Abstract
Colubridae represents the most phenotypically diverse and speciose family of snakes, yet no well-assembled and annotated genome exists for this lineage. Here, we report and analyze the genome of the garter snake, Thamnophis sirtalis, a colubrid snake that is an important model species for research in evolutionary biology, physiology, genomics, behavior, and the evolution of toxin resistance. Using the garter snake genome, we show how snakes have evolved numerous adaptations for sensing and securing prey, and identify features of snake genome structure that provide insight into the evolution of amniote genomes. Analyses of the garter snake and other squamate reptile genomes highlight shifts in repeat element abundance and expansion within snakes, uncover evidence of genes under positive selection, and provide revised neutral substitution rate estimates for squamates. Our identification of Z and W sex chromosome-specific scaffolds provides evidence for multiple origins of sex chromosome systems in snakes and demonstrates the value of this genome for studying sex chromosome evolution. Analysis of gene duplication and loss in visual and olfactory gene families supports a dim-light ancestral condition in snakes and indicates that olfactory receptor repertoires underwent an expansion early in snake evolution. Additionally, we provide some of the first links between secreted venom proteins, the genes that encode them, and their evolutionary origins in a rear-fanged colubrid snake, together with new genomic insight into the coevolutionary arms race between garter snakes and highly toxic newt prey that led to toxin resistance in garter snakes.
Keywords: Thamnophis sirtalis, co-evolution, venom, vision, sex chromosomes
Introduction
The order Squamata, comprised of lizards and snakes, represents a major portion of amniote vertebrate diversity with over 10,000 species that span ∼200 Myr of evolutionary history (Uetz and Etzold 1996; Kumar et al. 2017). Among squamates, snakes exhibit many unique and often extreme adaptations, including potent venoms and limblessness, making them of particular interest in studies of amniote evolution and biology. Recent studies have further revealed interesting and divergent features of snake genomes, including unique genomic GC isochore structure (Castoe 2013), evidence for pervasive and rapid positive selection (Castoe 2013), multiple origins of sex chromosomes (Gamble et al. 2017), and complex multigene family evolution in genes associated with sensory systems (Vandewege et al. 2016; Schott et al. 2018), venom (Vonk et al. 2013; Yin et al. 2016), and toxin resistance (McGlothlin et al. 2014). Genomic resources are now available for several snake species, including the Burmese python (Python molurus bivittatus) (Castoe 2013), King cobra (Ophiophagus hannah) (Vonk et al. 2013), Boa constrictor (Bradnam et al. 2013), and the five-pace viper (Deinagkistrodon acutus) (Yin et al. 2016). However, currently available genomic resources for snakes leave the single most diverse and speciose family of snakes, Colubridae, surprisingly underrepresented. Only one colubrid draft genome has been published to date (the corn snake, Pantherophis gutattus) (Ullate-Agote et al. 2014), but the highly fragmented state of this assembly limits its use and value for comparative genomic analyses.
Among colubrids, garter snakes (genus Thamnophis) are arguably the most well-studied lineage and have emerged as an important vertebrate model for research spanning a broad range of biological questions. For example, garter snakes have served as one of the only nonmammalian model systems for understanding the development of behavior, which has shown that snakes develop unique and consistent personalities that are shaped by experience with ecological factors including their interactions with predators, prey, and chemical stimuli (Herzog and Burghardt 1986, 2010). Garter snakes have also proved to be valuable models for a wide range of physiological questions, including those related to reproduction (Mendonca and Crews 1996), exercise (Jayne and Bennett 1989), and aging (Robert and Bronikowski 2010). They have also been important models for studying snake vision systems to examine the effects of fossorial ancestry on the morphology, physiology, and genetics of vision in snakes (Schott et al. 2016, 2018), and to study how decreased snake visual acuity may lead to enhanced chemosensation and pheromonal communication and behavior (Kubie et al. 1978; Crews et al. 1984; Mason and Crews 1986).
Like many other colubrids, garter snakes are capable of producing a relatively mild venom that is used to subdue prey (Vest 1981; Hayes and Hayes 1985). In contrast to other venomous snake lineages such as cobras (Elapidae) and rattlesnakes (Viperidae) that produce extremely potent venom in specialized glands with delivery through hollow anterior fangs, colubrid venom is produced in a homologous gland (Duvernoy’s gland) and delivered through grooved posterior maxillary teeth, which has led to the term “rear-fanged” for describing venomous colubrid snakes. Several studies have used available snake genomes to study the variation and evolution of venom gene families across snake species, but have been limited to either strictly nonvenomous or highly venomous species (Vonk et al. 2013; Reyes-Velasco et al. 2015; Yin et al. 2016). The genome of a rear-fanged venomous colubrid species such as the garter snake would provide a more complete representation of the diversity of venom systems in snakes, offering increased potential to understand the evolution of this extreme and medically relevant feature of snake biology. In addition to producing mildly toxic venoms, some species of garter snakes are also remarkable for having evolved resistance to the toxic compounds of their prey, specifically in toxic newts of the genus Taricha. Given this unique adaptation, garter snakes have become an important model for studying the evolution of toxin resistance and coevolutionary dynamics of predator–prey arms races (Brodie and Brodie 1999; Brodie et al. 2002; Feldman et al. 2009; McGlothlin et al. 2016).
Here, we sequenced and annotated the genome of the garter snake (Thamnophis sirtalis) and used this genome and related resources to address broad questions related to vertebrate genome evolution and garter snake biology. We examined molecular evolution across gene families that underlie unique and interesting biological features of garter snakes related to toxin resistance and venom, as well as olfactory and visual systems. We also compared evolutionary patterns of genome composition and structure (repeat content, microRNAs, and sex chromosomes) in garter snakes in the context of other amniote genomes, identified genes that have likely been under positive selection in the garter snake and colubrids in general, and estimated neutral substitution rates across 4-fold degenerate sites for the garter snake and other squamates reptiles.
Materials and Methods
Genome Sequencing and Annotation
All animal procedures were conducted with registered IACUC protocols (see supplementary Methods, Supplementary Material online). Garter snake genome sequencing libraries were constructed from a single adult female T.sirtalis collected in Benton County, Oregon. A genome size of 1.9 Gbp, previously estimated using flow cytometry (Vinogradov 1998), was used for the construction of libraries. We created and sequenced multiple whole-genome shotgun libraries on the Illumina platform, generating ∼72× genome coverage, which was subsequently used for assembly with ALLPATHS-LG (Gnerre et al. 2011), scaffolding with L_RNA_scaffolder (Xue et al. 2013), and SSPACE (Boetzer et al. 2011), and gapfilling using a custom script. This 6.0 version has been screened and cleaned of contaminating contigs and all contigs 200 bp or smaller were removed. The genome assembly was annotated by NCBI using their automated Eukaryotic Genome Annotation Pipeline (Thibaud-Nissen et al. 2013). To generate an updated estimate of genome size, kmers were extracted using jellyfish v2.2.3 (Marçais and Kingsford 2011) and genome size estimates based on both 19mers and 23mers were generated using GCE v1.0.0 (Liu et al. 2013). The completeness of the T. sirtalis genome and five other squamate genomes (three lizards: Pogona vitticeps, Anolis carolinensis, Ophiosaurus gracilis; and two snakes: Python bivittatus, O.hannah) was assessed by comparing their annotated gene sets to benchmarking sets of universal single-copy vertebrate orthologs using BUSCO v1.1b1 (Simão et al. 2015), which included 3,023 vertebrate genes. The NCBI-generated protein FASTA files for T. sirtalis and P. bivittatus contain both multiple isoforms and seemingly duplicated proteins (i.e., two or more proteins with identical sequence and description in protein FASTA file, identical coordinates in GFF file, but unique protein identifiers). We therefore filtered the protein FASTA files for these two species to contain only the longest isoform if multiple were present and only one protein entry in the case of duplicated entries, and used these filtered, nonredundant sets of proteins for BUSCO analysis. Genomic repeat elements were annotated using RepeatMasker v4.0.6 (Smit et al. 2015) using de novo repeat libraries derived from RepeatModeler v1.0.9 (Smit et al. 2014) analysis of the genome assembly, combined with repeat libraries from other snake genomes (Castoe 2013; Yin et al. 2016). Distributions of gene, exon, and intron number and length were generated using custom Python scripts. See supplementary Methods 1.1–1.3, Supplementary Material online for additional details.
Analyses of Positive Selection on Protein-coding Genes
To infer patterns of positive selection in protein coding genes in squamate reptiles, including the branch leading to T. sirtalis, we generated orthologous coding sequence alignments for 27 species, including 18 reptiles and nine mammals (supplementary table S2, Supplementary Material online), following a previous established protocol (McGaugh et al. 2015). Notably, our alignments contained transcriptomes for several species from the Colubridae family of snakes: African house snake (Lamprophis fuliginosus) and two species of garter snakes Thamnophiscouchii and Thamnophis elegans (the latter of which is represented by two separate transcriptomes for two distinct ecotypes), in addition to T. sirtalis. Alignments were used to identify evidence for positive selection in the codeml program in PAML v4.7 (Yang 2007). We used a likelihood-ratio test (LTR) and three separate branch-site models to test for evidence of positive selection. The three branch-site models varied in which branch was placed in the foreground and included: 1) the branch to the family Colubridae, 2) the branch to the Thamnophis genus, and 3) the branch to T. sirtalis. Functional characteristics of genes with significant evidence of positive selection were analyzed using Ingenuity Pathway Analysis (IPA) Comparison Analysis (Krämer et al. 2014). Enrichment of functional categories was determined using Fisher’s exact tests comparing genes with evidence of positive selection in each of the three branch site models to the IPA knowledge base, and was considered significant when P < 0.05. See supplementary Methods 1.4, Supplementary Material online for additional details.
Estimating Neutral Substitution Rates for Squamate Reptiles
We inferred neutral substitution rates across squamate reptiles by estimating rates of substitution at 4-fold degenerate third codon positions of orthologous protein-coding genes. We used a Perl script provided in Castoe (2013) to identify and sample 4-fold degenerate sites for seven squamates (glass lizard, Ophisaurus gracilis; green anole, A.carolinensis; bearded dragon, P.vitticeps; king cobra, O.hannah; garter snake, T.sirtalis; cottonmouth, Agkistrodon piscivorus; and Burmese python, P.molurus) and three mammal outgroups (chimpanzee, Pan troglodytes; human, Homo sapiens; and mouse, Mus musculus); these data were subsampled from the larger 1:1 orthologous gene alignments used for analyses of positive selection in this study (see supplementary Methods 1.4, Supplementary Material online for details). We estimated branch-specific substitution rates using a relaxed log-normal clock model in BEAST v2.4.7 (Bouckaert et al. 2014) by calibrating the divergence times of the 9 internal nodes (see supplementary table S4, Supplementary Material online for node calibrations and prior distributions) and constraining the topology to (((GlassLizard, (Anole, Pogona)),(((Cobra, Thamno), CottonMouth), Python)),((Chimp, Human), Mouse)). Two BEAST analyses were conducted for 100 million generations each, sampling parameter values every 10,000 generations. We discarded the first 25% of runs as burn-in (25 million) and confirmed convergence to the posterior by comparing the likelihood values and effective samples sizes (ESS >1000 for all parameters).
Sex Chromosomes
To identify sex chromosome-specific scaffolds in the T. sirtalis genome, we used BLAST to locate six gene fragments known to have both Z and W-specific alleles in snakes (Matsubara et al. 2006; Vicoso et al. 2013; Laopichienpong et al. 2017). Maximum likelihood phylogenetic analyses on these gene fragments from several squamates were used to verify sex linkage, which would produce a pattern where Z and W alleles from distinct species cluster together phylogenetically. We also used sex-specific amplification of the T. sirtalis gene fragments by PCR to confirm that these scaffolds were sex-linked gametologous alleles and not paralogs or misassemblies. Fluorescent in situ hybridization (FISH) of a Bkm-like repeat, (GATA)n, was used to identify the W chromosome in the karyotype of a female T. sirtalis. See supplementary Methods 1.5, Supplementary Material online for details.
Identification and Analysis of microRNA and Associated mRNA Targets
The sequence-specific microRNA prediction (SMIRP) tool was used for the identification of microRNAs that are specific to T. sirtalis (Peace et al. 2015), and these predictions were tested in a number of ways, as described in supplementary Methods 1.6, Supplementary Material online. Inferred mature microRNAs were putatively matched to target coding sequences in the T. sirtalis genome using miRanda v.3.3a (Enright et al. 2003), using the parameters described in the supplementary Methods 1.6, Supplementary Material online.
Analyses of Visual Gene Loss and Opsin Expression Localization
To identify the full repertoire of visual genes in the garter snake genome, we used BLAST (Altschul et al. 1990) to search the complete genome sequence, as well as the genomes of the king cobra (O.hannah) and Burmese python (P.molurus bivittatus), using a library of visual and opsin genes compiled from previous studies of vertebrate vision (Schott et al. 2017, 2018). Genes that could not be recovered in any of the three snake genomes were presumed to be lost. Gene loss in snakes was compared with losses in mammals, geckos, and crocodilians, three groups with hypothesized nocturnal or dim-light ancestry. Under approved animal protocols at the University of Toronto, we conducted immunohistochemistry to identify the location of opsin gene expression across cells in the T. sirtalis retina based on previously published methods (described in Schott et al. 2016; Bhattacharyya et al. 2017). See supplementary Methods 1.7, Supplementary Material online for additional experimental details.
Annotation and Phylogenetic Analysis of Olfactory Receptors (ORs)
To infer patterns of expansion and loss in ORs in snakes, we identified and analyzed OR genes from the garter snake and other reptile genomes. OR annotations from the green anole and Burmese python were collected from Vandewege et al. (2016). In addition to the garter snake genome, three additional squamate genomes were included: The bearded dragon (P.vitticeps) (Georges et al. 2015) and two snakes, the B.constrictor (Bradnam et al. 2013) and king cobra (O.hannah) (Vonk et al. 2013). Known OR amino acid sequences from the human, chicken, zebra fish, clawed frog, green anole, python, softshell turtle, painted turtle, saltwater crocodile, gharial and American alligator were used to query each genome using TBlastN. Predicted OR genes were categorized into intact genes, pseudogenes and truncated genes as defined in Vandewege et al. (2016). We aligned intact OR sequences and estimated phylogenetic trees for all OR sequences as described in supplementary Methods 1.8, Supplementary Material online.
Venom Gene and Protein Analysis
To identify venom gene homologs in the T. sirtalis genome, we used tBLASTx based on a query set of known venom genes from diverse snake species, and secondarily tested orthology by inferring phylogenies of sets of T. sirtalis genes that showed homology to known venom genes. We then used RNAseq data from eight different T. sirtalis tissues (Duvernoy’s gland, brain, kidney, liver, lung, ovary, upper digestive tract, and lower digestive tract) to estimate patterns of gene expression across tissues for venom gene orthologs. To characterize the protein composition of Thamnophis venom, and compare this to inferences based on RNAseq, we extracted and analyzed crude venom from T. sirtalis and T. elegans using SDS-PAGE and MALDI-TOF mass spectrometry, under approved animal protocols at the University of Northern Colorado (#9204). See supplementary Methods 1.9, Supplementary Material online for additional details.
Analysis of Tetrodotoxin (TTX) Resistance in Voltage-gated Sodium Channels
Garter snakes are well known for their resistance to TTX in their prey, which at the molecular level derives from resistant voltage-gated sodium channels. Because automated annotation approaches performed poorly on these genes, we manually predicted and extracted sequences of all sodium channel alpha subunit (SCNA) paralogs from the T. sirtalis genome and other published genomes, including three snakes (B.constrictor, O.hannah, and P.molurus bivittatus), two lizards (A.carolinensis and O.gracilis), a bird (Gallus gallus), a turtle (Chrysemys picta), the human, and as an outgroup, a frog (Xenopus tropicalis). We focused on the evolution of three voltage-gated sodium channels previously undescribed in T. sirtalis: SCN5A (Nav1.5), SCN10A (Nav1.8), and SCN11A (Nav1.9). We aligned these related paralogs using MUSCLE (Edgar 2004), built a gene tree using RAxML (Stamatakis 2014), and performed ancestral sequence reconstruction using the codeml program in PAML v4.8 (Yang 2007).
Results and Discussion
Genome Assembly and Annotation
The garter snake represents the first fully annotated genome available for any colubrid snake and is available at NCBI: version Thamnophis_sirtalis-6.0, BioProject PRJNA294278, GenBank accession GCA_001077635.2. The scaffolded genome assembly is 1.42 Gbp in length (including gaps), with an N50 contig size of 10.4 kb and scaffold N50 size of 647.6 kb. The T. sirtalis genome annotation contains ∼85% of BUSCO genes, making it more complete than the glass lizard (Ophisaurus; 84%) and king cobra (Ophiophagus; 71%) genomes, but less complete than the genomes of the python and other two lizards assessed (Anolis and Pogona), which each contained at least 95% of vertebrate BUSCOs (supplementary fig. S1, Supplementary Material online). Kmer-based estimation of genome size resulted in estimates of 1.19 Gbp and 1.27 Gbp (mean = 1.23 Gbp) using 19mers and 23mers, respectively. These estimates are substantially smaller than the estimate derived from flow cytometry (1.9 Gbp; Vinogradov 1998) but are similar to the total assembly length (1.42 Gbp), suggesting that kmer-based estimates are reasonable and likely more accurate than the original flow cytometry estimate.
Genome Structure and Evolution
Genomic Repeat Landscapes
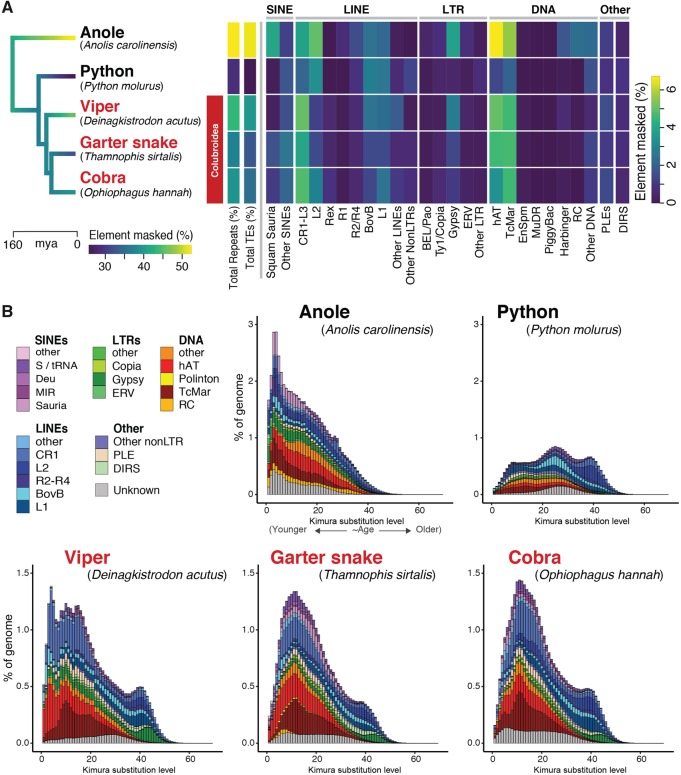
The genomic landscapes of repeat elements in squamate reptiles show remarkable variation in repeat abundance among species and lineages when compared with other vertebrate genomes (Castoe et al. 2011; Castoe 2013; Adams 2016). On the basis of limited studies, repetitive content appears to vary substantially between snake lineages in particular, despite low variation in genome size across snakes (Gregory 2018). We characterized the repeat landscape in the garter snake genome in relation to other squamate reptile genomes and estimate that readily identifiable repeat elements comprise 37.12% of the garter snake genome (ca. 417.1 Mbp), with TEs and microsatellites (SSRs) accounting for 32.9% and 2.8% of the assembled genome, respectively. Comparison of the garter snake repeat element landscape to those of other squamate reptiles reveals lineage- and species-specific patterns of repeat element evolution and expansion (fig. 1). Like other squamate genomes, the garter snake genome is dominated by LINE (nonLTR) elements of the CR1-like family (5.5% of the genome), as well as Gypsy LTR elements (1.37%), and hAT (4.6%) and Tc1-Mariner (TcMar; 4.7%) DNA transposons (fig. 1A). Overall, the genomes of colubroid snakes (Deinagkistrodon, Thamnophis, Ophiophagus in fig. 1) are similar in repeat element content and relative abundance, characterized by higher total repeat and TE content compared with the noncolubroid python genome, and all have notably higher CR1-L3 LINE and hAT and TcMar DNA transposon content (fig. 1A). The garter snake genome exhibits several unique repeat landscape features, such as an expansion of SINEs and subfamilies of DNA transposons that are otherwise absent or very rare in snakes. Our analyses detected recent and highly lineage-specific expansion of Deu SINEs, which are absent in the anole and python and are 6- and 2-fold more abundant in the garter snake than in the viper and cobra, respectively. Additionally, Polinton DNA elements are detected only in the garter snake assembly, and Rolling Circle (RC) DNA elements show a recent expansion in the garter snake that is on average 5-fold greater than the other snake species analyzed (fig. 1B). Together, analyses of repeat element content highlight the highly active, diverse, and dynamic nature of snake repeat element landscapes, particularly in colubroid snakes.
Fig. 1.
—Comparison of genomic repeat element landscapes across squamate reptiles genomes. (A) Summary of the repeat element content of the genome of Thamnophis sirtalis in comparison to other squamate reptiles. Branches on the time-calibrated consensus phylogeny are colored according to the estimated rate of genomic transposable element evolution. The associated heatmap shows the total repeat element and transposable element genomic content (%) for each taxon, as well as the genome coverage (% masked) of major components of the repeat element landscape. (B) Kimura 2-parameter distance-based TE copy divergence analysis. Genome abundance (% of genome; y-axis) is shown for each transposable element type and is clustered according to the CpG-corrected Kimura distance (K-value from 0 to 70; x-axis) from each type’s consensus sequence. Elements with low K-values are the least divergent from their respective consensus sequences, and likely represent more recent transposition events, whereas elements with higher K-values represented more degenerated copies that were likely inserted in the past.
Comparison of Gene Feature Distributions
Because few direct comparisons of annotated gene features across amniotes are available, we used the garter snake annotation to compare major characteristics of gene annotations across major amniote lineages. The garter snake genome contains a similar number of genes (20,186) compared with the human (21,407), although the median gene length of the garter snake, 13,384 base pairs (bp), is substantially lower that of the human (23,247.50 bp; supplementary table S1, Supplementary Material online). The chicken, with the smallest genome of the species analyzed, shows a relatively greater number of small genes (<∼5,000 bp) and a relatively lower number of large genes (>∼20,000 bp) compared with the other three species (supplementary fig. S2A, Supplementary Material online). The human, with the largest genome of the species analyzed, possesses a greater number of genes >∼40,000 bp compared with the other three species (supplementary fig. S2A, Supplementary Material online). Despite the variation in genome size across these species, the distributions of exon length are remarkably similar, although humans appear to possess a slightly greater number of exons between ∼1,000 and 3,000 bp (supplementary fig. S2B, Supplementary Material online). Furthermore, the median exon length is very similar in all four species (supplementary table S1, Supplementary Material online). Distributions of intron length are highly similar only for the garter snake and anole, whereas chicken shows a greater number of small introns (<∼2,000 bp) and lower number of larger introns (>∼3,000 bp) and the human shows a generally lower number of small introns (<∼3,000 bp) and slightly greater number of large introns (>∼65,000; supplementary fig. S2C, Supplementary Material online). Accordingly, the garter snake and anole have similar median intron lengths (1,439 and 1,351 bp, respectively), whereas chicken introns are generally smaller (median = 777 bp) and human introns are generally larger (median = 1,638 bp; supplementary table S1, Supplementary Material online).
Evidence of Positive Selection in the Garter Snake Genome
Previous estimates of molecular evolution based on a small number of snake genomes showed that rates of protein evolution are accelerated in snake lineages (Castoe 2013). Specifically, these analyses identified many genes with evidence of positive selection, some of which included genes involved in the cardiovascular system, multiple signaling pathways, cell cycle control, and lipid and protein metabolism, among others. With the added sampling of the garter snake genome and other snake genomic data, we investigated whether genes with evidence of positive selection were enriched for particular functions at three levels of taxonomic organization, specifically along the branch leading to Colubridae, as well as branches leading to the genus Thamnophis and the branch leading to T.sirtalis. We identified four genes (EMC7, WNT9A, STK40, and PICK1) that were significant for the branch-site model when placing colubrids in the foreground (supplementary table S3, Supplementary Material online). The last of these, PICK1, a synaptic protein, is notable for its interaction with both nonvoltage gated sodium channels (BNC1) as well as acid-sensing ion channel (ASIC) (Hruska-Hageman et al. 2002). We identified 10 genes that were significant when the branch to the Thamnophis genus was placed in the foreground. Mutations in many of these (e.g., TBCD, SULF2, EXT2) are associated with neurological developmental syndromes (supplementary table S3, Supplementary Material online; Jaglin and Chelly 2009; Wall et al. 2009; Wade et al. 2013). Finally, we identified eight genes with significant evidence of positive selection when placing only T.sirtalis in the foreground (supplementary table S3, Supplementary Material online).
In the IPA Comparison Analysis (Krämer et al. 2014), focusing first on the genes with evidence of positive selection in colubrids, PICK1 stood out as having functionality in cell morphology, organ morphology, and organismal development. PICK1 also interacts with cation channels (e.g., BNC1, ASIC) that are important in the central nervous system as well as peripheral sensory neurons (Hruska-Hageman et al. 2002), and recent research identified an association between a snake venom toxin (a Texas coral snake toxin) and ASICs, specifically where ASIC1a and b had strong toxin-evoked responses (Bohlen et al. 2011). Focusing in on the Thamnophis genus, we identified significant enrichment for genes involved in proline biosynthesis and in cancer (Fisher’s exact test, P < 0.05) (Bernard et al. 2001; Székely et al. 2008). Finally, at the species level, T.sirtalis genes with evidence of positive selection were particularly prominent in cancer and cell death (apoptosis phenotypes). Snakes are of interest in cancer biology because reptiles, in general, have lower incidences of cancer than mammals (Effron et al. 1977) and because within reptiles, snakes have the highest incidences of cancer (crocodilians and turtles have the lowest) (Garner et al. 2004). We also identified genes, such as ANO5, that are associated with skeletal and cardiac muscle, specifically with diseases such as muscle dystrophy and gnathodiaphyseal dysplasia (i.e., bone calcification defect) (Tsutsumi et al. 2004). Additionally, PK3D (phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit delta), which is associated with the venom production cycle in pit vipers (Kerchove et al. 2008), showed evidence of positive selection on the branch leading to T. sirtalis specifically.
Neutral Substitution Rate Estimates
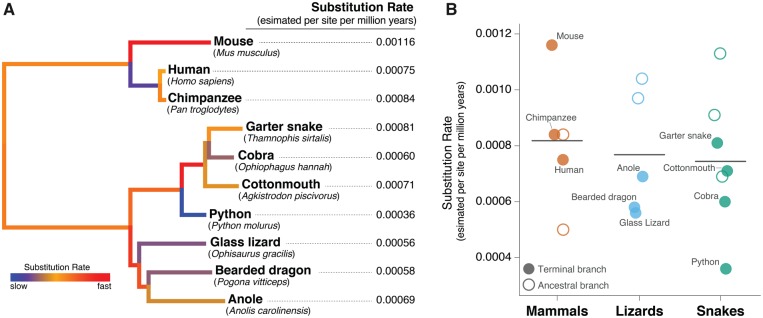
Branch-specific estimates of neutral substitution rates across 4-fold degenerate sites indicate that squamate reptiles have an average substitution rate (0.00076 per site per million year) that is similar to mammals (0.0008) (fig. 2). Consistent with other recent estimates (Green et al. 2014), we find that the high substitution rate of the mouse represents an outlier among vertebrates. Snakes appear to have slightly lower substitution rates (Avg. = 0.00074) compared with lizards (Avg. = 0.00077), due largely to the python, which had the lowest estimated rate in our data set. Notably, the garter snake had the highest estimated substitution rate among all squamate lineages (fig. 2). Overall, our estimates of substitution rates (for both squamates and mammals) using 4-fold degenerate sites were generally similar to recent substitution rate estimates for these lineages (Green et al. 2014). Variation in substitution rates has been linked to variation in recombination rate, efficacy of DNA repair, and life history traits such as body size or generation time (Martin and Palumbi 1993; Bromham 2002; Bromham and Penny 2003). Garter snakes are members of the most speciose family of snakes, the Colubridae, and future studies of substitution rates across this group may be useful in linking such rates with other features of snake biology and evolution (Tollis et al. 2018).
Fig. 2.
—Estimated neutral substitution rates of squamate reptiles. (A) Branch-specific estimates of substitution rates at 4-fold degenerate third-codon positions of coding genes. (B) Jitter plot of 4-fold degenerate site substitution rate estimates for mammals, lizards, and snakes, including all terminal and ancestral branches allocated to a clade. Terminal branches are shown as closed circles and are labeled with the corresponding species, whereas ancestral branches are shown as open circles. The horizontal lines denote the average substitution rate estimate for each clade.
Sex Chromosomes
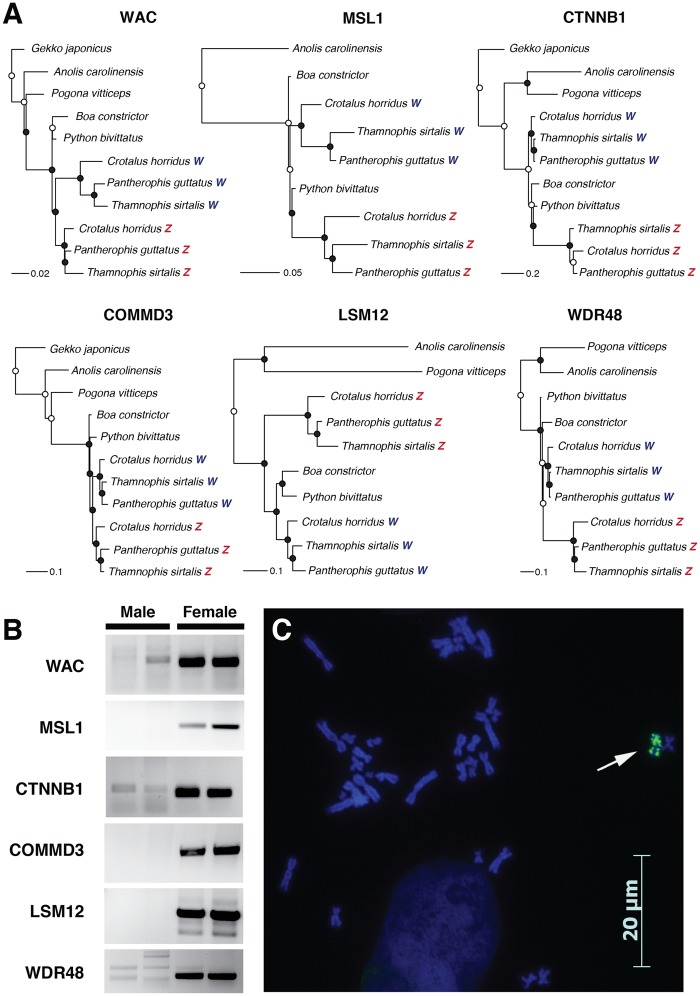
Snakes are important model systems for studying the evolution of sex chromosomes, due to the variation in sex-chromosome differentiation across lineages (Matsubara et al. 2006), and the recent discovery that sex chromosomes appear to have evolved multiple times and led to both XY and ZW systems (Gamble et al. 2017). Caenophidian snakes (which includes the family Colubridae) possess ZZ/ZW sex chromosomes derived from the same ancestral autosomes (Matsubara et al. 2006; Ohno 1967; Vicoso et al. 2013; Rovatsos et al. 2015), with males possessing two similar Z chromosomes and females possessing one Z and one W chromosome. Over time, limited recombination between the Z and W has led to degeneration of the snake W, resulting in the loss of numerous functional genes, the evolution of W-specific alleles, and accumulation of an excess of repetitive sequences (Matsubara et al. 2006; O’Meally et al. 2010; Vicoso et al. 2013; Rovatsos et al. 2015; Laopichienpong et al. 2017). Because a heterogametic female T. sirtalis was sequenced, we suspected that some genome scaffolds would be W-specific. BLAST of six genes with distinct Z and W alleles in snakes (Matsubara et al. 2006; Vicoso et al. 2013; Laopichienpong et al. 2017) revealed Z and W-specific scaffolds for each gene. To confirm that these scaffolds were W-specific, we first aligned fragments of Z and W scaffolds identified in the T. sirtalis genome assembly with sex chromosome sequences from two additional snake species with homologous ZZ/ZW, the corn snake (Pantherophis guttatus) and timber rattlesnake (Crotalus horridus). We used two snake species with XX/XY sex chromosomes, B.constrictor and P.molurus bivittatus (Gamble et al. 2017), and several lizard species as outgroups. Phylogenetic analyses of the aligned sequences using maximum likelihood recovered distinct clades for caenophidian Z and W alleles (fig. 3A). We then confirmed that putative T. sirtalis W scaffolds were indeed sex-linked gametologous alleles and not paralogs or misassemblies by designing PCR primers from the W scaffolds. Primers from all six loci amplified in a sex-specific manner in two male and two female T. sirtalis (supplementary table S5, fig. S3B, Supplementary Material online). Because a previously published T. sirtalis karyotype did not reveal heteromorphic sex chromosomes (Baker et al. 1972), we investigated whether the T. sirtalis W chromosome could be identified cytogenetically by fluorescent labelling repetitive DNA sequences. The Y and W sex chromosomes of many plants and animals show accumulation of Bkm-like satellite DNA (Jones and Singh 1985; Arnemann et al. 1986; Parasnis et al. 1999; O’Meally et al. 2010; Gamble et al. 2014). FISH of a Bkm-like repeat, (GATA)n, showed significant enrichment on a single chromosome in the female karyotype, presumably the W (fig. 3C). Together, the identification of Z and W scaffolds from the genome assembly and the FISH analyses reveal T. sirtalis has distinct sex chromosomes with significant sequence differences between the Z and W. The assembly of both Z and W scaffolds in the T. sirtalis genome illustrate its high quality and provide a new resource for further investigation into snake sex chromosome evolution.
Fig. 3.
—Identification of the W chromosome and sex-chromosome-linked genes in Thamnophis sirtalis. (A) Maximum likelihood trees of fragments of six genes with Z and W alleles in T. sirtalis. Circles at nodes represent bootstrap values: Black circles indicate bootstrap values >70; white circles indicate bootstrap values <70. (B) Female-specific amplification of PCR primers designed from T. sirtalis W scaffolds in two male and two female T. sirtalis samples for six genes. Primer sequences are in supplementary table S5, Supplementary Material online. (C) Fluorescent in situ hybridization (FISH) of a Bkm-like repeat, (GATA)n in a female T. sirtalis. Sex-specific hybridization on the W chromosome is indicated by an arrow.
Annotation and Analysis of MicroRNAs
MicroRNAs are short (18–23 nucleotide) noncoding RNAs that play central roles in the posttranscriptional regulation of mRNA transcripts (Ambros 2004; Bartel 2004). To establish a repertoire of small regulatory RNAs within the T. sirtalis genome, we identified 203 precursor (189 conserved and 14 novel; supplementary fig. S3, Supplementary Material online) and associated mature microRNAs from T. sirtalis (supplementary file S1, Supplementary Material online). Nucleotide frequencies within mature microRNAs were found to be highly similar between the garter snake and king cobra genomes when compared with human, with a high frequency of uracil base at position 9, just outside of the 2–8 seed region responsible for target binding (supplementary fig. S4, Supplementary Material online). A total of 576 mRNA transcripts were predicted to be targets of the novel T. sirtalis mature microRNAs (supplementary file S1, Supplementary Material online). Notably, the novel T. sirtalis microRNA, tsi-miR-novel4, was found to target several components of phosphoinositide metabolism; namely, phosphoinoside-3-kinase, regulatory subunit 5 (PIK3R5, also called p101), and inositol polyphosphate-4-phosphoatase (INPP4A). Interestingly, PI3K is responsible for the production of phosphatidylinositol 3, 4, 5-triphosphate (PIP3) from the phosphorylation of phosphatidylinositol 4, 5-bisphosphate (PIP2). PIP2 is a key lipid signaling molecule involved in the signaling cascade that is responsible for initiating the venom production cycle in snake secretory cells following the release and depletion of venom (Jansen and Foehring 1983; Kerchove et al. 2008). We speculate that snake-specific microRNAs may play a role in venom production cycle, perhaps by regulating the metabolism of PIP2. As such, it is likely that T. sirtalis-specific microRNA are involved in specific cellular pathways that contribute to unique facets of its biology.
Evolution of Genes Related to Vision, Olfaction, and Venom
Several multigene families are of particular interest in snakes due to their association with unique aspects of snake biology. Previous work in the python and cobra has shown significant expansions of gene families in terminal snake branches, including gene families involved in chemoreception (i.e., vomeronasal, olfactory, and ephrin-like receptors) and vision (i.e., opsin genes), which are associated with the utility of the snake tongue and the hypothesis of a fossorial ancestor to all snakes, respectively. Additionally, genes families involved in the production of snake venom proteins are important for understanding one of the most extreme adaptations of the snake lineage. Here, we characterize the size and composition of these gene families in the garter snake and provide additional insight into the evolution of visual, olfactory, and venom systems in snakes.
Evolution of Visual Systems and Related Genes in Snakes
It has been proposed that a period of visual degeneration occurred during the origin of snakes due to a hypothesized fossorial phase, although alternate theories include marine or nocturnal ancestry (Vidal and Hedges 2004; Davies et al. 2009; Simões et al. 2015; Schott et al. 2018). This period of visual degeneration is supported by many morphological changes to the snake eye and the loss of 11 opsin genes that are otherwise present in squamate reptiles (Castoe 2013; Schott et al. 2017, 2018). This loss is similar to mammals, which have independently also lost 11 opsin genes, presumably following the hypothesized nocturnal phase in early mammalian evolution (Hall et al. 2012; Angielczyk and Schmitz 2014; Bickelmann et al. 2015). Recently, Schott et al. (2018) analyzed gene loss and patterns of selection in snake phototransduction genes and found evidence to support a dim-light, but not fossorially adapted, origin of snakes. However, numerous other genes are involved in visual perception in vertebrates and it is unknown how these genes were affected by the hypothesized visual degeneration of ancestral snakes. The increasing number of reptilian genomes allows us to reconstruct where in squamate evolution visual genes may have been lost. Here, we examined the losses of visual genes in snakes and compared those to losses in mammals and other vertebrate clades with nocturnal ancestry.
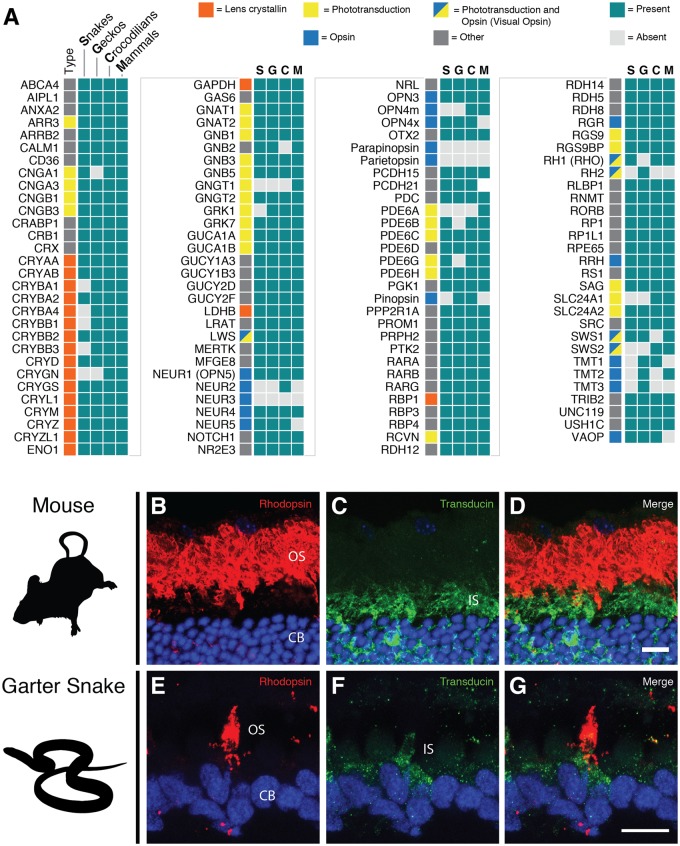
Snakes have lost 17 visual genes (20 including the single gene lost in all squamates and two genes lost in all reptiles), which is more than any other group with nocturnal or dim-light ancestry (fig. 4A, supplementary fig. S5, Supplementary Material online). In comparison, 11 genes have been lost in the lineage leading to mammals, all of which are opsins (fig. 4A). Placental mammals have lost three additional opsins, increasing the total number of lost opsin genes beyond that in snakes (supplementary fig. S5, Supplementary Material online). Geckos, another group with a hypothesized highly nocturnal ancestry (Walls 1942; Gamble et al. 2015), have specifically lost 11 visual genes, 7 of which were, independently, also lost in snakes (fig. 4A). Crocodilians have lost the fewest visual genes (five), all of which were opsins (fig. 4A).
Fig. 4.
—Visual system gene loss across amniote vertebrates and rod-like characteristics of some cone cells in the all cone retina of Thamnophis sirtalis. (A) Presence of 119 visual and opsin genes in snakes and other ancestrally nocturnal groups. (B–G) Immunohistochemical staining of mouse (control, B–D) and T. sirtalis (E–G) 20 µm transverse retinal cryosections with rhodopsin (RET-P1) and rod-specific transducin (K20) antibodies. Rhodopsin is found in a small subset of cone-like cells mainly in the outersegment (OS) of the photoreceptor (E). Rod transducin (F) is also found in the inner segments (IS) of these same photoreceptors (G). The nuclei of the photoreceptor cells (CB) is stained in blue, rhodopsin (RET-P1) staining is shown in red, and rod-specific transducin (K20) staining is shown in green. Scale bar = 10 µm.
Our analyses of the visual gene complement in T. sirtalis and other snakes, when compared with other groups with nocturnal/dim-light ancestry, strongly supports dim-light ancestry in the lineage leading to snakes. Opsins appear to be highly susceptible to loss in nocturnal groups, with each group analyzed showing the loss of two visual opsins: A middle-wavelength opsin (RH1 or RH2) and a short-wavelength opsin (either SWS1 or SWS2). We did not observe the loss of more than two visual opsins. Loss of nonvisual opsins was substantial in all groups, with many of the same genes being lost independently along different lineages. Loss of phototransduction genes was rare, and only occurred in snakes and geckos, whereas the loss of lens crystallin genes was largely unique to snakes, with the exception of a single loss in geckos. The snake lens differs substantially from other squamates in that it is rigid and spherical and is moved back and forth instead of being compressed during accommodation (Walls 1942). The loss of five lens crystallins genes may be related to these differences. The recruitment and evolution of new, snake-specific lens crystallins may also have occurred but has yet to be studied. Other than the increased loss of lens crystallins, gene loss in snakes closely resembles losses in mammals and geckos, which supports the hypothesis of dim-light ancestry in snakes.
Evidence of transmuted, cone-like rod photoreceptors in T. sirtalis
In the vertebrate eye, two types of photoreceptors are responsible for absorbing light for visual perception: Cones, which function in bright light and for color vision, and rods, which function in dim-light. While most animals have both rods and cones, enabling them to see in a wide range of light conditions, some squamates, including snakes, have only one type of photoreceptor, leading to an all-cone or all-rod retina (Walls 1942; Underwood 1970). Recently, it has been shown that the apparent all-cone retina of a diurnal garter snake, Thamnophis proximus, evolved not through loss of the rods, but rather through transmutation (i.e., evolutionary modification) of the rods to resemble the appearance and function of cones (Schott et al. 2016). This provided the first molecular support for Walls’ (1942) transmutation theory proposed over 75 years ago. This has since been demonstrated in a second colubrid, the pine snake (Pituophis melanoleucus), with additional support for a cone-like function of rod opsin that is expressed in transmuted photoreceptors (Bhattacharyya et al. 2017). It is still unknown, however, how widespread this particular mechanism is in the development of all-cone retinas.
Despite an apparent all-cone retina (Sillman et al. 1997), T. sirtalis maintains all rod phototransduction genes within its genome, with the exception of rhodopsin kinase (GRK1), which was lost in all snakes, including those with retinas containing both rods and cones (e.g., Python). To determine if rod genes are actually expressed in the photoreceptors, we performed immunohistochemistry on frozen sections of T. sirtalis retina and visualized them with fluorescent confocal imaging. A small subset of photoreceptors in the all-cone retina of T. sirtalis show rhodopsin protein in the outer segment and rod transducin in the inner segment of the same photoreceptor (fig. 4B). The combination of the intact rhodopsin gene in the genome and the presence of rhodopsin protein in the outer segment of photoreceptors, along with a downstream member of the rod phototransduction cascade suggests that T. sirtalis, like T. proximus, has transmuted, cone-like rods (Schott et al. 2016). These findings support the hypothesis that all-cone retinas in diurnal snakes evolved not through the loss of rods, but rather through evolutionary modification of the rods to resemble and function as cones. Recent work examining opsin expression in a variety of snakes suggests that this mechanism may be widespread (Simoes et al. 2016), but additional work is needed to determine if this is the sole mechanism that produced all-cone retinas in snakes, and conversely, to determine the mechanisms behind the evolution of all-rod retinas.
ORs
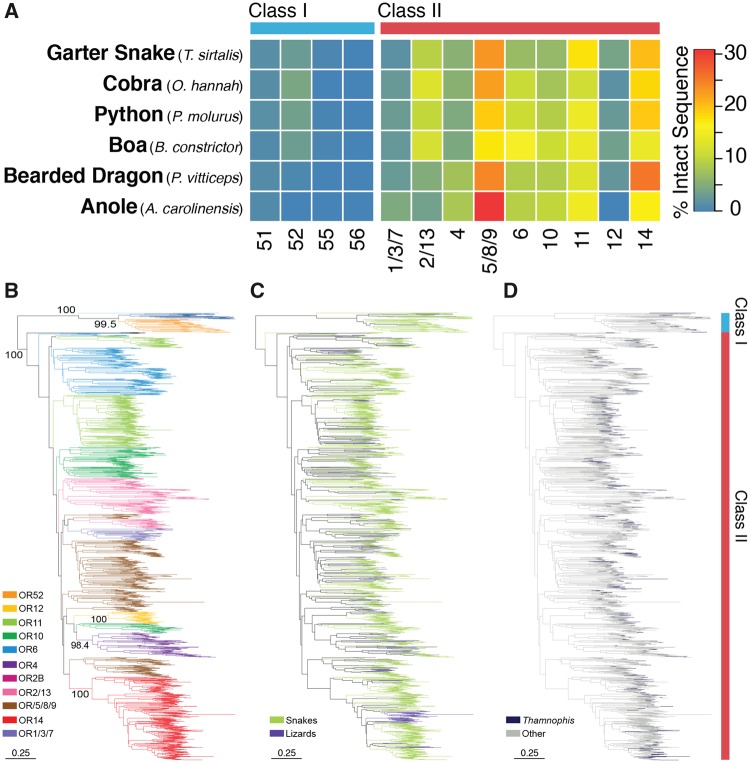
Snakes possess a greater reliance on olfaction than their more visually oriented lizard ancestors (Mason 1992; Mason and Parker 2010; Dehara et al. 2012; Castoe 2013; Vandewege et al. 2016), which suggests that changes in OR repertoires between lizards and snakes may play an important role in the evolution and tuning of snake sensation. A comparison among six representative squamate reptile species identified several striking differences between the OR repertoires of snakes and lizards, most notably that the two lizards possess substantially fewer OR genes than snakes (supplementary table S6, Supplementary Material online). The two lizards, the bearded dragon and anole, are visual insectivores and likely do not rely heavily on a sense of smell. In contrast, the high abundance of ORs in snakes is likely linked to the reliance of snakes on detecting odorants to find prey and mates (Mason and Parker 2010). Yet, despite the copy number variation observed between snakes and lizards, the overall composition of the OR repertoire was relatively similar (fig. 5A), with several exceptions. For example, there was a slightly higher abundance of Class I, subfamily 52 ORs in snakes (fig. 5A). Class I ORs are hypothesized to have a higher affinity for aquatic odorants and are typically uncommon in terrestrial vertebrates, excluding turtles (Wang et al. 2013). To visualize the expansion and contraction history of ORs, we estimated the phylogeny of fully intact ORs for squamates in our analysis (fig. 5B–D). Within most subfamilies, there were clear snake specific expansions (fig. 5B and C), although no obvious expansions appear to take place specifically within the lineage leading to Thamnophis (fig. 5D). Additionally, Thamnophis had the smallest OR repertoire among snakes (supplementary table S6, Supplementary Material online), with almost 180 fewer OR genes than the Burmese python. These results agree with a previous study (Vandewege et al. 2016) that suggested significant expansions of ORs during snake evolution. However, here we provide new evidence that no major changes in snake OR repertoires appear to have occurred early in snake evolution, indicating that differential expansions of ORs do not seem to be associated with shifts in life history or diversity among snakes.
Fig. 5.
—Genomic content and evolutionary history of olfactory receptors in squamate reptiles. (A) A heatmap representing the relative percentage of intact olfactory receptor sequences belonging to each subfamily for each species. (B–D) Neighbor joining tree of 1872 OR amino acid sequences: (B) Branches are colored by annotated subfamily; (C) as either snake or lizard; (D) garter snake terminal branches are contrasted from the remaining species.
Venom and venom encoding genes
Venom is a key adaptation for many colubroid snakes that facilitates the capture of prey. While the majority of medically important venomous snakes is restricted to the families Viperidae and Elapidae, venom originated early in squamate evolution and, consequently, most advanced snakes are venomous to some degree (Fry et al. 2008). This includes many rear-fanged venomous species of colubrid snakes, such as T. sirtalis. Although venom genes and gene families have been extensively studied in snakes, only a few studies have examined them in the context of a high-quality reference genome (Vonk et al. 2013; Reyes-Velasco et al. 2015; Yin et al. 2016; Aird et al. 2017). Additionally, the lack of genomic resources for rear-fanged venomous colubrid snake species has prevented detailed analysis of the genes involved in this venom system specifically. We were therefore motivated to examine a subset of known venom gene families in the garter snake and analyze the composition of its secreted venom to provide important context for the evolution of venom repertoires in rear-fanged snake species.
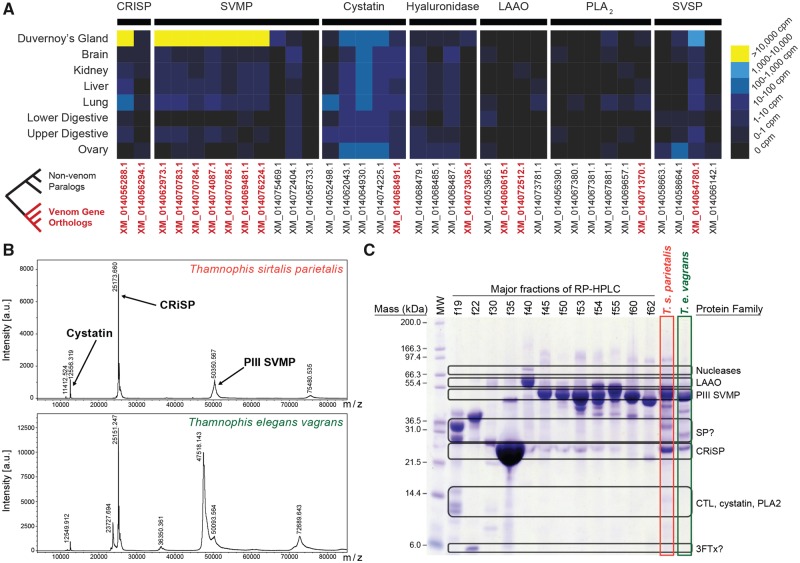
We analyzed the T. sirtalis genome for evidence of venom homologs related to the cysteine-rich secretory protein (CRISP), snake venom metalloproteinase (SVMP), cystatin, hyaluronidase, L-amino acid oxidase (LAAO), phospholipase A2 (PLA2), and snake venom serine protease (SVSP) toxin families. Phylogenetic reconstructions of genes identified in the T. sirtalis genome and related homologs from other snake and vertebrate species revealed a total of 15 garter snake genes nested within snake venom gene clades (supplementary figs. S6–S12, Supplementary Material online), indicative of a role in venom (based on forming a phylogenetic cluster with known snake venom toxins). We identified multiple paralogs for the SVMP (seven), CRISP (two), and LAAO (two) toxin families. In addition, we found single orthologous venom genes in the garter snake genome for cystatin, hyaluronidase, SVSP and PLA2 toxin families. We investigated linkage among venom genes based on our assembly for the garter snake and found that both CRISP genes are located on the same scaffold in the assembly and are separated by two long noncoding RNA genes, while all cystatins (including the venom paralog) are located on different scaffolds. Interestingly, we find evidence for duplications in two distinct clades of SVMP in T. sirtalis (supplementary fig. S12, Supplementary Material online), and three of the seven identified SVMPs are located on the same scaffold and in tandem, despite not all being members of the same clade. However, the annotation of one of these SVMPs (XM_014070785) is questionable because this gene and a second SVMP have exons that appear to be interspersed but not overlapping, potentially representing an erroneous assembly or misannotation of this region. A fourth gene (XM_014070779) in this region is also annotated as a SVMP, but this gene did not produce a significant BLAST hit to known SVMPs, and was therefore excluded from analyses. In sum, there remains some question as to the number of SVMP paralogs in the garter snake genome, which may be between six and eight; regardless, the SVMP toxin family encodes more paralogs in the T. sirtalis genome than any of the other detected toxin types. To summarize patterns of venom gene family evolution across squamate reptiles, we compared the total number of genes identified for the SVMP, CRISP, and cystatin toxin families in the garter snake genome to numbers identified previously in the genomes of two venomous snakes, two nonvenomous snakes, and a nonvenomous lizard (Yin et al. 2016). Notably, we find that the genome of the garter snake, a lineage not often thought of as venomous, contains similar numbers of genes in these venom families as do snakes that are considered highly venomous (e.g., pitvipers; supplementary fig. S13, Supplementary Material online).
To assess whether these putative toxin genes contribute to the venom secretions of T. sirtalis, we compared their expression levels in the transcriptomes of various tissues, including the venom gland (Duvernoy’s gland). There is growing evidence that many venom toxins have evolved from genes that are co-expressed in at least a few different tissue types, and following their recruitment, exhibit significantly higher levels of transcription in the venom gland than other tissues (Vonk et al. 2013; Hargreaves et al. 2014; Junqueira-de-Azevedo et al. 2015; Reyes-Velasco et al. 2015). We compared the expression level of the venom homologs identified in the T. sirtalis genome in eight different tissues: The venom gland, brain, kidney, liver, lung, ovaries and the lower and upper segments of the digestive tract. Of the 15 genes encoding venom toxins found in the garter snake, 13 were expressed in the venom gland; however, only eight of those genes exhibited high levels of expression consistent with their use as venom toxins (>10,000 counts per million [CPM]) (fig. 6A). These eight toxins consisted of the seven SVMP genes (10,013–119,661 CPM) and one CRISP (157,558 CPM; shown in yellow in fig. 6A). In contrast, the remaining CRISP gene showed very low expression (0.42 CPM), comparable to expression levels in other tissues (0.07–0.66 CPM). Twelve of the 15 venom genes were expressed in more than two tissue types, with only hyaluronidase restricted to expression solely in the venom gland (albeit at low levels; 3.73 CPM; fig. 6A). In contrast, all eight of the toxins with high expression levels in the venom gland were also expressed in at least five of the seven other tissue types sampled, and in each case venom gland expression was >1,000 fold greater than elsewhere (fig. 6A). These findings of co-expression across tissue types, with high expression in the venom gland, are consistent with current hypotheses of venom evolution (Vonk et al. 2013; Hargreaves et al. 2014; Junqueira-de-Azevedo et al. 2015; Reyes-Velasco et al. 2015).
Fig. 6.
—Transcriptomic and proteomic analysis of Thamnophis venom. (A) Tissue-specific expression of venom gene homologs in Thamnophis sirtalis. IDs of orthologous venom genes are red and bolded. (B) MALDI-TOF mass spectrum of T. sirtalis parietalis (top) and Thamnophis elegans vagrans (bottom) venom, using a mass window of 4–85 kDa. Note that only one minor peak, at 12.55 kDa (cystatin), and two major peaks, at 25 kDa (CRISP proteins) and 50 kDa (SVMP PIII), are observed. (C) 12% acrylamide NuPAGE SDS-PAGE gel of major RP-HPLC fractions of 1.0 mg T. sirtalis parietalis venom on a Jupiter C18 column, as well as total venom from T. s. parietalis and T. e. vagrans, indicating major protein families represented. See supplementary figure S12, Supplementary Material online for additional details. LAAO, L-amino acid oxidase; SVMP, snake venom metalloproteinase; SP, serine protease; CRISP, cysteine-rich secretory protein; CTL, C-type lectin; PLA2, phospholipase A2; 3FTx, three-finger toxins.
To link patterns of venom gene transcription with venom proteins, we analyzed venom protein content using MALDI-TOF-MS and RP-HPLC of crude venom, which demonstrated that the venom of T. sirtalis consists primarily (>80%) of two protein families: CRISPs and SVMPs (fig. 6B and C). Consistent with transcriptomic data, CRISPs and SVMPs are highly abundant in the Duvernoy’s venom gland (fig. 6A and B), and the SVMPs in particular appear to have multiple proteoforms that elute at different times following RP-HPLC (fig. 6C, supplementary fig. S14, Supplementary Material online). CRISPs and SVMPs (particularly PIII SVMPs) are common and often abundant components of rear-fanged snake venoms (Mackessy 2002; Junqueira-de-Azevedo et al. 2016; Mackessy and Saviola 2016; Pla et al. 2017). Many rear-fanged snake venom SVMPs have been shown to promote haemorrhage, suggesting an important role in feeding ecology, but the biological activities of most CRISPs are poorly defined at best, and their role in envenomation is unknown. Analysis of minor protein peaks by SDS-PAGE and MALDI-TOF-MS confirms the presence of numerous protein families (fig. 6B, supplementary fig. S14, Supplementary Material online), most of which contribute to total venom protein by <1% each (e.g., LAAO; supplementary fig. S15, Supplementary Material online) and are transcribed in the Duvernoy’s venom gland of T. sirtalis at low to moderate levels (fig. 6A). Their contribution to envenomation sequelae, if any, is therefore likely to be minimal relative to the highly expressed SVMPs and CRISPs. Anecdotal support of this conclusion is provided by a case of human envenomation by T. s. sirtalis; following extended contact by the snake, the bite resulted in minor localized edema and ecchymosis, likely due to the effects of SVMPs, but resolved quickly (Hayes and Hayes 1985).
Evolution of TTX Resistance
Thamnophis sirtalis is among the few vertebrates capable of withstanding high concentrations of TTX found in newts of the genus Taricha (Brodie and Brodie 1999; Brodie et al. 2002; Feldman et al. 2009). Populations of garter snakes and newts in western North America show a rough match between snake resistance and newt toxicity, a pattern indicative of an ongoing coevolutionary arms race (Brodie et al. 2002; Hanifin et al. 2008). TTX binds to specific amino acids in the four outer pore regions (“P-loops”) of voltage-gated sodium channels (Nav, encoded by genes in the family SCNA), blocking ion conduction and impairing physiological function (Fozzard and Lipkind 2010). Garter snake resistance to TTX is known to derive from evolutionary changes in these P-loops (Geffeney et al. 2002, 2005; Feldman et al. 2009; McGlothlin et al. 2014).
Previous work has shown that T. sirtalis possesses at least three TTX-resistant Nav channels: The skeletal muscle channel Nav1.4 (encoded by the gene SCN4A), and two peripheral nerve channels Nav1.6 (SCN8A, which is also expressed in the brain) and Nav1.7 (SCN9A) (Geffeney et al. 2005; McGlothlin et al. 2014). In contrast, three channels found only in the central nervous system (Nav1.1, 1.2, and 1.3, encoded by SCN1A, 2A, and 3A, respectively) (Catterall et al. 2005) and thus shielded by the blood–brain barrier (Zimmer 2010), do not appear to be TTX-resistant (McGlothlin et al. 2014). Peripheral nerves evolved TTX resistance significantly earlier than did muscle, with resistant Nav1.7 evolving at least 170 Ma in nonsnake squamates and resistant Nav1.6 evolving ∼40 Ma in three different snake families (McGlothlin et al. 2016). Resistance in Nav1.4 evolved within T. sirtalis and is polymorphic, with more highly resistant alleles found in populations that overlap with more toxic newts (Geffeney et al. 2005; Feldman et al. 2010; Hague et al. 2017). At least four other snake species have also evolved TTX resistance in Nav1.4 due to interactions with toxic amphibians (Feldman et al. 2009, 2012), all of which occur in lineages with resistant Nav1.6 and 1.7 (McGlothlin et al. 2016).
The three other members of the voltage-gated sodium channel, Nav1.5 (found in cardiac muscle and encoded by the gene SCN5A), 1.8, and 1.9 (both found in small sensory neurons and encoded by SCN10A and 11A, respectively) have not been previously described in T. sirtalis. These three channels are closely related, arising from tandem gene duplications before the mammal-reptile split (Dib-Hajj et al. 1999; Zakon et al. 2011; Widmark et al. 2011). In mammals, all three channels display TTX resistance due to the substitution of a non-aromatic residue for an aromatic one in the domain I (DI) P-loop (Backx et al. 1992; Satin et al. 1992; Akopian et al. 1996; Cummins et al. 1999; Leffler et al. 2005). Previously, TTX resistance of these channels was thought to be ubiquitous in amniotes, with resistance arising before the gene duplication events, but recent work on Nav1.5 has called this into question (Vornanen et al. 2011). Therefore, reconstructing the history of these genes is important for a complete understanding of physiological resistance to TTX in T. sirtalis. Because each of these three channels is expressed in the periphery and thus potentially exposed to ingested TTX, we predict that all three paralogs should be resistant to TTX in garter snakes. This resistance may have either been inherited from an ancient common ancestor or evolved more recently in response to selection imposed by toxic prey.
From the current genome assembly, we obtained partial or complete coding sequences for all nine SCNA genes. Here, we focus on the three genes previously undescribed in T. sirtalis; additional detail on the location of annotated SCNA genes can be found in Supplementary Information 2.1. We found evidence suggesting that Nav1.5, Nav1.8, and Nav1.9 are all either moderately or strongly resistant to TTX in T. sirtalis (supplementary table S7, Supplementary Material online). Nav1.5 has a cysteine in DI (Y371C) that is known to confer ∼3,000-fold resistance to TTX in mammalian Nav1.5 and pufferfish Nav1.4a (Backx et al. 1992; Satin et al. 1992; Perez-Garcia et al. 1996; Penzotti et al. 1998; Venkatesh et al. 2005). (For each substitution we discuss, the ancestral amino acid refers to mammalian Nav1.4, a TTX-sensitive channel, and the position number refers to the location for the specific T. sirtalis paralog.) DI also has an arginine (N374R) known to provide 2-fold resistance to TTX (Terlau et al. 1991). A substitution in domain III (DIII; D1425S) occurs at a site known to influence TTX binding, but has unknown effects. Taken together, the presence of these amino acids suggests that the T. sirtalis Nav1.5 should be blocked only at micromolar to millimolar concentrations of TTX (as opposed to nanomolar for TTX-sensitive channels).
Surprisingly, Nav1.8 DI does not have Y348S as in mammals, but does possess N351R, homologous to the arginine in Nav1.5 DI. DIII has a threonine (M1376T) known to provide ∼15-fold resistance, which also occurs in several pufferfish channels (Jost et al. 2008) and in the garter snake T.couchii Nav1.4 (Feldman et al. 2009). This suggests that T. sirtalis Nav1.8 can probably function up to TTX concentrations of about 1 µM. Nav1.9 is likely to be extremely TTX-resistant, due to Y362S (∼7,000-fold; Leffler et al. 2005) and N365R in DI. A DII serine (E767S) and a DIV glutamate (G1479E) may provide additional TTX resistance, although these particular substitutions have not been tested. Thus, as in mammals, T. sirtalis Nav1.9 is probably resistant to TTX in the millimolar range.
Ancestral reconstructions confirm that extreme resistance in Nav1.5 and Nav1.9 is ancient, dating to the common ancestor of mammals and reptiles (∼320 Ma; fig. 7; Hedges et al. 2015). All species we examined had a DI substitution providing micromolar resistance in Nav1.9. In Nav1.5, Y371C also dates to an ancient common ancestor, although some groups, including Gallus and Anolis, have lost this substitution. In contrast, ancestral Nav1.8 shows only mild resistance via N351R. Although several lineages have independently evolved increased resistance in this channel, the M1376T found in T. sirtalis is unique. On the basis of our reconstruction, this substitution apparently evolved sometime within the last 50 my, after the split between colubrid and elapid snakes (fig. 7; Zheng and Wiens 2016).
Fig. 7.
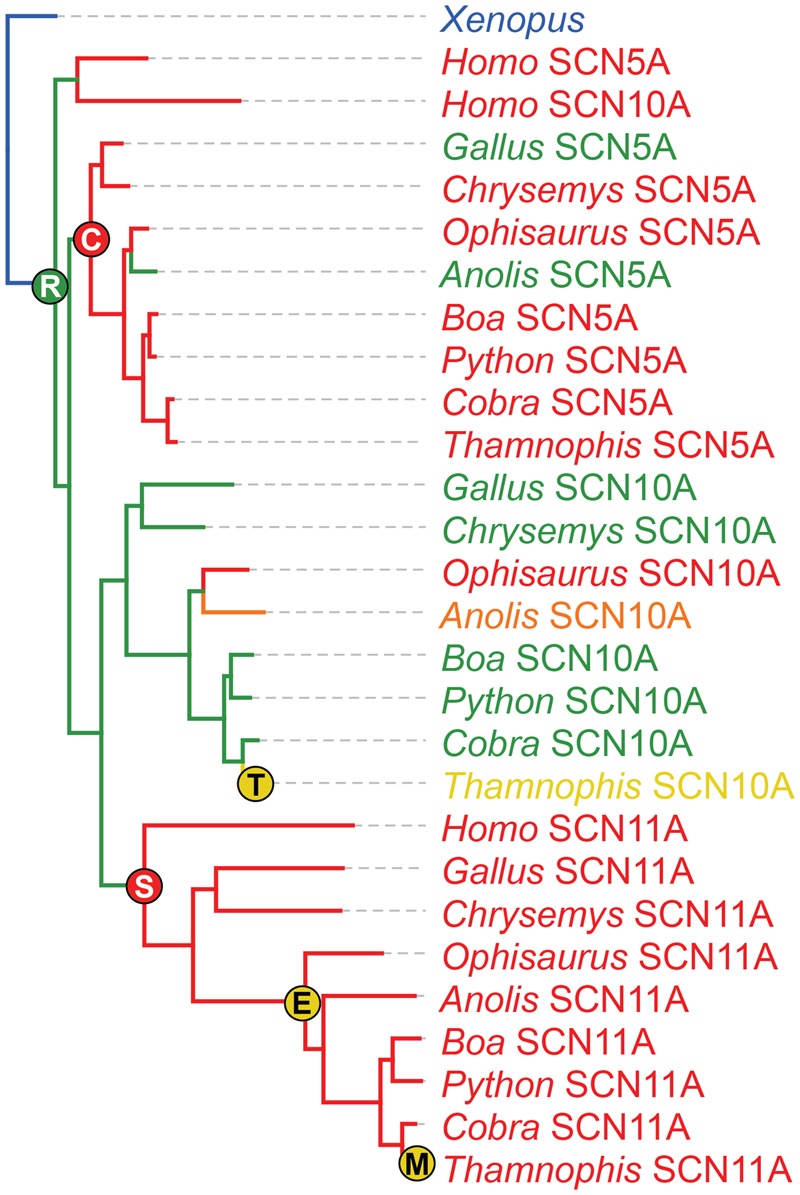
—Evolutionary reconstruction of TTX resistance in SCN5A (Nav1.5), SCN10A (Nav1.8), and SCN11A (Nav1.9) across amniote vertebrates. Tips and branches are color-coded by predicted TTX resistance, with warmer colors representing stronger resistance (see supplementary table S7, Supplementary Material online for details). Resistance-conferring substitutions found in Thamnophis sirtalis are plotted at their origin, with warmer colors indicating stronger resistance (supplementary table S7, Supplementary Material online). A nonresistant SCN5A-like paralog from Xenopus is used as the outgroup.
Combined with previous work, these results indicate that TTX resistance in T. sirtalis involves six out of the nine genes in the SCNA family, with only the brain channels Nav1.1, Nav1.2, and Nav1.3 remaining completely TTX sensitive. Snakes were predisposed to evolve physiological TTX resistance because they inherited strong TTX resistance in three channels—Nav1.5, Nav1.7, and Nav1.9—from their common ancestor. Later, Nav1.6 evolved TTX resistance in three families, perhaps in response to eating amphibians, facilitating evolution of resistance in Nav1.4 via co-evolutionary arms races with amphibians in five species (McGlothlin et al. 2016). Our results show that a third channel, Nav1.8, although mildly resistant to TTX for 300 Myr, has shown an evolutionary increase in TTX resistance within the last 50 Myr, which may have also been selected for by an amphibian diet. Additional research is needed to determine the timing of this substitution and the selective factors involved.
Conclusion
Our analysis of the garter snake genome, together with the genomes of other amniotes, provides new insight into vertebrate genome structure, function, and evolution, and provides genomic context to key aspects of garter snake biology. Our analyses of genome evolution and structure reveal major shifts in repeat element landscape composition and expansion in the garter snake and other colubroid snakes, and identify both snake- and garter snake-specific microRNAs that may play a role in unique features of snake biology such as venom. We identified a small number of genes under positive selection in garter snakes and colubrids in general, and estimates of neutral substitution rates across 4-fold degenerate sites suggest that the rate of substitutions in snakes is similar to that in mammals, and generally slightly lower than that of lizards. Identification of Z and W sex chromosome-specific scaffolds in the garter snake genome assembly highlight the value of this genome resource for future studies of sex chromosome evolution. Evolutionary analyses of gene families involved in snake sensory systems reveal patterns of gene loss in visual gene families that strongly support dim-light ancestry in snakes, suggesting that the all-cone retina of Thamnophis evolved through transmutation of rods into cone-like cells. Additionally, these analyses provide evidence for a major expansion of OR repertoires early in snake evolution, but with no further expansions associated with subsequent diversification or life history shifts in the snake lineage. Finally, we provide one of the first detailed analyses to characterize and link the protein composition of venom, the genes that encode these proteins, and their evolutionary origins in a rear-fanged venomous colubrid species, together with new insight into the evolution of TTX resistance in some garter snakes in response to toxic newt prey. Collectively, our analyses demonstrate a broad spectrum of genomic adaptations linked to the many extreme and unique features of snakes, while also highlighting the variation in genomic features among snakes that together demonstrate why snakes represent intriguing and valuable model systems for diverse research questions.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the production sequencing group at the McDonnell Genome Institute for assistance with genome sequencing, assembly and annotation, and John Abramyan for assistance with SCNA annotations in the chicken and turtle. The work was supported by funding from the National Human Genome Research Institute at National Institutes of Health [5U54HG00307907 to W.C.W.]; the National Science Foundation [DEB-1655571 and IOS-1655735 to T.A.C.]; a Natural Sciences and Engineering Research Council of Canada Discovery Grant [to B.S.W.C., K.B.S., and K.K.B.]; an Academia Sinica award [to C.H.K.]; a Minnesota Supercomputing Institute award [to S.E.M.]; and a Grand Challenges in Biology Postdoctoral Fellowship [to C.N.P.].
Literature Cited
- Adams RH. 2016. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 59(5):295–310. [DOI] [PubMed] [Google Scholar]
- Aird SD, et al. 2017. Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genome Biol Evol. 9(10):2640–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian AN, Sivilotti L, Wood JN.. 1996. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 379(6562):257–262. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Ambros V. 2004. The functions of animal microRNAs. Nature 431(7006):350–355. [DOI] [PubMed] [Google Scholar]
- Angielczyk KD, Schmitz L.. 2014. Nocturnality in synapsids predates the origin of mammals by over 100 million years. Proc R Soc Lond Ser B: Biol Sci. 281(1793):20141642.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnemann J, et al. 1986. Clustered GATA repeats (Bkm sequences) on the human Y chromosome. Hum Genet. 73(4):301–303. [DOI] [PubMed] [Google Scholar]
- Backx PH, et al. 1992. Molecular localization of an ion-binding site within the pore of mammalian sodium channels. Science 257(5067):248–252. [DOI] [PubMed] [Google Scholar]
- Baker RJ, Mengden GA, Bull JJ.. 1972. Karyotypic studies of thirty-eight species of North American snakes. Copeia 1972(2):257–265. [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. [DOI] [PubMed] [Google Scholar]
- Bernard MA, et al. 2001. Diminished levels of the putative tumor suppressor proteins EXT1 and EXT2 in exostosis chondrocytes. Cytoskeleton 48(2):149–162. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya N, et al. 2017. Cone-like rhodopsin expressed in the all cone retina of the colubrid pine snake as a potential adaptation to diurnality. J Exp Biol: Jeb. 156430:2418–2425. [DOI] [PubMed] [Google Scholar]
- Bickelmann C, et al. 2015. The molecular origin and evolution of dim-light vision in mammals. Evolution 69(11):2995–3003. [DOI] [PubMed] [Google Scholar]
- Boetzer M, et al. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Bohlen CJ, et al. 2011. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 479(7373):410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comp Biol. 10(4):e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, et al. 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience 2(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie ED, Brodie ED.. 1990. Tetrodotoxin resistance in garter snakes: an evolutionary response of predators to dangerous prey. Evolution 44(3):651–659. [DOI] [PubMed] [Google Scholar]
- Brodie ED III, Brodie ED Jr. 1999. Predator–prey arms races. Bioscience 49(7):557–568. [Google Scholar]
- Brodie ED Jr, Ridenhour BJ, Brodie ED III. 2002. The evolutionary response of predators to dangerous prey: hotspots and coldspots in the geographic mosaic of coevolution between newts and snakes. Evolution 56:2067–2082. [DOI] [PubMed] [Google Scholar]
- Bromham L. 2002. Molecular clocks in reptiles: life history influences rate of molecular evolution. Mol Biol Evol. 19(3):302–309. [DOI] [PubMed] [Google Scholar]
- Bromham L, Penny D.. 2003. The modern molecular clock. Nat Rev Genet. 4(3):216.. [DOI] [PubMed] [Google Scholar]
- Castoe TA. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 110:20645–20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, et al. 2011. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol Evol. 3:641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J.. 2005. Nomenclature and structure–function relationships of voltage-gated calcium channels, international union of pharmacology. XLVIII. Pharmacol Rev. 57(4):411–425. [DOI] [PubMed] [Google Scholar]
- Crews D, et al. 1984. Hormonal independence of courtship behavior in the male garter snake. Horm Behav. 18(1):29–41. [DOI] [PubMed] [Google Scholar]
- Cummins TR, et al. 1999. A novel persistent tetrodotoxin-resistant sodium current in SNS-null and wild-type small primary sensory neurons. J Neurosci. 19(24):RC43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WL, et al. 2009. Shedding light on serpent sight: the visual pigments of henophidian snakes. J Neurosci. 29(23):7519–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehara Y, et al. 2012. Characterization of squamate olfactory receptor genes and their transcripts by the high-throughput sequencing approach. Genome Biol Evol. 4(4):602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj SD, et al. 1999. Coding sequence, genomic organization, and conserved chromosomal localization of the mouse gene Scn11a encoding the sodium channel NaN. Genomics 59(3):309–318. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effron M, Griner L, Benirschke K.. 1977. Nature and rate of neoplasia found in captive wild mammals, birds, and reptiles at necropsy. J Natl Cancer Inst. 59(1):185–198. [DOI] [PubMed] [Google Scholar]
- Enright AJ, et al. 2003. MicroRNA targets in Drosophila. Genome Biol. 5(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Brodie ED, Pfrender ME.. 2009. The evolutionary origins of beneficial alleles during the repeated adaptation of garter snakes to deadly prey. Proc Natl Acad Sci USA. 106(32):13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Pfrender ME.. 2010. Genetic architecture of a feeding adaptation: garter snake (Thamnophis) resistance to tetrodotoxin bearing prey. Proc R Soc Lond Ser B: Biol Sci. 277:3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED Jr, Brodie ED III, Pfrender ME.. 2012. Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes. Proc Natl Acad Sci U S A. 109(12):4556–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozzard HA, Lipkind GM.. 2010. The tetrodotoxin binding site is within the outer vestibule of the sodium channel. Mar Drugs. 8(2):219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, et al. 2008. Evolution of an arsenal structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol Cell Proteomics. 7(2):215–246. [DOI] [PubMed] [Google Scholar]
- Gamble T, et al. 2017. The discovery of XY sex chromosomes in a boa and python. Curr Biol. 27(14):2148–2153.e2144. [DOI] [PubMed] [Google Scholar]
- Gamble T, Geneva AJ, Glor RE, Zarkower D.. 2014. Anolis sex chromosomes are derived from a single ancestral pair. Evolution 68(4):1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T, Greenbaum E, Jackman TR, Bauer AM.. 2015. Into the light: diurnality has evolved multiple times in geckos. Biol J Linn Soc. 115(4):896–910. [Google Scholar]
- Garner MM, Hernandez-Divers SM, Raymond JT.. 2004. Reptile neoplasia: a retrospective study of case submissions to a specialty diagnostic service. Vet Clin North Am Exot Anim Pract. 7(3):653–671. [DOI] [PubMed] [Google Scholar]
- Geffeney S, Ruben PC, Brodie ED Jr, Brodie ED III. 2002. Mechanisms of adaptation in a predator–prey arms race: TTX resistant sodium channels. Science 297(5585):1336–1339. [DOI] [PubMed] [Google Scholar]
- Geffeney SL, et al. 2005. Evolutionary diversification of TTX-resistant sodium channels in a predator–prey interaction. Nature 434(7034):759–763. [DOI] [PubMed] [Google Scholar]
- Georges A, et al. 2015. High-coverage sequencing and annotated assembly of the genome of the Australian dragon lizard Pogona vitticeps. Gigascience 4:45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346(6215):1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2018. Animal Genome Size Database. http://www.genomesize.com, Last accessed January 12, 2018.
- Hague MTJ, Feldman CR, Brodie ED, Brodie ED.. 2017. Convergent adaptation to dangerous prey proceeds through the same first-step mutation in the garter snake Thamnophis sirtalis. Evolution 71(6):1504–1518. [DOI] [PubMed] [Google Scholar]
- Hall MI, Kamilar JM, Kirk EC.. 2012. Eye shape and the nocturnal bottleneck of mammals. Proc R Soc Lond Ser B: Biol Sci. 279(1749):4962–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanifin CT, Brodie ED, Brodie ED.. 2008. Phenotypic mismatches reveal escape from arms-race coevolution. PLoS Biol. 6(3):e60–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves AD, Swain MT, Hegarty MJ, Logan DW, Mulley JF.. 2014. Restriction and recruitment—gene duplication and the origin and evolution of snake venom toxins. Genome Biol Evol. 6(8):2088–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes WK, Hayes FE.. 1985. Human envenomation from the bite of the eastern garter snake, Thamnophis s. sirtalis (Serpentes: colubridae). Toxicon 23(4):719–721. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 32(4):835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog HA Jr, Burghardt GM.. 1986. Development of antipredator responses in snakes: I. Defensive and open-field behaviors in newborns and adults of three species of garter snakes (Thamnophis melanogaster, T. sirtalis, T. butleri). J Comp Psychol. 100(4):372–379. [Google Scholar]
- Herzog HA, Burghardt GM.. 2010. Development of antipredator responses in snakes III. Stability of individual and litter differences over the first year of life. Ethology 77(3):250–258. [Google Scholar]
- Hruska-Hageman AM, Wemmie JA, Price MP, Welsh MJ.. 2002. Interaction of the synaptic protein PICK1 (protein interacting with C kinase 1) with the non-voltage gated sodium channels BNC1 (brain Na+ channel 1) and ASIC (acid-sensing ion channel). Biochem J. 361(3):443.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin XH, Chelly J.. 2009. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 25(12):555–566. [DOI] [PubMed] [Google Scholar]
- Jansen DW, Foehring RC.. 1983. The mechanism of venom secretion from Duvernoy's gland of the snake Thamnophis sirtalis. J Morphol. 175(3):271–277. [DOI] [PubMed] [Google Scholar]
- Jayne BC, Bennett AF.. 1989. The effect of tail morphology on locomotor performance of snakes: a comparison of experimental and correlative methods. J Exp Zool. 252(2):126–133. [Google Scholar]
- Jones KW, Singh L.. 1985. Snakes and the evolution of sex chromosomes. Trends Genet. 1:55–61. [Google Scholar]
- Jost MC, et al. 2008. Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family. Mol Biol Evol. 25(6):1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira-de-Azevedo ILM, et al. 2015. Venom-related transcripts from Bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol Biol Evol. 32(3):754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira-de-Azevedo ILM, Campos PF, Ching ATC, Mackessy SP.. 2016. Colubrid venom composition: an-omics perspective. Toxins 8(8):230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchove CM, et al. 2008. α1-adrenoceptors trigger the snake venom production cycle in secretory cells by activating phosphatidylinositol 4, 5-bisphosphate hydrolysis and ERK signaling pathway. Comp Biochem Physiol A: Mol Integr Physiol. 150(4):431–437. [DOI] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J Jr, Tugendreich S.. 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubie JL, Vagvolgyi A, Halpern M.. 1978. Roles of the vomeronasal and olfactory systems in courtship behavior of male garter snakes. J Comp Physiol Psychol. 92(4):627. [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Laopichienpong N, et al. 2017. Evolutionary dynamics of the gametologous CTNNB1 gene on the Z and W chromosomes of snakes. J Hered. 108(2):142–151. [DOI] [PubMed] [Google Scholar]
- Leffler A, et al. 2005. Pharmacological properties of neuronal TTX-resistant sodium channels and the role of a critical serine pore residue. Pflügers Archiv. 451(3):454–463. [DOI] [PubMed] [Google Scholar]
- Liu B, et al. 2013. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv 1308:2012. [Google Scholar]
- Mackessy SP. 2002. Biochemistry and pharmacology of colubrid snake venoms. J Toxicol: Toxin Rev. 21(1–2):43–83. [Google Scholar]
- Mackessy SP, Saviola AJ.. 2016. Understanding biological roles of venoms among the Caenophidia: the importance of rear-fanged snakes. Integr Comp Biol. 56(5):1004–1021. [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27(6):764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP, Palumbi SR.. 1993. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci U S A. 90(9):4087–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RT. 1992. Reptilian pheromones In: Gans C, Crews D, editors. Hormones, brain, and behavior. Biology of the reptilia. Physiology E, Vol. 18 Chicago: University of Chicago Press; p. 114–228. [Google Scholar]
- Mason RT, Crews D.. 1986. Pheromone mimicry in garter snakes In: Duvall D, Muller-Schwarze D, Silverstein RM, editors. Chemical signals in vertebrates 4. New York, NY: Plenum Publishing Corporation. [Google Scholar]
- Mason RT, Parker MR.. 2010. Social behavior and pheromonal communication in reptiles. J Comp Physiol A. 196(10):729–749. [DOI] [PubMed] [Google Scholar]
- Matsubara K, et al. 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci USA. 103(48):18190–18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, et al. 2015. Rapid molecular evolution across amniotes of the IIS/TOR network. Proc Natl Acad Sci U S A. 112(22):7055–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin JW, et al. 2014. Parallel evolution of tetrodotoxin resistance in three voltage-gated sodium channel genes in the garter snake Thamnophis sirtalis. Mol Biol Evol. 31(11):2836–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin JW, et al. 2016. Historical contingency in a multigene family facilitates adaptive evolution of toxin resistance. Curr Biol. 26(12):1616–1621. [DOI] [PubMed] [Google Scholar]
- Mendonca MT, Crews D.. 1996. Effects of ovariectomy and estrogen replacement on attractivity and receptivity in the red sided garter snake (Thamnophis sirtalis parietalis). J Comp Physiol A. 178(3):373–381. [DOI] [PubMed] [Google Scholar]
- O’Meally D, et al. 2010. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 18(7):787–800. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin: Springer. [Google Scholar]
- Parasnis AS, et al. 1999. Microsatellite (GATA) n reveals sex-specific differences in papaya. Theor Appl Genet. 99(6):1047–1052. [Google Scholar]
- Peace RJ, Biggar KK, Storey KB, Green JR.. 2015. A framework for improving microRNA prediction in non-human genomes. Nucleic Acids Res. 43:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzotti JL, Fozzard HA, Lipkind GM, Dudley SC.. 1998. Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the Na+ channel outer vestibule. Biophys J. 75(6):2647–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garcia MT, Chiamvimonvat N, Marban E, Tomaselli GF.. 1996. Structure of the sodium channel pore revealed by serial cysteine mutagenesis. Proc Natl Acad Sci U S A. 93(1):300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla D, et al. 2017. What killed Karl Patterson Schmidt? Combined venom gland transcriptomic, venomic and antivenomic analysis of the South African green tree snake (the boomslang), Dispholidus typus. Biochim Biophys Acta (BBA)-Gener Subj. 1861(4):814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Velasco J, et al. 2015. Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol Biol Evol. 32(1):173–183. [DOI] [PubMed] [Google Scholar]
- Robert KA, Bronikowski AM.. 2010. Evolution of senescence in nature: physiological evolution in populations of garter snake with divergent life histories. Am Nat. 175(2):147–159. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Vukić J, Lymberakis P, Kratochvíl L. 2015. Evolutionary stability of sex chromosomes in snakes. Proc R Soc Lond Ser B: Bio Sci. 282(1821): 20151992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satin J, et al. 1992. A mutant of TTX-resistant cardiac sodium channels with TTX-sensitive properties. Science 256(5060):1202–1205. [DOI] [PubMed] [Google Scholar]
- Schott RK, et al. 2016. Evolutionary transformation of rod photoreceptors in the all-cone retina of a diurnal garter snake. Proc Natl Acad Sci U S A. 113(2):356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott RK, et al. 2017. Targeted capture of complete coding regions across divergent species. Genome Biol Evol. 9(2):398–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott RK, et al. 2018. Shifts in selective pressures on snake phototransduction genes associated with photoreceptor transmutation and dim-light ancestry. Mol Biol Evol. 35(6):1376–1389. [DOI] [PubMed] [Google Scholar]
- Sillman AJ, et al. 1997. The photoreceptors and visual pigments of the garter snake (Thamnophis sirtalis): a microspectrophotometric, scanning electron microscopic and immunocytochemical study. J Comp Physiol. 181(2):89–101. [DOI] [PubMed] [Google Scholar]
- Simão FA, et al. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Simões BF, et al. 2015. Visual system evolution and the nature of the ancestral snake. J Evol Biol. 28(7):1309–1320. [DOI] [PubMed] [Google Scholar]
- Simoes BF, et al. 2016. Multiple rod–cone and cone–rod photoreceptor transmutations in snakes: evidence from visual opsin gene expression. Proc R Soc B. 283:20152624.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. 2014. RepeatModeler Open-1.0. 2008–2010. http://repeatmasker.org. Last accessed February 2017.
- Smit AFA, Hubley R, Green P.. 2015. RepeatMasker Open-4.0. 2013–2015. Institute for Systems Biology. http://repeatmasker.org, Last accessed February 2017.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely G, et al. 2008. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53(1):11–28. [DOI] [PubMed] [Google Scholar]
- Terlau H, et al. 1991. Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett. 293(1–2):93–96. [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen F, et al. 2013. Eukaryotic genome annotation pipeline. In: The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK169439/
- Tollis M, et al. 2018. Comparative genomics reveals accelerated evolution in conserved pathways during the diversification of anole lizards. Genome Biol Evol. 10(2):489–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S, et al. 2004. The novel gene encoding a putative transmembrane protein is mutated in gnathodiaphyseal dysplasia (GDD). Am J Hum Genet. 74(6):1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Etzold T.. 1996. The EMBL/EBI reptile database. Herpetol Rev. 27:175. [Google Scholar]
- Ullate-Agote A, Milinkovitch MC, Tzika AC.. 2014. The genome sequence of the corn snake (Pantherophis guttatus), a valuable resource for EvoDevo studies in squamates. Int J Dev Biol. 58:881–888. [DOI] [PubMed] [Google Scholar]
- Underwood G. 1970. The eye In: Gans C, editor. Biology of the reptilia. New York: Academic Press; p. 1–97. [Google Scholar]
- Vandewege MW, et al. 2016. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol Evol. 8(3):470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, et al. 2005. Genetic basis of tetrodotoxin resistance in pufferfishes. Curr Biol. 15(22):2069–2072. [DOI] [PubMed] [Google Scholar]
- Vest DK. 1981. The toxic Duvernoy's secretion of the wandering garter snake, Thamnophis elegans vagrans. Toxicon 19(6):831–839. [DOI] [PubMed] [Google Scholar]
- Vicoso B, et al. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11(8):e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal N, Hedges SB.. 2004. Molecular evidence for a terrestrial origin of snakes. Proc R Soc Lond Ser B: Biol Sci. 271(Suppl. 4):S226–S229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. 1998. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry 31(2):100–109. [DOI] [PubMed] [Google Scholar]
- Vonk FJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 110(51):20651–20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornanen M, Hassinen M, Haverinen J.. 2011. Tetrodotoxin sensitivity of the vertebrate cardiac Na+ current. Mar Drugs. 9(11):2409–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade A, et al. 2013. Proteoglycans and their roles in brain cancer. FEBS J. 280(10):2399–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DP, et al. 2009. Comparative analysis of neurological disorders focuses genome-wide search for autism genes. Genomics 93(2):120–129. [DOI] [PubMed] [Google Scholar]
- Walls GL, editor. Biol Symposia. 1942. [Google Scholar]
- Wang Z, et al. 2013. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 45(6):701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmark J, Sundström G, Ocampo Daza D, Larhammar D.. 2011. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol. 28(1):859–871. [DOI] [PubMed] [Google Scholar]
- Xue W, et al. 2013. L_RNA_scaffolder: scaffolding genomes with transcripts. BMC Genomics. 14:604.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yin W, et al. 2016. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat Commun. 13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon HH, Jost MC, Lu Y.. 2011. Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol Biol Evol. 28(4):1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wiens JJ.. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol Phylogen Evol. 94:537–547. [DOI] [PubMed] [Google Scholar]
- Zimmer T. 2010. Effects of tetrodotoxin on the mammalian cardiovascular system. Mar Drugs. 8(3):741–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.