ABSTRACT
LncRNA H19 is involved in the development of multiple cancers. Here, we firstly provide new evidence that H19 can induce LIN28B, a conserved RNA binding protein, to accelerate lung cancer growth through sponging miR-196b. Abundance in LIN28B was observed in clinical lung cancer samples. A positive link was observed between H19 and LIN28B in clinical lung cancer samples. In lung cancer cells, H19 was capable of increasing LIN28B expression. Mechanistically, miR-196b directly targeted LIN28B to inhibit LIN28B expression. H19 was capable of promoting LIN28B expression through sequestering miR-196b. Functionally, H19-increased LIN28B conferred the cell proliferation of lung cancer. Our finding indicates that H19 depresses miR-196b to elevate LIN28B, resulting in accelerating cell proliferation in lung cancer.
KEYWORDS: H19, LIN28B, miR-196b, growth, lung cancer
Introduction
The studies have proven that there exist lncRNAs in multiple processes including chromatin modification, genomic reprogramming, and differentiation [1–3]. LncRNA H19 is firstly identified imprinting gene and plays important roles in cancers [4–6]. In multiple cancers including breast, kidney, stomach, ovarian, esophagus, and lung, overexpressed H19 promotes tumorigenesis and development [7–13]. But, it still remains mysterious about the role of H19 in lung cancer.
LIN28A and LIN28B, as two pro-oncogenes, are two homologous members of highly conserved RNA binding protein LIN28 family. In some cancers, high LIN28A or LIN28B is relevant to advanced disease stage and poor prognosis [14]. LIN28B is also involved in many oncogenic signaling networks and is reported as oncogenic stem-cell factor [15,16]. High levels of LIN28B are observed in CD166-positive NSCLC tumor-initiating cells [15]. LIN28B is also required for cell growth in lung adenocarcinoma [14]. The miR-200c/LIN28B signal could be a useful therapeutic candidate in NSCLC cells of acquired EGFR-TKI resistance and harboring epithelial-mesenchymal transition (EMT) property. C-MYC-activated LIN28B is found in many human and mouse tumor models [17,18]. LIN28B is involved in inflammation/cancer link through the positive regeneration of NF-κB activation and let-7-mediated IL-6 depression [19]. It has been reported that some microRNAs could directly bind 3ʹUTR of LIN28B mRNA and repress its translation [20–25]. However, it still needs to be further investigated about how post-transcription of LIN28B is regulated.
In current study, our finding presents the role of H19/miR-196b-associated LIN28B in lung cancer growth. We reveal that H19 induces LIN28B expression by depressing miR-196b in the acceleration of lung cancer growth. Our finding indicates a new mechanism by which lncRNA H19 augments lung cancer progression.
Results
H19 is positively associated to LIN28B in clinical lung cancer tissues
Lots of studies reveal that H19 takes pivotal part in the development of multiple cancers [4–6,26,27]. Given that LIN28B is a pro-oncogene, we speculated that H19 could regulate LIN28B to enhance lung cancer progression. To investigate the significant role of LIN28B during lung cancer progression, we evaluated the expression of LIN28B in clinical lung tumor samples and peritumor samples by real time-PCR assay. As shown in `Figure 1(a) (Wilcoxon’s signed-rank test, p < 0.001) and Supplementary Figure S1, our data manifested that LIN28B levels were higher in 32 cases of clinical lung tumor samples relative to the corresponding nontumorous tissues. And then, the obviously positive correlation between H19 and LIN28B was revealed in human lung tumor samples through real time-PCR assay (Figure 1(b), Pearson’s correlation, R = 0.7025, p < 0.001). We further validate the expression of H19 and LIN28B, and the correlation between H19 and LIN28B using the TCGA database (Supplementary Figure S2A-S2C). We find that in lung cancer the abundance of H19 is positively related to abundant LIN28B.
Figure 1.
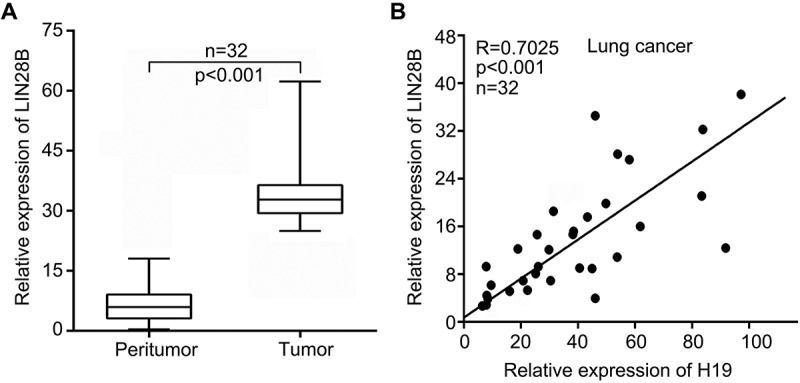
H19 is positively associated with LIN28B in human lung cancer samples. (a) LIN28B expression was detected by qRT-PCR in human lung cancer tissues and paired noncancerous tissues (Wilcoxon’s signed-rank test). (b) Correlation between H19 with LIN28B was analyzed by qRT-PCR in human lung cancer samples (Pearson’s correlation coefficient, R = 0.7025, P < 0.001).
H19 enhances LIN28B level in lung cancer
In the further investication, we focused on the function of H19 in LIN28B regulation. Our finding demonstrated that H19 significantly upregulated LIN28B in lung cancer cells A549 and H1299. The introduction of H19 in cells was evaluated by real time-PCR assay (Figure 2(a,b)). Moreover, silencing of H19 led to LIN28B decrease in the cells. We confirmed the successful siH19 transfection (Figure 2(c,d)). Taken together, we reveal that in lung cancer cells LIN28B can be unregulated by H19.
Figure 2.
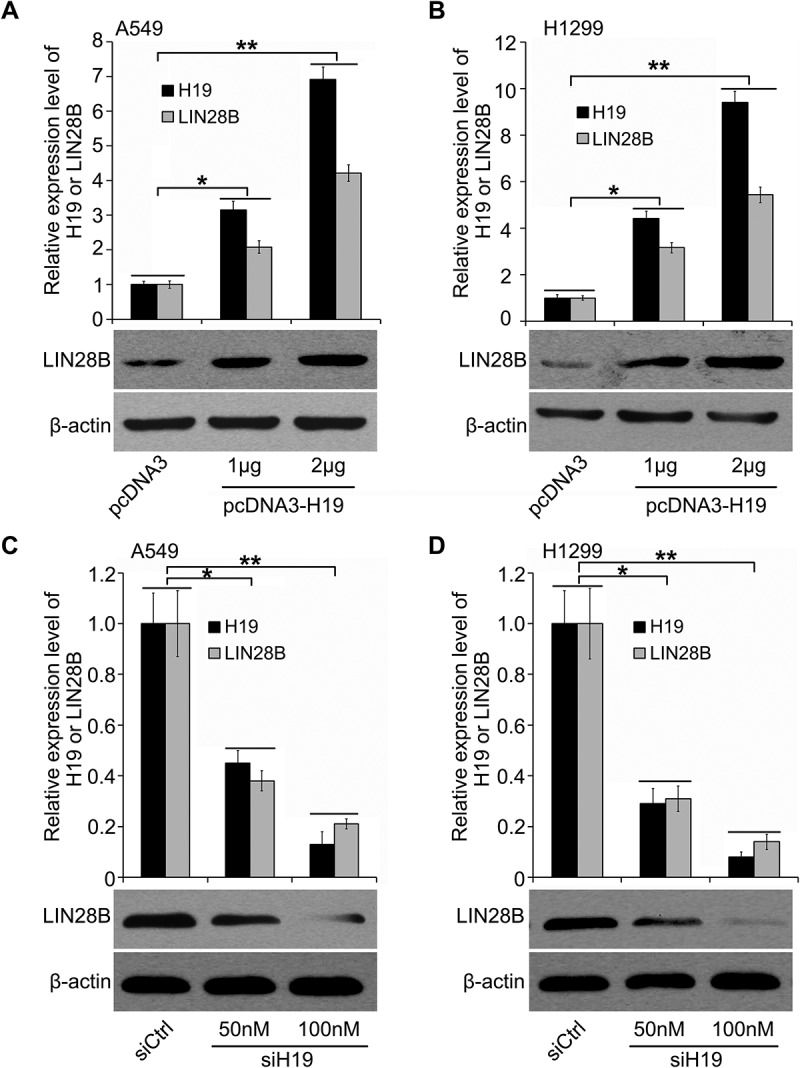
In lung cancer cells H19 induces LIN28B expression. (a, b) The modulation of H19 on LIN28B was determined by qRT-PCR and Western blotting in lung cancer A549 and H1299 cells. (c, d) The effect of H19 siRNA on LIN28B expression was investigated by Western blotting or qRT-PCR. *P < 0.05, **P < 0.01.
MiR-196b is capable of directly targeting LIN28B to control its expression
For lncRNAs has been reported to restrain miRNAs as a molecular sponge, we are wondering whether miRNAs are involved in H19-inducing LIN28B. At first, we searched for potential miRNA candidates by starBase v2.0 (http://starbase.sysu.edu.cn/). Among them, miR-196b was of most interest because of its tumor suppressive role. As indicated in Figure 3(a), using TargetScan (http://www.targetscan.org/) we found that miR-196b might bind to LIN28B mRNA 3'UTR. Furthermore, luciferase reporter analysis showed that miR-196b inhibited the activities of pGL-LIN28B-wt (wild-type miR-196b binding site of LIN28B mRNA 3'UTR cloned into pGL3-control) in a dosage-dependent manner but not pGL-LIN28B-mut (mutant miR-196b binding site of LIN28B mRNA 3'UTR cloned into pGL3-control) (Figure 3(a)). The inhibitor of miR-196b treatment could obtain the opposite results (Figure 3(b)), indicating the targeting of miR-196b on LIN28B. Finally, miR-196b was able to decrease the LIN28B level and then the miR-196b inhibitor treatment showed the inverse data (Figure 3(c,d)). We show that LIN28B is a target gene of miR-196b.
Figure 3.
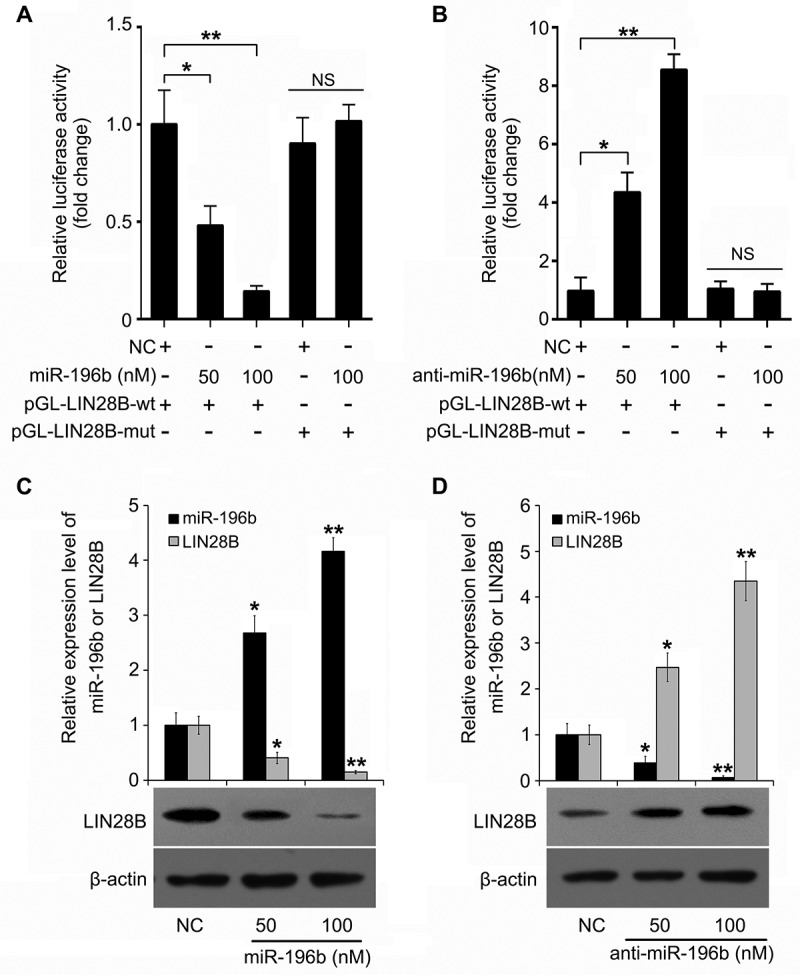
MiR-196b suppresses LIN28B expression via directly targeting LIN28B mRNA 3ʹUTR. (a, b) In A549 cells regulation of miR-196b (or anti-miR-196b) on pGL-LIN28B-wt and pGL-LIN28B-mut was tested by using luciferase reporter gene assays. (c, d) Effect of miR-196b (or anti-miR-196b) on LIN28B expression was examined by Western blotting or qRT-PCR. N.S.: no significance, *P < 0.05, **P < 0.01.
H19 induces LIN28B via suppressing mir-196b
Taken a step further, we analyzed the regulation of miR-196b by H19. We applied Bielefeld Bioinformatics Service (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) and found the potential interaction of H19 with miR-196b by complementary base-pairing reactions (Figure 4(a,b)). Moreover, we generated a mutant of H19 (H19-196b-mut) in which sixteen nucleotides of H19 for binding with miR-196b were substituted. Our data indicated that miR-196b-suppressed pGL-LIN28B luciferase activities were abolished by H19, whereas H19-196b-mut could not induce the similar result (Figure 4(c)). Next, in A549 and H1299 cells we confirmed H19-regulating LIN28B at the protein level by immunoblotting analysis (Figure 4(d)). We imply that H19 is able to upregulate LIN28B through miR-196b downregulation.
Figure 4.
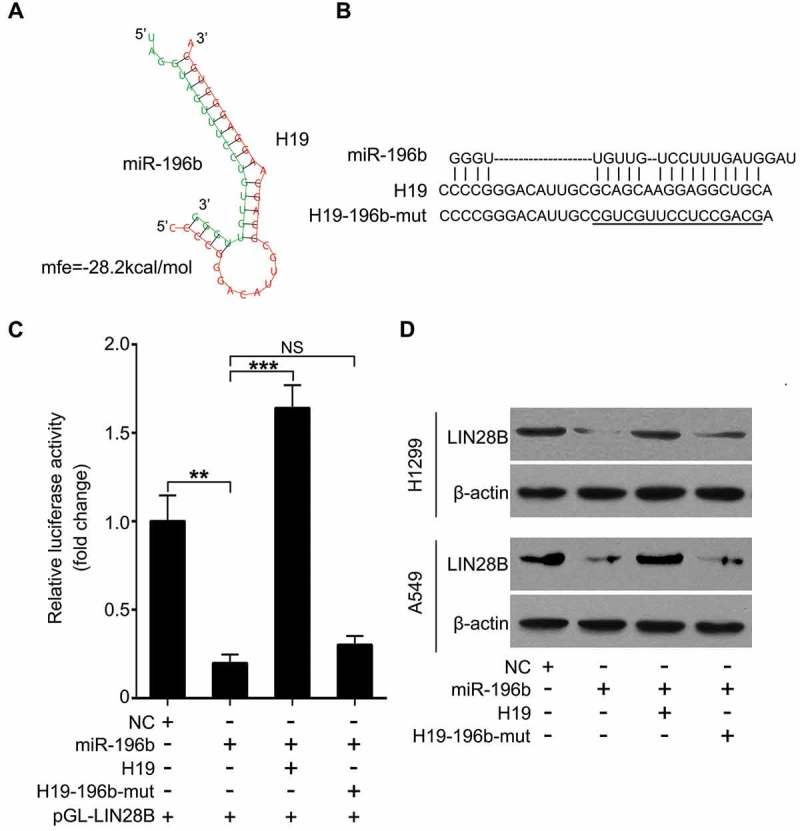
H19 is capable of elevating LIN28B expression through inhibiting miR-196b. (a, b) Bioinformatics prediction of interaction of H19 with miR-196b through complementary base-pairs was shown. The mutant of sequence of H19 binding with miR-196b was indicated. (c) Luciferase activities of pGL-LIN28B were analyzed by luciferase reporter gene assays in miR-196b and/or H19 (or H19-206-mut) transfected A549 cells. (d) In miR-196b and/or H19 (or H19-206-mut) treated cells, LIN28B expression was assessed by Western blotting. N.S.: no significance, **P < 0.01, *** P < 0.001.
H19-enhanced cell proliferation is induced by miR-196b-targeting LIN28B in lung cancer
To chase the function of the H19/miR-196b/LIN28B cascade, we tested cell proliferation of lung cancer by MTT analysis. We found that H19 promotes cell proliferation of lung cancer. However, LIN28B knockdown or miR-196b introduction could restrain the proliferation (Figure 5(a,b); Supplementary Figure S3A and S3B). Next, we observed that both H19 and LIN28B all could promote the cell colony formation, whereas miR-196b transfection or LIN28B silence disrupted H19-increased colony formation (Figure 5(c)). The overexpression or RNA interference efficiency of H19, siLIN28B or miR-196b was confirmed by Western blotting or qRT-PCR (Supplementary Figure S3C and S3D). Collectively, our finding supports that H19/miR-196b/LIN28B signal quickens cell proliferation in lung cancer.
Figure 5.
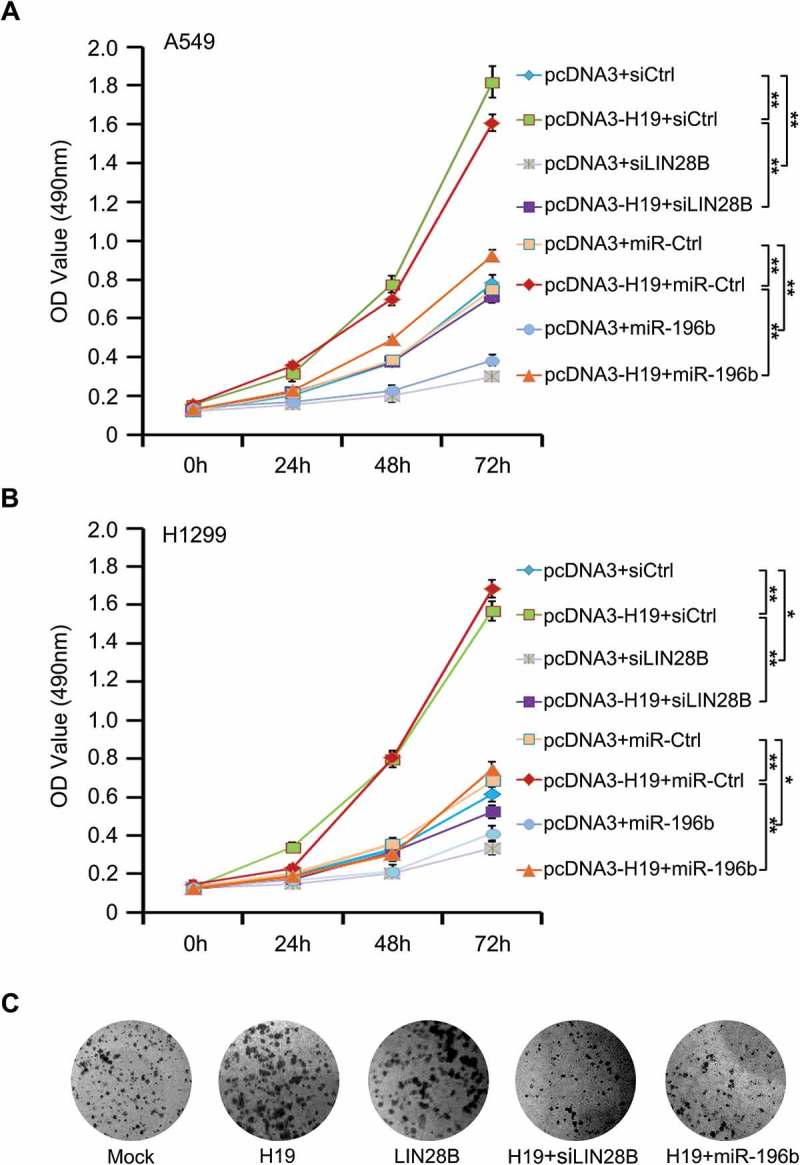
MiR-196b/LIN28B signal mediates H19-accelerating cell proliferation in lung cancer. (a, b) Proliferation was tested by MTT in A549 and H1299 cells with indicated plasmids or siRNAs. (c) Colony formation of A549 cells was counted post-transfection with indicated plasmids or siRNAs.
Discussion
As a firstly found imprinting gene, lncRNA H19 takes pivotal parts in the development of many cancers [4–6,26,27]. Overexpressed H19 is able to promote tumorigenesis and development in several cancers including breast, stomach, kidney, esophagus, ovarian, and lung [7–13]. Highly expressed H19 is also frequently observed in some human cancers, such as bladder carcinoma or head and neck squamous carcinoma [28–32]. However, the molecular mechanism by which H19 functions in lung cancer progression remains largely unclear.
RNA binding proteins (RBPs) LIN28A or LIN28B can function as proto-oncogenes. Overexpressed LIN28A or LIN28B is relevant to advanced disease stage and bad prognosis in multiple tumor types [14]. Highly expressed LIN28B was first identified in liver cancer [33]. Overexpressed LIN28B is frequently found and related to the development of many cancers [34,35]. Silencing of LIN28A or LIN28B induces tumor depression in human xenograft mouse models [36].Great evidence reveals that LIN28A and LIN28B play key role in cancer stem cell formation in some cancers [37]. Highly expressed LIN28B together with glycine decarboxylase are required for NSCLC tumorigenesis and growth [15]. However, the regulation of H19 on LIN28B in lung cancer development need to be further explored.
For the investigation of the role of H19 in LIN28B modulation in lung cancer, we tested the expression of H19 and LIN28B in clinical lung cancer tissues. Our data showed that overexpressed LIN28B and a positive association between H19 and LIN28B in lung cancer samples. Notably, H19 was able to upregulte LIN28B in lung cancer cells. In the next study, we focused on how H19 played a role in LIN28B regulation. MicroRNAs (miRNAs) take key parts in gene regulation at the post-transcriptional level [38–40]. Interestingly, H19 can antagonize let-7 to enhance HMGA2-mediated EMT, leading to cell migration and invasion in pancreatic cancer [41]. In colorectal cancer H19 serves as the sponges of miR-138 and miR-200a to induce epithelial to mesenchymal transition [42]. H19 is able to depress miR-29a or miR-140 to regulate glioma angiogenesis or growth [43,44]. The stem cell maintenance of breast cancer can be increased by H19/let-7/LIN28 cascade signaling [45]. Overexpressed LIN28A/LIN28B and decreased let-7 is frequently found in resistance to multiple cancer therapies [46]. Previous study showed that H19 was able to sponge some microRNAs including let-7, miR-138 and miR-200a to induce LIN28B [42,45,47]. In our study, we found that LIN28B was a target gene of miR-196b. Our finding further implied that H19 induced LIN28B by sponging miR-196b. Functionally, H19/miR-196b/LIN28B signal promoted cell proliferation in lung cancer. Accordingly, all these investigations indicate that H19 can upregulate LIN28B via regulation of multiple miRNAs.
We here reveal a role of H19-elavated LIN28B by inhibiting miR-196b in lung cancer progression. H19 is able to induce LIN28B expression in lung cancer. For the mechanism investigation, H19 restrains tumor suppressive miR-196b which directly targets LIN28B, resulting in LIN28B overexpression in lung cancer. We provide a novel evidence to understand how lung cancer is mediated by H19.
Materials and methods
Patient samples
Freshly frozen lung cancer tissues and paired noncancerous tissues used in this study were obtained from the Second Hospital of Jilin University (the Second Hospital of Jilin University, China). Patient information was summarized in Table S1. All patients were diagnosed with lung cancer and gave written consents. Research ethics committee at the Second Hospital of Jilin University (the Second Hospital of Jilin University, China) approved study protocol.
Cell lines
RPMI Medium 1640 with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen, USA) was utilized to culture lung cancer cells A549 and H1299. The cell incubator was applied to keep an atmosphere of 5% CO2 and a temperature of 37°C for all cell growth.
Real-time PCR
RNA from human clinical tissues or lung cancer cells was extracted by TRIzol Reagent (Invitrogen, USA) and was applied to perform reverse transcription using SuperScript™ IV Reverse Transcriptase (ThermoFisher Scientific, USA). Polyadenylating total RNA induced by poly (A) polymerase (Ambion, USA) was used to analyze the miR-196b level. TransStart Top Green qPCR SuperMix (TransGen Biotech, China) for real time-PCR was utilized. The transcription changes of H19 (or LIN28B) and miR-196b were normalized by comparing with GAPDH and U6, respectively. Primers used in this study were included in Table S2.
Transfection
Lipofectamine 2000 for cell transfection was bought from Invitrogen Company (USA). RiboBio (Guangzhou, China) help us to synthesize miR-196b, NC, anti-miR-196b, siH19 or siLIN28B. Sequences were included in Table S2.
Immunoblotting
RIPA lysis buffer was used to lyse cells. After running on SDS-PAGE and transferring onto polyvinylidene fluoride membranes, protein samples were incubated with primary and then second antibodies. The primary antibodies were demonstrated as described: anti-LIN28B (Santa Cruz Biotechnology, USA) and anti-β-actin (NeoMarkers, USA). ECL Western Blotting Substrate (Solarbio, Beijing, China) was used for detecting the immune complexes.
Reporter system
Cells (3 × 104/well) were seeded on twenty-four-well plates. pRL-TK plasmid (Promega, USA) and indicated plasmids (or miRNAs) were co-transfected into cells after cells were cultured for twenty-four hours. And then, luciferase activities were analyzed after the transfection was performed for forty-eight hours. Relative luciferase activities were normalized to pRL-TK.
Cell proliferation evaluation
A549 cells (5 × 104 cells/well) were cultured on ninety-six-well plate. Then, cells were incubated overnight and collected after the transfection was applied for twenty-four, forty-eight, and seventy-two hours. After transfection, was evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) analysis was utilized for testing cell proliferation ability.
Analysis of colony forming
The transfected cells were trypsinized and cultured in 6-well plate (1000 cells/well). The medium was replaced every three days. After growth for 14 days, cells were washed with PBS for at least 2 times and fixed in methanol for 20 min at room temperature; finally cells were stained with crystal violet. The number of colonies was counted and the colony forming efficiency was determined with the formula: colony forming efficiency = number of colonies counted/number of cells plated × 100%.
Statistical analysis
The statistical significance was assessed by comparing mean values (± SD) using a 2-tailed Student’s t-test. The significance was set as ***p < 0.001, **p < 0.01 and *p < 0.05. The association of H19 with LIN28B in clinical lung cancer tissues was determined by Pearson’s correlation coefficient.
Author contributions
ZA designed the experiments and drafted the manuscript. JR and JLF performed the experiments and drafted the manuscript. TGM, BDY and RG performed the experiments. DW designed the experiments and revised the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang Y, Tycko B.. Monoallelic expression of the human H19 gene. Nat Genet. 1992;1:40–44. [DOI] [PubMed] [Google Scholar]
- [5].Moulton T, Crenshaw T, Hao Y, et al. Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat Genet. 1994;7:440–447. [DOI] [PubMed] [Google Scholar]
- [6].Dugimont T, Curgy JJ, Wernert N, et al. The H19 gene is expressed within both epithelial and stromal components of human invasive adenocarcinomas. Biol Cell. 1995;85:117–124. [DOI] [PubMed] [Google Scholar]
- [7].Lottin S, Adriaenssens E, Dupressoir T, et al. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–1895. [DOI] [PubMed] [Google Scholar]
- [8].Berteaux N, Lottin S, Monte D, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. The J Biol Chem. 2005;280:29625–29636. [DOI] [PubMed] [Google Scholar]
- [9].Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. [DOI] [PubMed] [Google Scholar]
- [10].Wang L, Cai Y, Zhao X, et al. Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma. 2015;62:412–418. [DOI] [PubMed] [Google Scholar]
- [11].Huang C, Cao L, Qiu L, et al. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2015;10:291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhu Z, Song L, He J, et al. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol. 2015;8:10082–10091. [PMC free article] [PubMed] [Google Scholar]
- [13].Wang L, Sun Y, Yi J, et al. Targeting H19 by lentivirus-mediated RNA interference increases A549 cell migration and invasion. Exp Lung Res. 2016;42:346–353. [DOI] [PubMed] [Google Scholar]
- [14].Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang WC, Shyh-Chang N, Yang H, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. [DOI] [PubMed] [Google Scholar]
- [16].Schnepp RW, Khurana P, Attiyeh EF, et al. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell. 2015;28:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chang TC, Zeitels LR, Hwang HW, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jiang X, Huang H, Li Z, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Iliopoulos D, Hirsch HA, Struhl K.. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li X, Zhang J, Gao L, et al. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2012;19:378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A. 2013;110:E2362–E2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhong X, Li N, Liang S, et al. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem. 2010;285:41961–41971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang L, Zhang LF, Wu J, et al. IL-1beta-mediated repression of microRNA-101 is crucial for inflammation-promoted lung tumorigenesis. Cancer Res. 2014;74:4720–4730. [DOI] [PubMed] [Google Scholar]
- [24].Borrego-Diaz E, Powers BC, Azizov V, et al. A potential regulatory loop between Lin28B: miR212in androgen-independent prostate cancer. Int J Oncol. 2014;45:2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sangiao-Alvarellos S, Manfredi-Lozano M, Ruiz-Pino F, et al. Changes in hypothalamic expression of the Lin28/let-7 system and related microRNAs during postnatal maturation and after experimental manipulations of puberty. Endocrinology. 2013;154:942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ariel I, Lustig O, Schneider T, et al. The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology. 1995;45:335–338. [DOI] [PubMed] [Google Scholar]
- [27].Yoshimizu T, Miroglio A, Ripoche MA, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A. 2008;105:12417–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kondo M, Suzuki H, Ueda R, et al. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–1198. [PubMed] [Google Scholar]
- [29].Elkin M, Shevelev A, Schulze E, et al. The expression of the imprinted H19 and IGF-2 genes in human bladder carcinoma. FEBS Lett. 1995;374:57–61. [DOI] [PubMed] [Google Scholar]
- [30].Hibi K, Nakamura H, Hirai A, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–482. [PubMed] [Google Scholar]
- [31].Kim HT, Choi BH, Niikawa N, et al. Frequent loss of imprinting of the H19 and IGF-II genes in ovarian tumors. Am J Med Genet. 1998;80:391–395. [DOI] [PubMed] [Google Scholar]
- [32].el-Naggar AK, Lai S, Tucker SA, et al. Frequent loss of imprinting at the IGF2 and H19 genes in head and neck squamous carcinoma. Oncogene. 1999;18:7063–7069. [DOI] [PubMed] [Google Scholar]
- [33].Guo Y, Chen Y, Ito H, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. [DOI] [PubMed] [Google Scholar]
- [34].Wang T, Wang G, Hao D, et al. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol Cancer. 2015;14:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou Y, Liang H, Liao Z, et al. miR-203 enhances let-7 biogenesis by targeting LIN28B to suppress tumor growth in lung cancer. Sci Rep. 2017;7:42680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Piskounova E, Polytarchou C, Thornton JE, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Viswanathan SR, Daley GQ. Lin28: a microRNA regulator with a macro role. Cell. 2010;140:445–449. [DOI] [PubMed] [Google Scholar]
- [38].Hagan JP, Croce CM. MicroRNAs in carcinogenesis. Cytogenet Genome Res. 2007;118:252–259. [DOI] [PubMed] [Google Scholar]
- [39].Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Connerty P, Ahadi A, Hutvagner G. RNA binding proteins in the miRNA pathway. Int J Mol Sci. 2015;17(1). pii: E31. doi: 10.3390/ijms17010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9169. [DOI] [PubMed] [Google Scholar]
- [42].Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jia P, Cai H, Liu X, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381:359–369. [DOI] [PubMed] [Google Scholar]
- [44].Zhao H, Peng R, Liu Q, et al. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016;610:1–7. [DOI] [PubMed] [Google Scholar]
- [45].Peng F, Li TT, Wang KL, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Balzeau J, Menezes MR, Cao S, et al. The LIN28/let-7 pathway in cancer. Front Genet. 2017;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sureban SM, May R, Weygant N, et al. XMD8-92 inhibits pancreatic tumor xenograft growth via a DCLK1-dependent mechanism. Cancer Lett. 2014;351:151–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


