Abstract
The spatial arrangement of COPII coat protein subunits was analyzed by crosslinking to an artificial membrane surface and by electron microscopy of coat proteins and coated vesicle surfaces. The efficiency of COPII subunit crosslinking to phospholipids declined in order of protein recruitment to the coat: Sar1p > Sec23/24p ≫ Sec13/31p. Deep-etch rotary shadowing and electron microscopy were used to explore the COPII subunit structure with isolated proteins and coated vesicles. Sec23/24 resembles a bow tie, and Sec13/31p contains terminal bilobed globular structures bordering a central rod. The surface structure of COPII vesicles revealed a coat built with polygonal units. The length of the side of the hexagonal/pentagonal units is close to the dimension of the central rod-like segment of Sec13/31. Partially uncoated profiles revealed strands of Sec13/31p stripped from the vesicle surface. We conclude that the coat subunits form layers displaced from the membrane surface in reverse order of addition to the coat.
Keywords: coat proteins‖liposome‖endoplasmic reticulum membrane‖transport vesicle
Intracellular trafficking of proteins along the secretory pathway is mediated by the sequential movement of transport vesicles between successive membrane organelles (1). The transport vesicles are generated by the action of coat proteins (1, 2). To date, three classes of coated vesicles are well characterized. COPII-coated vesicles that mediate the anterograde transport from the endoplasmic reticulum to the Golgi apparatus, COPI-coated vesicles that mediate the retrograde transport from the Golgi apparatus to the endoplasmic reticulum and the transport between the Golgi cisternae, and clathrin-coated vesicles that mediate several transport steps in the late secretory pathway (1, 2). Clathrin-coated vesicles can be classified into at least two distinct subclasses, one of which, the clathrin/AP-1 coat, comprises three components: a small GTP-binding protein (Arf1), the AP-1 adaptor, and clathrin. The COPII coat is assembled from another set of three components: the small GTP-binding protein (Sar1), the Sec23/24p complex, and the Sec13/31p complex. Unlike these two coats, the COPI coat is formed from two separable constituents, Arf and coatomer.
We have analyzed COPII vesicles as a model to understand how transport vesicles are formed during the protein transport process. Our previous in vitro analysis with purified coat proteins indicated that vesicle budding is initiated by the association of the GTP form of Sar1p to the membrane and followed by binding of Sec23/24p and then Sec13/31p, which induce the formation of COPII transport vesicles (3, 4). Here we analyzed biochemically the distance of each protein subunit from the membrane surface and morphologically the surface structures of each coat protein complex and COPII-coated vesicles. Based on the observations, we suggest a spatial order of the subunits in the assembled COPII coat.
Materials and Methods
Purified COPII proteins were prepared as described previously (3, 4). Amino-reactive homobifunctional crosslinkers and N-hydroxysuccinimidobiotin (NHS-biotin) were purchased from Pierce.
Microsomes were prepared from Saccharomyces cerevisiae RSY445 (MATα, gal2, leu2-3, 112, ura3-52, trp1-289, his4-579, prb1, pep4∷URA3). Budding of COPII vesicles from isolated microsomes, purified coat proteins, and the incubation with an ATP regeneration system and guanylylimidodiphosphate (GMP-PNP) was carried out as described (3). After the budding reaction, vesicles were separated from microsomes by centrifugation and purified by sucrose density gradient centrifugation. In brief, 16 0.4-ml budding reactions were centrifuged at 10,000 × g for 4 min, and the supernatants were collected. The pooled supernatants were loaded on a sucrose gradient containing 0.7 ml of 2.2 M sucrose in B88 [20 mM Hepes-KOH/0.25 M sorbitol/0.15 M KOAc/1 mM Mg(OAc)2, pH 6.8] and 1.5 ml of 0.45 M sucrose in B88, and the gradient was centrifuged for 2 h at 50,000 rpm in a Beckman SW55 Ti rotor. Crude vesicles that sedimented between the 0.7 and 2.2 M sucrose layers were collected and mixed with 3 volumes of 2.2 M sucrose in B88. The suspension was placed on the bottom of a centrifuge tube and overlayed with 0.6 ml of 1.6 M sucrose in B88, 1 ml each of 1.4 or 1.2 M sucrose in B88, and 1.1 ml of 0.8 M sucrose in B88, and the gradient was centrifuged at 50,000 rpm in a Beckman SW55 Ti rotor for 16.5 h. After centrifugation, gradients were fractionated from the top. Fractions containing COPII vesicles were kept on ice until the preparation of electron microscopy samples.
Crosslinking of coat proteins with lipids in the COPII vesicles generated from synthetic liposomes was carried out as follows. Liposomes consisting of a modified “major-minor mix” (4) containing 23 mol % 1-oleoyl-2-{12-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoethanolamine (NBD-PE) were incubated with COPII coat proteins and GMP-PNP for 30 min at 30°C in a 20-μl reaction. The resulting solution was mixed with 2 μl of crosslinkers in a solution of dimethyl sulfoxide with or without NHS-biotin for 30 min on ice, and then the excess crosslinker was quenched by mixing 3 μl of 0.25 M Tris/1.9 M glycine, pH 8.2 and incubated at room temperature for 30 min. Proteins in this reaction were separated by SDS/PAGE and transferred to a polyvinylidene difluoride membrane, and the fluorescence of crosslinked NBD-PE was recorded by using a STORM860 image analyzer at a setting of 950 V. After recording, biotinylated proteins on the membrane were probed with a streptavidin-alkaline phosphatase conjugate (Pierce), and the activity of alkaline phosphatase was detected by using the Vistra ECF substrate (Amersham Pharmacia). The fluorescent product was detected by a STORM860 image analyzed at a setting of 650 V. Thereafter, proteins on the membrane were visualized by colloidal gold staining.
Visualization of the coat proteins and isolated coated vesicles by rotary shadowing was carried out essentially as described (5, 6). In brief, COPII proteins adsorbed onto either freshly cracked mica flakes pretreated with polylysine and fixed with glutaraldehyde or onto an untreated or a polylysine-coated glass surface were frozen quickly and freeze-dried. Platinum replicas of the proteins were made by rotary shadowing. In some cases, proteins adsorbed on mica or glass were fixed with 2% glutaraldehyde before freeze-drying. In the case of COPII vesicles, gradient-isolated vesicles, either unfixed or fixed with glutaraldehyde in solution, were adsorbed on mica or polylysine-coated glass and processed as described above.
Results
Biochemical Analysis to Define the Relative Distance of Coat Subunits from the Membrane Surface.
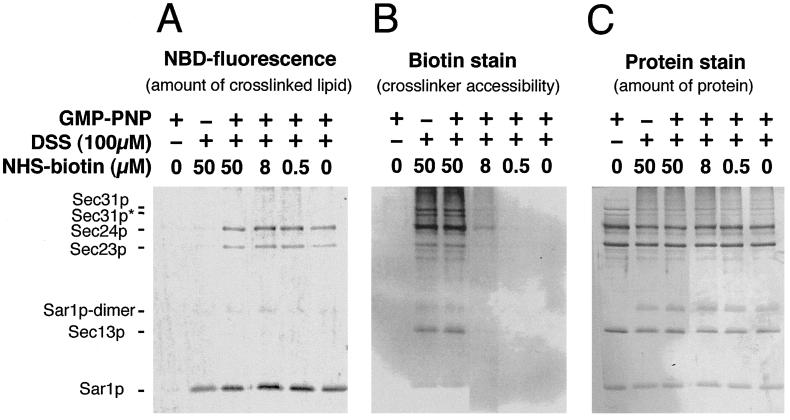
To assess the relative position of each COPII subunit with respect to the membrane in the assembled coat, we examined the crosslinking of coat subunits to phospholipid in the membrane. We chose an amino-reactive crosslinker, because phosphatidylethanolamine and phosphatidylserine, which are major phospholipids in biomembranes, contain amino groups that are good targets for crosslinking. We used COPII-coated membranes generated from synthetic liposomes to manipulate the composition of the membranous lipids. Liposomes consisting of 23 mol % of NBD-PE were subjected to a COPII budding reaction in the presence and absence of GMP-PNP. Inclusion of NBD-PE up to 23 mol % does not affect the binding of COPII to liposomes (7). After the reaction, the whole mixture was incubated with amine-reactive bifunctional crosslinkers. Samples were resolved on an SDS/PAGE and evaluated for NBD fluorescence (Fig. 1A) and total protein (Fig. 1C).
Figure 1.
Crosslinking of COPII proteins and membrane lipids in the synthetic COPII vesicles generated from liposomes. Liposomes containing NBD-PE were incubated with COPII-coat proteins for 30 min at room temperature in the presence and absence of GMP-PNP. After incubation, the whole reaction mixture was incubated with various combinations of disuccinimidyl suberate (DSS, 100 μM final concentration) and NHS-biotin. After quenching the amine-reactive reagents, proteins in the incubation mixture were resolved by SDS/PAGE, and the NBD fluorescence of the proteins, biotinylation, and total protein was visualized as described in Materials and Methods. (A) NBD fluorescence represents NBD-PE crosslinked by disuccinimidyl suberate (DSS) to COPII proteins. (B) NHS-biotin labeling of reactive amino groups on all proteins in each sample. (C) Colloidal gold staining of all proteins in each sample. Sec31p* indicates a degradation product of Sec31p present in our preparation.
We found that crosslinking of Sar1p and NBD-PE with a homobifunctional and amine-reactive crosslinker, disuccinimidyl suberate, was not strictly GMP-PNP-dependent, although crosslinking was enhanced in the presence of GMP-PNP (Fig. 1A, lanes 2 and 3). In contrast, the crosslinking between Sec23/24p and NBD-PE strictly depended on GMP-PNP (Fig. 1A, lanes 2 and 3). No crosslinking of Sec13/31p with NBD-PE was observed. The use of crosslinkers with different spacer arm lengths such as disuccinimidyl tartarate (0.64 nm) or ethylenglycol bis(succinimidylsuccinate) (1.61 nm) did not affect the crosslinking efficiency between NBD-PE and coat proteins (data not shown).
To rule out a possibility that the lower crosslinking of Sec13/31p with NBD-PE was caused by the absence of accessible amino groups, we included various concentrations of an amine-reactive biotinylation reagent, NHS-biotin, in the crosslinking reaction. The spacer length of NHS-biotin (1.35 nm) is similar to that of disuccinimidyl suberate (1.14 nm). Biotinylated COPII proteins were detected by using a streptavidin-alkaline phosphatase enzyme stain on a blot of the SDS/PAGE (Fig. 1B). Both Sec13/31p and Sec23/24p were biotinylated efficiently in the presence of 50 mM NHS-biotin (Fig. 1B, lanes 2 and 3), whereas Sar1p was only weakly reactive. The intensity of the signals for both Sec13/31p and Sec23/24p was decreased at lower concentrations of NHS-biotin (Fig. 1B, lanes 4 and 5). Sec24p was more reactive than Sec23p in respect to biotinylation and crosslinking to phospholipid. This result may be caused by a greater number of exposed reactive amino groups on Sec24p.
These observations indicate that the distance between the amino groups of assembled Sar1p and Sec23/24p approach the head groups of phospholipids on the surface of a liposome more closely than do those of Sec13/31p. Thus, Sar1 and Sec23/24p may separate and bridge the membrane surface from an outer scaffold layer of Sec13/31p.
Morphological Inspection of COPII Coat Protein Subunits.
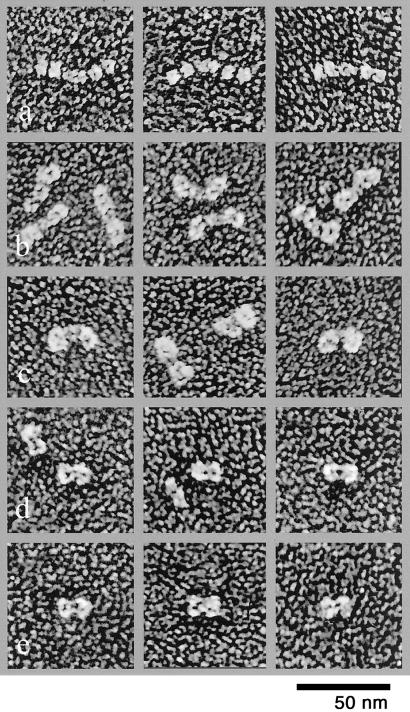
The biochemical observation described above suggests that the spatial order of the coat protein subunits reflects the sequence in which they are assembled in a COPII coat: Sar1p, Sec23/24p, and then Sec13/31p. Accordingly, Sec13/31p should cover the surface of a COPII-coated vesicle. To test this, we first analyzed the morphology of individual coat protein subunits by quick-freeze/deep-etch electron microscopy. Sar1p appeared as small globular particles (data not shown). The Sec23/24p complex resembles a bow tie of ≈18-nm length and 12-nm width (n = 60) with a central knot of ≈4-nm diameter (Fig. 2). Because the thickness of the platinum coat is 1–2 nm, the size of the Sec23/24p complex is ≈16 nm in length and 10 nm in width consisting of symmetrically joined units. Sec23p and Sec24p are ≈24% identical in sequence (8, 9); thus the shape of the complex is consistent with a heterodimer of symmetrical subunits. Detailed crosslinking and analytical gel filtration and ultracentrifugation analyses have documented the heterodimeric nature of Sec23/24p (10).
Figure 2.
Structure of Sec13/31p and Sec23/24p. (A) Images of unfixed Sec13/31p. (B) Briefly fixed (0.2% glutaraldehyde for 30 s) Sec13/31p. (C) More completely fixed (0.3% glutaraldehyde for 2 min) Sec13/31p. (D) Unfixed Sec23/24p. (E) Fixed Sec23/24p.
The unfixed Sec13p/31p complex contains two sets of globular domains of ≈8-nm diameter (n = 45), each bordering a central connecting rod (Fig. 2). Correcting for the platinum thickness, we estimate a central rod dimension of ≈12-nm length and 4-nm width (n = 45) and four globular terminal domains of ≈6-nm diameter each. The overall size of this complex is larger than expected for a heterodimer (11, 12). Rather, the images are more consistent with a protein complex of 300–400 kDa, for example, of a heterotetramer with two Sec13p and two Sec31p subunits. Recently, this assignment has been documented by detailed crosslinking and gel electrophoretic analysis (10).
The globular structures of Sec13/31p appeared to be joined by hinge-like linkers, because the orientations of the cylindrical and globular domains and between different globular domains varied significantly (Fig. 2B and ref. 10). Images of aldehyde-fixed Sec13p/31p showed a different structure than the unfixed samples (Fig. 2 A–C). In this case, two large domains were attached to each side of the central rod. The angles of the side domains and the central rod varied in images, suggesting that the junctions are flexible.
Surface Structure of the COPII-Coated Vesicles.
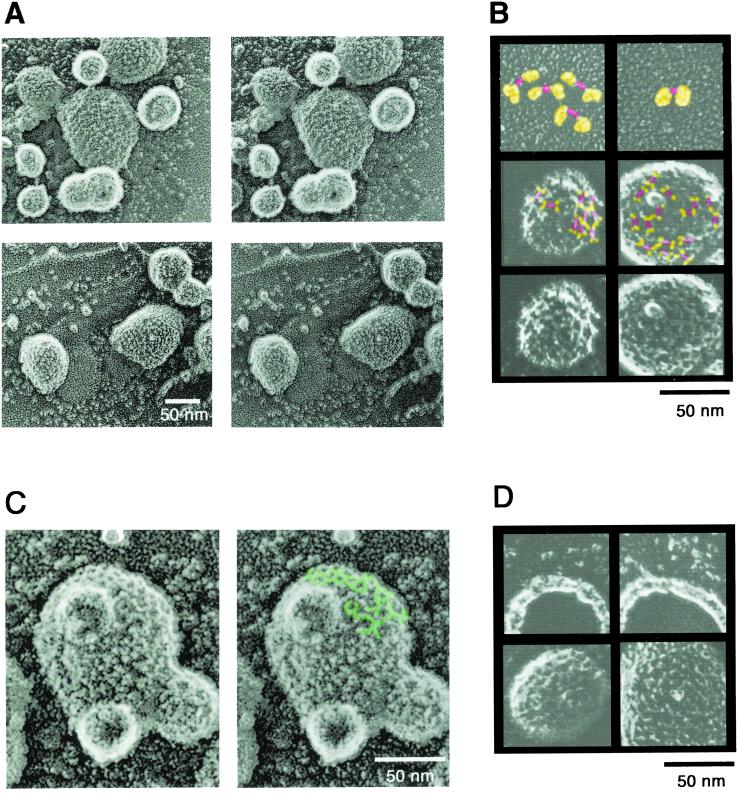
COPII vesicles generated in vitro with purified coat proteins, yeast microsomes, and GMP-PNP and purified by sucrose density gradient centrifugation were adsorbed on the surface of freshly cracked mica flakes. The flakes then were frozen, deep-etched, and rotary-shadowed with platinum (5, 6). Fig. 3A shows representative stereo images of the vesicles. Unlike clathrin-coated vesicles, the surface structure of the COPII-coated vesicles contained tightly packed ridges and spikes and lacked wide grooves and big holes. Clear straight ridges were observed frequently. The lengths of each ridge were shorter than the overall length of the Sec13/31p complex; however, the length of the ridge (≈10 nm) was similar to that of the central rod of the Sec13/31p (Fig. 3B, marked with red). Both ends of the ridge formed a Y shape (Fig. 3B, marked with yellow), which may correspond to the globular domains of the Sec13/31p complex. In some places on the vesicles there were clear mesh-like ridges with polygonal lattices, possibly the combination of pentagonal and hexagonal units (Fig. 3C). The lengths of the ridges of these polygonal units are 8–12 nm, some of which have an open side.
Figure 3.
Morphology of the COPII vesicle generated from microsomes. (A) Stereo view of COPII vesicles adsorbed on freshly cracked mica. (B) Comparison of the structure of fixed Sec13/31p and the surface image of the vesicle. The central rod of the Sec13/31p complex is marked red. The flexible domains of aldehyde-fixed Sec13/31p complex or the upper two lines of the Y-shaped structures on the vesicle are marked yellow. First row, aldehyde-fixed Sec13/31 complex; second row, COPII vesicles with possible Sec13/31p marked with colors; third row, unmarked images of COPII vesicles for comparison. (C) Mesh-like polygonal structure of the COPII coat. Typical polygonal mesh on a high magnification stereo image of a COPII vesicle is marked green. (D) Comparison of the cross fractures of the COPII vesicle and intact coated vesicle. Images of the cross fracture (upper row) and intact coated vesicles (lower row) from the same experiment are shown.
Cross fractures of the coated vesicles were observed occasionally in the same images (Fig. 3D). The thickness of the cross-fractured vesicles with platinum decoration was 10–14 nm with no apparent uncoated spaces. Thus, the thickness of the COPII coat ranges from 8 to 12 nm, which is smaller than the sum of the thickness of Sec23/24p and Sec13/31p (14 nm).
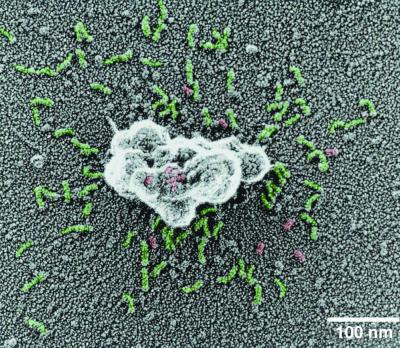
Surface images of the COPII vesicles adsorbed on glass plates revealed many coat subunits shed from the vesicles (Fig. 4). Most of the shed proteins appear to be Sec13/31p. The surface structure of vesicles on glass was different from mica-adsorbed vesicles, exposing ridges that appear to resemble the bow-tie structure of Sec23/24p. We interpret this structure to mean that Sec13/31p occupies the outermost layer of the coat covering an inner layer of Sec23/24p.
Figure 4.
Sec13/31p shed from the coat. Purified COPII vesicles were adsorbed on a glass plate and processed for electron microscopy. Green, Sec13/31p; red, Sec23/24p.
Discussion
Phospholipid-coat protein crosslinking experiments support the view that the COPII coat consists of layers that reflect the order of assembly of the COPII subunits: Sar1p, Sec23/24p, and then Sec13/31p. Chemical crosslinking to phospholipids confirms, as shown by direct binding measurements (4), that Sar1p associates with the bilayer. Sec23/24p, which is recruited to liposomes bearing Sar1-GTP and acidic phospholipids (4), is crosslinked less efficiently to phospholipid. Sec13/31p, which is recruited to liposomes bearing Sar1p and Sec23/24p (4), shows little if any crosslinking to phospholipid despite abundant and exposed reactive amino groups.
Quick-freeze/deep-etch electron microscopy provided images of the complexes that comprise the coat and the layers that contribute to the assembled structure. Sec23/24p, which is likely to be involved along with Sar1p in direct interaction with cargo proteins (13), forms a heterodimer of two conserved subunits that appear as bow-tie symmetrical structures (Fig. 2 D and E). Unfixed Sec13/31p complexes appear as linear, flexible, rod-like chains of 4-nm diameter globular domains, two on each side of a cylindrical core of 12-nm length.
Recently, a detailed electron microscopy analysis of negatively stained COPII subunits revealed structures similar in shape and dimension to those reported here using quick-freeze/deep-etch electron microscopy (10). However, the two techniques exposed a different appearance of the central portion of the Sec13/31p flexible rod. Negatively stained images showed a globular domain similar to the other segments of the chain of flexibly hinged globular domains, whereas quick-freeze/deep-etch images reveal a more extended cylindrical central domain.
Surface images of coated vesicles formed from endoplasmic reticulum membranes suggest a repeat structure with polygonal, but irregularly arranged faces of ≈10-nm length (Fig. 3). Similar repeat units were observed on coated vesicles formed from synthetic liposomes (4). In favorably oriented surface impressions, hexagonal repeat units were observed (Fig. 3C). Such an arrangement is compatible with flattened or curved coated surfaces found on large coated liposomes (4), coated vesicles (3), and coated tubules (14). Coated vesicles captured during apparent uncoating (Fig. 4) show abundant strands of Sec13/31p adhering to a glass surface with residual globular units, possibly representing Sec23/24p and Sar1p remaining bound to the coat residue on the vesicle surface.
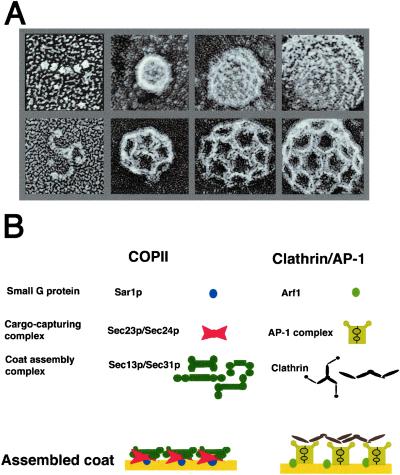
On the basis of the crosslinking experiments and deep-etch microscopy of the coat subunits and the assembled coat, we propose a layered structure, the dimensions of which are consistent with the width of the individual subunits and the average thickness of the coat seen in cross sections of COPII vesicles (Figs. 3D and 5). This layering superficially resembles the elements of the clathrin coat, which consists of the GTPase Arf1, an intermediate layer of adaptor protein, and a surface layer of clathrin triskelion, although the unit sizes of clathrin and COPII lattices are quite different (Fig. 5). The layers also conform to our previous conclusions that Sar1 and Sec23/24p serve primarily to detect and recruit membrane cargo proteins exposed on the surface of the endoplasmic reticulum membrane (13), whereas Sec13/31p serves a structural role to cluster Sec23/24p complexes and their bound cargo proteins (1). Given the flexibility of the globular domains that comprise the bulk of the strands of Sec13/31p, the COPII coat, unlike the more rigid clathrin coat, may accommodate less regularly shaped cargo molecules, possibly even rigid rods such as collagen, in tubular as well as spherical coated vesicles.
Figure 5.
Comparison of the assembly of COPII and clathrin coat. (A) Deep-etch structures of Sec13/31p, COPII vesicles, clathrin triskelion, and clathrin-coated vesicles. First row, Sec13/31p and COPII vesicles; second row, clathrin triskelion and clathrin-coated vesicles. (B) Comparison of the assembly models of COPII and clathrin/AP-1-coated vesicles.
Acknowledgments
We thank Bob Lesch for COPII proteins, Sebastian Springer for detailed discussions and encouragement, and members of the Schekman lab for providing a stimulating and lively working environment. Thanks to Robyn Roth of the Heuser laboratory for generating all the electron microscopic images presented in this report. This work was supported by the Howard Hughes Medical Institute (to R.S.), National Institutes of Health Grant GM29641 (to J.E.H.), Ministry of Education, Science and Culture, Japan, Grants 11660327 and 13460152 (to K.M.), and the Swiss National Science Foundation (to L.O.).
Abbreviations
- NHS-biotin
N-hydroxysuccinimidobiotin
- GMP-PNP
guanylylimidodiphosphate
- NBD-PE
1-oleoyl-2-{12-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoethanolamine
References
- 1.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 2.Kirchhausen T. Nat Rev Mol Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 3.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 4.Matsuoka K, Orci L, Amherdt M, Bednarek S Y, Hamamoto S, Schekman R, Yeung T. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 5.Heuser J E. J Mol Biol. 1983;169:155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- 6.Heuser J E. Prog Clin Biol Res. 1989;295:71–83. [PubMed] [Google Scholar]
- 7.Matsuoka K, Schekman R. Methods. 2000;20:417–428. doi: 10.1006/meth.2000.0955. [DOI] [PubMed] [Google Scholar]
- 8.Kurihara T, Hamamoto S, Gimeno R E, Kaiser C A, Schekman R, Yoshihisa T. Mol Biol Cell. 2000;11:983–998. doi: 10.1091/mbc.11.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimoni Y, Kurihara T, Ravazzola M, Amherdt M, Orci L, Schekman R. J Cell Biol. 2000;151:973–984. doi: 10.1083/jcb.151.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lederkremer G Z, Cheng Y, Petre B M, Vogan E, Springer S, Schekman R, Walz T, Kirchhausen T. Proc Natl Acad Sci USA. 2001;98:10704–10709. doi: 10.1073/pnas.191359398. . (First Published September 4, 2001; 10.1073/pnas.191359398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salama N R, Yeung T, Schekman R W. EMBO J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salama N R, Chuang J S, Schekman R W. Mol Biol Cell. 1997;8:205–217. doi: 10.1091/mbc.8.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Springer S, Schekman R. Science. 1998;281:698–700. doi: 10.1126/science.281.5377.698. [DOI] [PubMed] [Google Scholar]
- 14.Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]