç
Geleneksel prognostik faktörler, kronik lenfositik löseminin (KLL) klinik davranışındaki heterojenitenin nedenini açıklamak için yetersiz bulunmaktadır. Kriptokrom-1 (CRY-1) sirkadiyen ritmin sağlanması ve hücre proliferasyonu regülasyonunda gerekli olan bir sirkadiyen saat genidir. Biz, KLL’de CRY-1 gen ekspresyonunu değerlendirdik ve KLL’nin klinik seyri için prognostik gösterge olarak varsayılan rolünü irdeledik.
Gereç ve Yöntemler
Toplam 100 KLL hastası tanı sırasında kantitatif gerçek zamanlı ters transkripsiyon polimeraz zincir reaksiyonu ile CRY-1 gen ekspresyonu için incelendi ve ilk tedaviye kadar geçen zamanın (TFT) değerlendirilmesi için takip edildi.
Bulgular
KLL hastalarının %94’ünde tanı anında CRY-1 ekspresyonu mevcuttu. Medyan CRF-1 rölatif gen ekspresyon düzeyi (0,006) hastaları yüksek ve düşük ekspresyon gruplarına ayırmaktadır. Yüz hastanın 40’ı (%40) yüksek CRY-1, 54/100’ü (%54) düşük CRY-1, ve 6/100’ü (%6) tespit edilemez CRY-1 gen ekspresyonu gösterdi. Yüksek CRY-1 gen ekspresyonu CD38+, Zap-70+ ve çift CD38+Zap-70+ ekspresyonu; olumsuz/orta sitogenetik; mutasyona uğramamış immünoglobulin ağır zincir değişken bölge geni ve diffüz kemik iliği infiltrasyonu ile uyumlu idi. Yüksek CRY-1 gen ekspresyonu olan hasta grubu, düşük CRY-1 gen ekspresyonu olan hastalardan daha kısa TFT gösterdi. Cox oransal hazard regresyon modeli CRY-1 gen ekspresyonunu TFT için bağımsız prediktif olarak tanımlamıştır.
Sonuç
CRY-1, KLL hastalarında farklı olarak eksprese edilerek onları düşük-risk ve yüksek-risk gruplarına ayırmaktadır. CRY-1 gen ekspresyonu, KLL progresyonu için iyi-belirlenmiş standart prognostik planlamada ve KLL hastalarının takibinde hastalık progresyonu için prognostik bir gösterge olarak kullanılabilir.
Introduction
Chronic lymphocytic leukemia (CLL) is a lymphoproliferative neoplasm defined by proliferation and accumulation of morphologically mature, immunologically dysfunctional monoclonal B cells in the peripheral blood (PB), bone marrow (BM), and lymphatic tissues [1].
The clinical course of CLL is heterogeneous and difficult to predict; some patients may exhibit rapid disease progression while others may live for decades without requiring treatment [2]. Early treatment of the latter group would risk the development of therapy-related complications that might affect the quality of life and/or survival. Assigning markers that reliably stratify patients into good-risk or poor-risk disease groups could help in evaluating the potential benefit of early treatment and assist risk-adapted treatment strategies [3]. Traditional prognostic factors have proved insufficient to account for heterogeneity in the clinical behavior of CLL, indicating a need for further prognostic indicators that can better correlate with patients’ clinical outcome and survival [4,5].
The immunoglobulin heavy-chain variable-region (IgHV) gene mutational status correlates with clinical behavior and represents a robust prognostic indicator for CLL. However, IgHV gene sequencing is complicated and time-consuming for routine laboratories; hence, exploring an alternative for IgHV gene mutations is a priority [6].
The circadian machinery comprises various genes that show functional interplays with cell cycle regulators. Aberrant expression of circadian clock genes can therefore lead to aberrant expression of downstream target genes responsible for cell proliferation and apoptosis, resulting in the emergence of different types of cancers, including CLL [6,7,8,9].
The Cryptochrome-1(CRY-1) gene (12q23-q24.1) is a member of the circadian clock essential to the maintenance of circadian rhythm. In addition to its circadian function, it also acts as a transcriptional regulator for several genes with roles in cell metabolism and proliferation [10].
Accordingly, we aimed to evaluate CRY-1 gene expression in CLL patients and address the gene’s putative role as a prognostic indicator for the clinical course of CLL.
Materials and Methods
This prospective cohort study included 100 newly diagnosed and untreated CLL patients who attended the Hematology/Oncology Clinic of the Ain Shams University Hospitals. Patients were selected on the basis of standard clinical, hematologic, and immunophenotypic characteristics for the diagnosis of CLL [11]. These CLL patients comprised 64 males and 36 females; the male to female ratio was 1.8:1 and median age was 61 years (interquartile range (IQR): 55-68 years). All patients were followed after diagnosis for assessment of the time to first treatment (TFT) (median: 20 months; range: 10-24 months).
TFT is the time from diagnosis to the start of therapy or the last follow-up. The criteria for starting treatment are: (i) the existence of constitutional symptoms; (ii) progressive marrow failure indicated by anemia and/or thrombocythemia; (iii) autoimmune anemia and/or thrombocytopenia not responding to steroids; (iv) massive (more than 6 cm below the costal margin) or progressive splenomegaly; (v) massive nodes or nodal clusters (more than 10 cm in the longest diameter) or progressive lymphadenopathy; (vi) progressive lymphocytosis by more than 50% over 2 months or a doubling time of less than 6 months [12]. Informed consent was provided prior to patient enrollment. The study protocol was endorsed by the Ethical Committee for Human Research of Ain Shams University and complied with the Helsinki Declaration of 1975, as revised in 2002. The clinicopathologic characteristics of CLL patients at diagnosis are presented in Table 1.
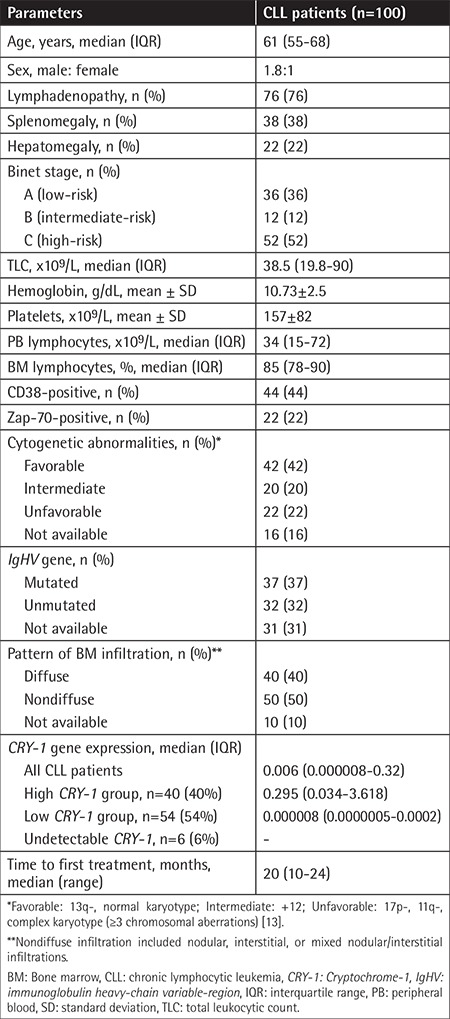
Table 1. Clinicopathologic characteristics of chronic lymphocytic leukemia patients at diagnosis.
Sampling
PB and BM samples were collected in K-EDTA (1.5 mg/mL) for morphologic, immunophenotypic, and molecular analyses and in lithium heparin for cytogenetic analysis. PB samples were used for flow cytometric immunophenotyping and real-time reverse-transcription polymerase chain reaction (qRT-PCR) for quantification of CRY-1 gene expression, while PB or BM samples, when available, were used for the cytogenetic analysis.
Flow Cytometric Immunophenotyping
Immunophenotyping using the standard panel for chronic lymphoproliferative disease (CD5, CD19, CD23, FMC7, CD20, CD38, CD79b, CD10, CD25, CD103, CD123, kappa, lambda, surface immunoglobulin G) (Beckman Coulter, Miami, FL, USA), along with ZAP-70 (BioLegend, San Diego, CA, USA), was performed with the EPICS XL Flow Cytometer (Coulter Electronics, Hialeah, FL, USA). The positivity threshold was defined as the expression of the marker by ≥30% of the B lymphocytes [13]; however, for CD38 and Zap-70 a cut-off value of ≥20% and ≥10% of the B lymphocytes was considered positive, respectively [14].
Fluorescence In Situ Hybridization
Probes for 13q-, 17p-, 11q-, and +12 (Vysis, Downers Grove, IL, USA) were used. Patients were stratified into cytogenetic-based risk groups as follows: favorable, 13q- or normal karyotype; intermediate, +12; and unfavorable, 17p-, 11q-, or complex karyotype (≥3 chromosomal aberrations) [13].
qRT-PCR for Quantification of CRY-1 Gene Expression
qRT-PCR amplification was done using gene expression sets for the CRY-1 gene (Homo sapiens; Hs00172734-m1 TaqMan® Gene Expression Assays), TaqMan b-actin control reagents for the b-actin reference gene, and TaqMan Universal PCR Master Mix (all from Applied Biosystems, Foster City, CA, USA). Total RNA was extracted from PB samples using a QIAamp® RNA blood kit (QIAGEN, Valencia, CA, USA), while cDNA was synthesized using a QuantiTectReverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. The cDNA was stored at -20 °C until it was used.
cDNA samples were employed in synthesizing PCR products using sequence-specific primers and TaqMan oligonucleotide fluorescence-labeled probes: FAM for the CRY-1 gene (Hs_CRY-1_1_FAM Quantifast Probe Assay, QIAGEN) and MAX for the b-actin gene (Hs_b-actin_1_MAX Quantifast Probe Assay, QIAGEN). Both probes were labeled with Iowa Black Fluorescent Quencher. Each PCR included all the necessary reagents and 50 ng of cDNA in a final volume of 25 µL. A negative control (cDNA replaced by nuclease-free water) was included in each assay. The reaction protocol comprised 40 cycles of heating at 95 °C for 5 min (hot-start PCR), then heating at 95 °C for 30 s (denaturation), and then heating at 60 °C for 30 s (annealing/extension). PCR and data analysis were carried out with Stratagene Mx3000P (Stratagene Inc., La Jolla, CA, USA). Undetectable CRY-1 was noted for cases in which cycle threshold (CT) values exceeded the 40th cycle.
CRY-1 expression levels in unknown samples were calculated by relative quantification using the ΔΔCT method, which relies on comparison of CT values of CRY-1 (target gene) to b-actin (reference gene) in unknown and normal calibrator samples. The results were expressed as the fold change in gene expression normalized to the reference gene and relative to the calibrator [15].
Statistical Analysis
Analysis of data was done using SPSS 17 for Windows 7 (SPSS Inc., Chicago, IL, USA). Categorical variables are presented as frequency and percentage, compared using the chi-square (χ2) test. Continuous variables are presented as mean ± standard deviation or median and IQR for parametric and nonparametric variables, respectively. Student and Mann-Whitney U tests were employed for comparing continuous parametric and nonparametric variables between two groups, respectively. Kaplan-Meier curves for TFT were plotted and the log-rank test was used to compare TFT distributions between high and low CRY-1 gene expression groups. Multivariate Cox proportional hazard regression analysis (hazard ratio, HR) was employed to identify the independent association of CRY-1 gene expression with TFT. Values of p<0.05 and p<0.01 were taken to be significant and highly significant, respectively.
Results
CRY-1 Gene Expression in CLL Patients at Diagnosis
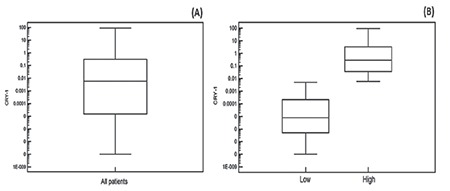
The CRY-1gene was expressed in 94 of 100 (94%) CLL patients at diagnosis (median: 0.006; IQR: 0.000008-0.32) (Table 1). The median CRY-1 relative gene expression level (0.006) was employed as the cut-off value for stratifying high and low CRY-1 gene expression groups. Accordingly, 40 of 100 (40%) CLL patients showed high CRY-1 gene expression (≥0.006) (median: 0.295; IQR: 0.034-3.618) and 54 of 100 (54%) CLL patients showed low CRY-1 gene expression (<0.006) (median: 0.000008; IQR: 0.0000005-0.0002). Six of 100 (6%) CLL patients had undetectable CRY-1 expression (Table 1, Figure 1).
Figure 1. Cryptochrome-1 gene expression in chronic lymphocytic leukemia patients at diagnosis: (A) all chronic lymphocytic leukemia patients, (B) low and high Cryptochrome-1 gene expression groups. CRY-1: Cryptochrome-1.
High and Low CRY-1 Gene Expression and Clinicopathologic Characteristics of CLL Patients at Diagnosis
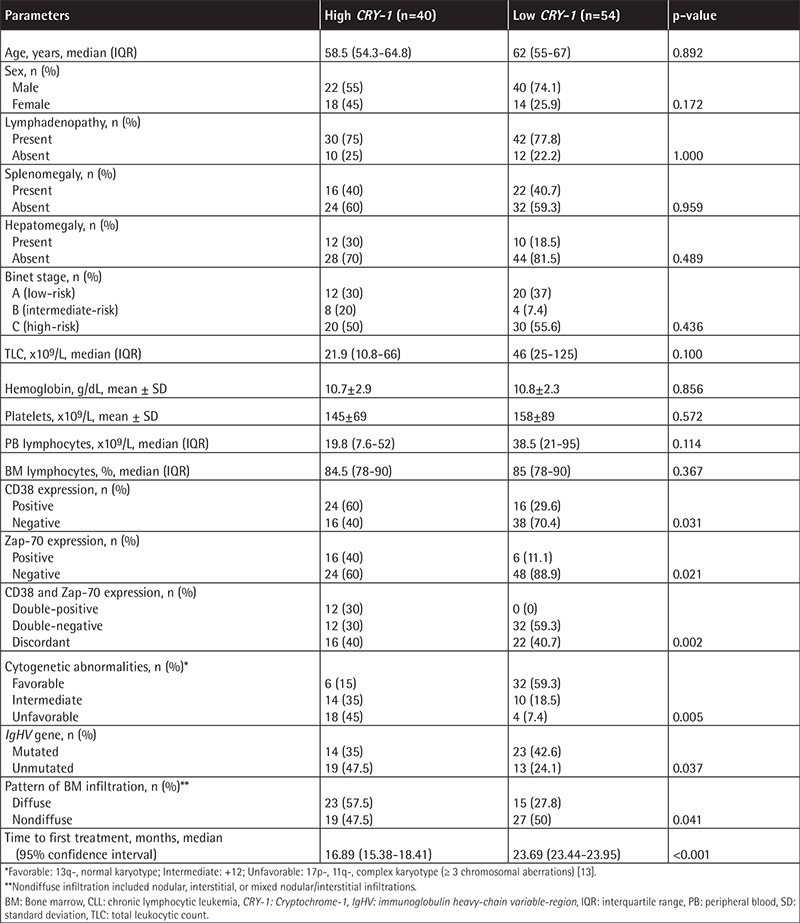
High CRY-1 gene expression was significantly related to CD38+, Zap-70+, and double CD38+Zap-70+ expression; unfavorable/intermediate cytogenetics; unmutated IgHV gene; and diffuse BM infiltration in trephine biopsy (p<0.05). On the contrary, low CRY-1 gene expression was significantly related to CD38-, Zap-70-, and double CD38-Zap-70- expression and favorable cytogenetics (p<0.05). No further significance was found between high and low CRY-1 groups regarding other studied clinicopathologic parameters (p>0.05) (Table 2).
Table 2. High and low Cryptochrome-1 gene expression and clinicopathologic characteristics of chronic lymphocytic leukemia patients at diagnosis.
CRY-1 Gene Expression and TFT of CLL Patients
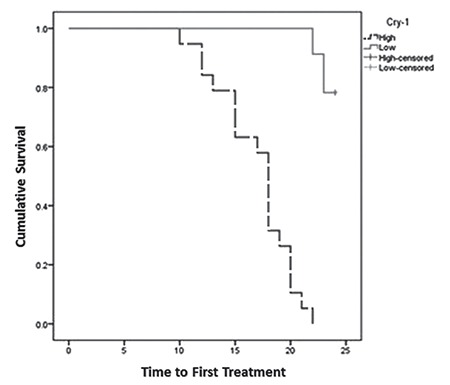
Using the Kaplan-Meier method, the TFT for the high CRY-1 gene expression group (median: 16.89 months; 95% confidence interval (CI): 15.38-18.41) was found to be significantly shorter than the TFT for the low CRY-1 gene expression group (median: 23.69 months; 95% CI: 23.44-23.95) (p<0.001) (Table 2, Figure 2).
Figure 2. Kaplan-Meier curve showing the time to first treatment for chronic lymphocytic leukemia patients based on Cryptochrome-1 gene expression levels. CRY-1: Cryptochrome-1.
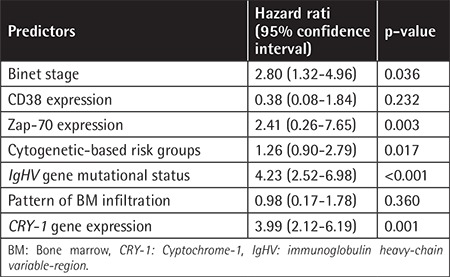
Multivariate Cox hazard regression analysis denoted CRY-1 gene expression to be independently predictive for TFT (HR: 3.99; 95% CI: 2.12-6.19; p=0.001). The same was also shown for IgHV mutational status (p<0.001), Binet stage, Zap-70, and cytogenetic-based risk groups (p<0.05) (Table 3).
Table 3. Predictors for earlier time to first treatment according to Cox proportional hazard regression model.
Discussion
Diverse circadian genes are known to be correlated with poor prognosis when aberrantly overexpressed in several cancers, including CRY-1 [16]. In this work, the CRY-1 gene was expressed in 94% of the studied CLL patients at diagnosis. The median relative CRY-1 gene expression level (0.006) was employed to stratify patients into high and low expression groups. High CRY-1 was related to CD38+, Zap-70+, and double CD38+Zap-70+ expression while low CRY-1 was associated with CD38-, Zap-70-, and double CD38-Zap-70- expression and discordant CD38/Zap-70 was comparable between the high and low CRY-1 gene expression groups. Similarly, a significant concordance was previously shown between elevated CRY-1 transcripts and high-risk patients, as defined by CD38+ and/or unmutated IgHV genes, compared with their low-risk counterparts, as defined by CD38- and/or mutated IgHV genes [6,10,17]. Moreover, elevated CRY-1 transcripts in high-risk patients were found to be associated with Zap-70+ and double CD38+Zap-70+ expression [6,17].
Unlike our findings, cases of discordant CD38/Zap-70 expression (intermediate-risk) showed CRY-1 gene expression levels comparable to those of the high-risk group in a previous study [17]. This discrepancy might be attributed to two major pitfalls that could hamper the prognostic use of CD38: first, the debate about the threshold that indicates CD38+ expression for defining patient prognosis, and second, the probability that CD38 expression may be unstable and differ over time, provoking the concern that CD38 may be an unreliable marker in CLL [18]. Another possible explanation is the variation in the positivity threshold for Zap-70 in the literature [6,14,17]. In accordance with Deaglio et al. [14], we used a cut-off value of ≥10% to indicate Zap-70+ expression. The threshold values for CD38 and Zap-70 used by Deaglio et al. [14] were functional thresholds, below which CD38-mediated Zap-70 tyrosine phosphorylation was undetectable.
We found an association between high CRY-1 gene expression and both unfavorable/intermediate cytogenetic abnormalities and diffuse BM infiltration. In accordance, high CRY-1 gene expression was detected in association with 17p- and +12, while low CRY-1 gene expression was found in a case with 13q- as the sole abnormality [6]. Interestingly, since the CRY-1 gene is located at chromosome 12q23-q24.1, an increased copy number was detected in patients with +12 [10]. On the other hand, we could not find further association between CRY-1 gene expression levels and other studied clinicopathologic parameters. Yu et al. [19] also stated that neither age nor sex was associated with CRY-1 gene expression level.
In our study, CRY-1 was differentially expressed among CLL patients with mutated and unmutated IgHV genes, being overexpressed in the unmutated group. Other authors advocated the association between high CRY-1 gene expression and the unmutated IgHV gene [6,10,17]. A cut-off value of 0.090 for CRY-1 gene expression in CD19+ B cells of CLL patients was previously determined [6] and considered the best cut-off for segregating patients with mutated and unmutated IgHV genes (sensitivity: 95%; specificity: 92%; area under the curve: 0.963). The data of that study showed 92.8% concordance between CRY-1 expression and IgHV mutational status.
We observed a shorter median TFT in the group with high CRY-1 expression (16.89 months) compared with that with low CRY-1 gene expression (23.69 months). Lewintre et al. [6] reported that high CRY-1 gene expression was significantly related to shorter median progression-free survival of 63.2 months (95% CI: 48.2-78.2), compared to a median of 139 months (95% CI: 133.1-146.4) for the low CRY-1 gene expression group (p<0.0001). Using multivariate Cox hazard regression, we found that CRY-1 gene expression was independently predictive for TFT (p=0.001). Eisele et al. [17] employed univariate Cox hazard regression and reported that CRY-1 could predict the clinical outcome of CLL patients as measured by TFT.
Of note, Hanoun et al. [10] reported that the methylation status of the CRY-1 promoter revealed a considerable prognostic influence in CLL, whereby patients with hypermethylated CRY-1 promoters showed significantly longer treatment-free survivals compared with their hypomethylated counterparts. Unexpectedly, they reported comparable levels of CRY-1 mRNA in high-risk CLL and normal donor B cells. Thus, they postulated that expression differences of the CRY-1 gene in CLL could be attributed to an underexpression of CRY-1 in low-risk cases of CLL rather than an overexpression in the high-risk group.
Conclusion
The circadian clock gene CRY-1 is differentially expressed among CLL patients, stratifying them into low-risk and high-risk groups. CRY-1 gene expression could constitute a reliable prognostic indicator for CLL progression, complementing the role of standard well-established prognostic factors. Accordingly, CRY-1 gene expression could be employed as a prognostic indicator for disease progression during the initial prognostic work-up and follow-up for CLL patients.
Evaluation of CRY-1 expression with respect to the overall survival of CLL patients is warranted. Study of CRY-1 gene methylation status and stability of expression at different time points throughout the course of CLL represents an interesting area for future research. Clinical trials for assessment of therapeutic modalities targeting the CRY-1 gene in larger cohorts of CLL patients are worthwhile.
Footnotes
Ethics
Ethics Committee Approval: The study protocol was endorsed by the Ethical Committee for Human Research of Ain Shams University.
Informed Consent: Informed consent was provided prior to patient enrollment.
Authorship Contributions
Surgical and Medical Practices: D.M.H., D.S.E., M.M.A.; Concept: D.M.H., D.S.E.; Design: D.M.H., D.S.E.; Data Collection or Processing: M.M.A.; Analysis or Interpretation: D.M.H., D.S.E., M.M.A.; Literature Search: D.S.E., M.M.A.; Writing: D.M.H., D.S.E., M.M.A.
Conflict of Interest: The authors of this paper have no conflicts of interest, including specific financial interests, relationships, and/or affiliations relevant to the subject matter or materials included.
References
- 1.Adena M, Houltram J, Mulligan SP, Todd C, Malanos G. Modelling the cost effectiveness of rituximab in chronic lymphocytic leukaemia in first-line therapy and following relapse. Pharmacoeconomics. 2014;32:193–207. doi: 10.1007/s40273-013-0125-7. [DOI] [PubMed] [Google Scholar]
- 2.Shanafelt TD. Predicting clinical outcome in CLL: how and why. Hematology Am Soc Hematol Educ Program. 2009:421–429. doi: 10.1182/asheducation-2009.1.421. [DOI] [PubMed] [Google Scholar]
- 3.Rassenti LZ, Jain S, Keating MJ, Wierda WG, Grever MR, Byrd JC, Kay NE, Brown JR, Gribben JG, Neuberg DS, He F, Greaves AW, Rai KR, Kipps TJ. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–1930. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abruzzo LV, Barron LL, Anderson K, Newman RJ, Wierda WG, O’Brien S, Ferrajoli A, Luthra M, Talwalkar S, Luthra R, Jones D, Keating MJ, Coombes KR. Identification and validation of biomarkers of IgVH mutation status in chronic lymphocytic leukemia using microfluidics quantitative real-time polymerase chain reaction technology. J Mol Diagn. 2007;9:546–555. doi: 10.2353/jmoldx.2007.070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg JB, Volkheimer AD, Chen Y, Beasley BE, Jiang N, Lanasa MC, Friedman D, Vaccaro G, Rehder CW, Decastro CM, Rizzieri DA, Diehl LF, Gockerman JP, Moore JO, Goodman BK, Levesque MC. Clinical and molecular predictors of disease severity and survival in chronic lymphocytic leukemia. Am J Hematol. 2007;82:1063–1070. doi: 10.1002/ajh.20987. [DOI] [PubMed] [Google Scholar]
- 6.Lewintre EJ, Martín CR, Ballesteros CG, Montaner D, Rivera RF, Mayans JR, García-Conde J. Cryptochrome-1 expression: a new prognostic marker in B-cell chronic lymphocytic leukemia. Haematologica. 2009;94:280–284. doi: 10.3324/haematol.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salavaty A. Carcinogenic effects of circadian disruption: an epigenetic viewpoint. Chin J Cancer. 2015;34:375–383. doi: 10.1186/s40880-015-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 9.Rana S, Munawar M, Shahid A, Malik M, Ullah H, Fatima W, Mohsin S, Mahmood S. Deregulated expression of circadian clock and clock-controlled cell cycle genes in chronic lymphocytic leukemia. Mol Biol Rep. 2014;41:95–103. doi: 10.1007/s11033-013-2841-7. [DOI] [PubMed] [Google Scholar]
- 10.Hanoun M, Eisele L, Suzuki M, Greally JM, Hüttmann A, Aydin S, Scholtysik R, Klein-Hitpass L, Dührsen U, Dürig J. Epigenetic silencing of the circadian clock gene CRY1 is associated with an indolent clinical course in chronic lymphocytic leukemia. PLoS One. 2012;7:e34347. doi: 10.1371/journal.pone.0034347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, Kipps TJ; International Workshop on Chronic Lymphocytic Leukemia. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute- Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tecchio C, Nichele I, Mosna F, Zampieri F, Leso A, Al-Khaffaf A, Veneri D, Andreini A, Pizzolo G, Ambrosetti A. A proliferation-inducing ligand (APRIL) serum levels predict time to first treatment in patients affected by B-cell chronic lymphocytic leukemia. Eur J Haematol. 2011;87:228–234. doi: 10.1111/j.1600-0609.2011.01650.x. [DOI] [PubMed] [Google Scholar]
- 13.Cuneo A, Rigolin GM, Bigoni R, De Angeli C, Veronese A, Cavazzini F, Bardi A, Roberti MG, Tammiso E, Agostini P, Ciccone M, Della Porta M, Tieghi A, Cavazzini L, Negrini M, Castoldi G. Chronic lymphocytic leukemia with 6q- shows distinct hematological features and intermediate prognosis. Leukemia. 2004;18:476–483. doi: 10.1038/sj.leu.2403242. [DOI] [PubMed] [Google Scholar]
- 14.Deaglio S, Vaisitti T, Aydin S, Bergui L, D’Arena G, Bonello L, Omedé P, Scatolini M, Jaksic O, Chiorino G, Efremov D, Malavasi F. CD38 and ZAP-70 are functionally linked and mark CLL cells with high migratory potential. Blood. 2007;110:4012–4021. doi: 10.1182/blood-2007-06-094029. [DOI] [PubMed] [Google Scholar]
- 15.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 16.Birky TL, Bray MS. The contribution of circadian rhythms to cancer formation and mortality. Atlas Genet Cytogenet Oncol Haematol. 2014;18:133–145. [Google Scholar]
- 17.Eisele L, Prinz R, Klein-Hitpass L, Nückel H, Lowinski K, Thomale J, Moeller LC, Dührsen U, Dürig J. Combined PER2 and CRY1 expression predicts outcome in chronic lymphocytic leukemia. Eur J Haematol. 2009;83:320–327. doi: 10.1111/j.1600-0609.2009.01287.x. [DOI] [PubMed] [Google Scholar]
- 18.Ghia P, Guida G, Scielzo C, Geuna M, Caligaris-Cappio F. CD38 modifications in chronic lymphocytic leukemia: are they relevant? Leukemia. 2004;18:1733–1735. doi: 10.1038/sj.leu.2403504. [DOI] [PubMed] [Google Scholar]
- 19.Yu H, Meng X, Wu J, Pan C, Ying X, Zhou Y, Liu R, Huang W. Cryptochrome 1 overexpression correlates with tumor progression and poor prognosis in patients with colorectal cancer. PLoS One. 2013;8:e61679. doi: 10.1371/journal.pone.0061679. [DOI] [PMC free article] [PubMed] [Google Scholar]


