Abstract
Fringe modulates Notch signaling resulting in the establishment of compartmental boundaries in developing organisms. Fringe is a β3N-acetylglucosaminyltransferase (β3GlcNAcT) that transfers GlcNAc to O-fucose in epidermal growth factor-like repeats of Notch. Here we use five different Chinese hamster ovary cell glycosylation mutants to identify a key aspect of the mechanism of fringe action. Although the β3GlcNAcT activity of manic or lunatic fringe is shown to be necessary for inhibition of Jagged1-induced Notch signaling in a coculture assay, it is not sufficient. Fringe fails to inhibit Notch signaling if the disaccharide generated by fringe action, GlcNAcβ3Fuc, is not elongated. The trisaccharide, Galβ4GlcNAcβ3Fuc, is the minimal O-fucose glycan to support fringe modulation of Notch signaling. Of six β4galactosyltransferases (β4GalT) in Chinese hamster ovary cells, only β4GalT-1 is required to add Gal to GlcNAcβ3Fuc, identifying β4GalT-1 as a new modulator of Notch signaling.
Notch receptors are transmembrane glycoproteins that play key roles in cell fate decisions and growth control (1). Signal transduction by Notch requires binding to ligand on adjacent cells which induces a proteolytic cleavage of Notch extracellular domain (NECD), followed by a γ-secretase cleavage that releases Notch intracellular domain (NICD) into the cytosol. Translocation of NICD complex to the nucleus causes altered expression of numerous target genes. Fringe is a β3N-acetylglucosaminyltransferase (β3GlcNAcT) that adds GlcNAc to O-fucose on certain epidermal growth factor (EGF) repeats of NECD (2–4). In Drosophila, fringe positions Notch signaling at the dorsal/ventral boundary of the wing imaginal disk as a result of its ability to inhibit Serrate-induced, and to potentiate Delta-induced, Notch signaling (5, 6). In early oogenesis, and in developing legs and eye, fringe also positions Notch activation (7, 8). Mutation analyses have shown that the glycosyltransferase activity of fringe is essential for its effect on Notch signaling in Drosophila (2–4).
Like most glycosyltransferases, fringe functions in the Golgi (3, 4, 9), although it is also secreted. Manic (Mfng) and lunatic (Lfng) mammalian fringes expressed in different cultured cells modulate Jagged (Serrate)- and Delta-induced Notch signaling in a manner consistent with fringe functions in Drosophila (2, 9–11). In Chinese hamster ovary (CHO) cells expressing Mfng or Lfng, Jagged1-induced signaling by endogenous Notch is inhibited (2, 11). Structural studies of the O-fucose glycans released from NECD showed that fringe action generates a disaccharide (GlcNAcβ3Fuc), a trisaccharide (Galβ4GlcNAcβ3Fuc), and a tetrasaccharide (SAα3Galβ4GlcNAcβ3Fuc) on Notch (2). It seemed likely that either the mature O-fucose tetrasaccharide was required for the fringe effect or that the GlcNAc added by fringe was sufficient for the fringe effect, because of the masking of an O-fucose epitope important for Notch signaling. Here we use CHO glycosylation mutants defective in the addition of Gal or sialic acid (SA) to reveal that, contrary to expectations, the minimum O-fucose glycan that exhibits a fringe effect is the trisaccharide, Galβ4GlcNAcβ3Fuc. Thus, although fringe β3GlcNAcT activity is necessary, it is not sufficient for fringe to inhibit Jagged1-induced Notch signaling. Of the six mammalian β4GalTs that transfer Gal to GlcNAc in CHO cells (12), only β4galactosyltransferase-1 (β4GalT-1) was required to elongate the fringe disaccharide GlcNAcβ3Fuc, identifying β4GalT-1 as a new modulator of Notch signaling.
Materials and Methods
Constructs.
The constructs pMirb/MfngAP, pMirb/LfngAP, IgTag/Mfng-Fc, IgTag/Lfng-Fc, and CBF1-luciferase JH-26 with 8 C promoter binding factor (CBF1) binding sites were described (2, 9). pSVL/SGT encodes the short form of bovine β4GalT-1 (13), and pMirb/GDP-4,6-dehydratase (GDP-4,6-DH; ref. 14) was made by PCR amplification of human GDP-4,6-DH cDNA cloned in the pMT21 vector. The primers were 5′-CGCGAATTCCCGCCGCGGGACATGGC-3′ and 5′-CGCGGATCCGCTGCTCAGGCATTGGG-3′, digested with EcoRI/BamHI, and ligated into pMirb.
Cells and Transfection.
Lec1.3C (15), Lec2.6A (16), Lec8.3D (17), Lec13.6A (14, 18), and Lec20.15C (12) CHO glycosylation mutants derived from Pro−5 CHO cells were characterized. They are referred to as Lec1, Lec2, Lec8, Lec13, and Lec20, respectively. CHO cells were stably transfected with 1–2 μg of plasmid DNA by using Fugene 6 (Roche Diagnostics) and selected in 1.5 mg/ml of active G418 (Gemini Biological Products, Calabasas, CA). The alkaline phosphatase (AP) activity of medium was assayed to identify high expressers of fng.AP chimeric protein (2). L cells stably expressing Jagged1 (J1 cells; ref. 19) and L cells were sorted by using goat-anti-rat Jagged1 ECD Ab (R & D Systems) to obtain J1 cells expressing high levels of J1 and L cells expressing minimal J1. CHO cells endogenously express Notch1 (20) and Notch2 (11) and have a low level of transcripts for radical fringe (Rfng) and Lfng (11). We confirmed that Rfng and Lfng were expressed in our CHO cells by reverse-transcription (RT)-PCR, using degenerate primers based on human and mouse fringe sequences (see Supporting Information, which is published on the PNAS web site, www.pnas.org). No Mfng transcripts were detected by RT-PCR (see Supporting Information) or by northern analysis (11). Western blot analysis with fringe Abs (Santa Cruz Biotechnology) detected no specific signals, but structural analyses revealed O-fucose tetrasaccharide (20) consistent with a low level of endogenous fringe activity in CHO cells.
Structural Characterization of O-Fucose Glycans.
Cells were transiently transfected with 5 μg of pSectag2C/Hygro mouse Notch1 ECD fragments EGF19–23 (2) or EGF1–36 by using GenePORTER (Gene Therapy Systems, San Diego) and labeled for 24–48 h with 20 μCi of 5,6-3H-l-fucose (60 Ci/mmol; American Radiolabeled Chemicals, St. Louis). Secreted Notch1 EGF19–23 was purified by using Ni2+-agarose beads (Qiagen, Chatsworth, CA). Secreted Notch1 EGF1–36 was purified by using a HiTrap chelating column (Amersham Pharmacia) charged with Ni2+. [3H]fucose labeling, PNGase F digestion, β-elimination of O-glycans, and analysis of O-fucose glycans by gel filtration chromatography and high pH anion-exchange chromatography were described (20). O-fucose standards were derived from Lec1 3H-labeled O-fucose tetrasaccharide (SAα3Galβ4GlcNAcβ3Fucitol) digested with neuraminidase I (Glyko, Novato, CA) to generate the O-fucose trisaccharide (Galβ4GlcNAcβ3Fucitol) and β-galactosidase (Diplococcus pneumoniae, Roche Molecular Biochemicals) to generate the disaccharide (GlcNAcβ3Fucitol) as described (20). Monosaccharide (MS) is fucitol, the reduced form of fucose.
Site-Directed Mutagenesis.
The LfngDDA mutation was made on pCS2+/Lfng by changing codon 201 of mouse Lfng (21) with 5′-GGTTCTGCCACGTGGATGATGCCAACTACGTCAACC-3′ and 5′-GGTTGACGTAGTTGGCATCATCCACGTGGCAGAACC-3′ as primers, using site-directed mutagenesis (Stratagene). The LfngDDA ORF was subsequently amplified with primers 5′-CCCAAGCTTGCCGCCGGCCACCATGCTCC-3′ and 5′-GGAAGATCTGAAGATGGCGGAGCGAGG-3′, digested with HindIII/BglII, and cloned into IgTag vector. The MfngDDA mutation was made directly on pMirb/MfngAP by changing codon 144 of mouse Mfng (21) with 5′-GGTTCTGCCACGTGGATGATGCCAACTATGTGAACCC-3′ and 5′-GGGTTCACATAGTTGGCATCATCCACGTGGCAGAACC-3′ as primers. The MfngDDA ORF was amplified from pMirb/MfngAP.DDA with 5′-CCCAAGCTTGGGGGCCTGCTACCAATGC-3′ and 5′-GGAAGATCTGGGCGCTGCCAGCAGCGG-3′ as primers, digested with HindIII/BglII, and ligated into IgTag vector. All mutated sites were confirmed by direct sequencing.
Pea Lectin Binding.
Lec13 cells were transiently transfected with pMirb/GDP-4,6-DH by using Fugene 6. After 30 h, cells were fixed with 4% formaldehyde and blocked with 5% BSA in PBS/1 mM CaCl2/1 mM MgCl2/1 mM MnCl2. After 30 min at room temperature, 25 μg/ml of FITC-labeled pea lectin (Vector Laboratories) was added for 1 h at room temperature. After washing with PBS lacking cations, cells were photographed.
Preparation of Soluble Fringe and Glycosyltransferase Assay.
HEK-293T cells were transfected with 1 μg of DNA in 3 μl of Fugene 6. After 24 h, medium was replaced with 2 ml of Opti-mem (GIBCO), 1% FBS, antibiotics, and 0.1 mg/ml of CaCl2. Medium was collected 48 h later, and complete Mini EDTA-free protease inhibitors (CPI-EDTA; Roche Biochemicals) were added. The supernatant was clarified before concentration by using a spin device (30-kd cutoff, Millipore). Protein A/G Sepharose beads (30 μl; Santa Cruz Biotechnology) were added. After a 12-h rotation at 4°C, the beads were washed as described (2). The purity and concentration of bound fng.Fc was determined by densitometric analysis of Coomassie blue-stained SDS/PAGE gels compared with BSA standard. The GlcNAc-transferase assay was performed as described (2) by using 10 mM Fuc-O-pNp as substrate.
Notch Signaling Assay.
The coculture Notch signaling assay was performed as described (2). CHO cells in a 6-well plate were cotransfected with 200 ng of CBF1-luciferase plasmid JH-26, 100 ng of pRL-TK Renilla luciferase (Promega), and 1.5 μg of pMirb vector by using Fugene 6. After 16 h at 37°C, 2 × 106 J1 or parental L cells were overlayed. After another 32 h, luciferase and Renilla luciferase activities in lysates were measured (Promega). All experiments were performed twice and in duplicate. Background luciferase levels for CHO cells were ≈1, J1 coculture luciferase units ranged from ≈100 to 1,400 (average ≈200), and Renilla luciferase units ranged from ≈800 to 8,000 (average ≈2,500). Jagged1-dependent Notch activation of CBF1-luciferase is expressed as a ratio of normalized luciferase units in J1 vs. L cell cocultures.
FACS Analysis.
Cells (2 × 105) were washed with 4 ml of cold 2% BSA in Hanks' balanced salt solution (HBSS; Ca2+- and Mg2+-free; Mediatech, Herndon, VA) and resuspended with 200 μl of 2% BSA in HBSS containing either 2.5 μg of rabbit anti-NECD Ab (H131, Santa Cruz Biotechnology) or 2.5 μg of normal rabbit IgG (Sigma). After incubation at 4°C with gentle rocking for 45 min, cells were washed with 4 ml of cold 2% BSA in HBSS, resuspended with 200 μl of 2% BSA in HBSS containing 1 μg of FITC-labeled goat anti-rabbit IgG (H + L) (Zymed), and incubated in 4°C for 45 min. After washing with 4 ml of cold 2% BSA in HBSS, cells were resuspended with 500 μl of HBSS and subject to FACS analysis.
Results
Fringe β3GlcNAcT Activity Is Required for Modulation of Jagged1-Induced Notch Signaling.
Endogenous Notch signaling in CHO cells is induced 4–6-fold by coculture with cells expressing Jagged1 (2, 9, 11). CHO cells expressing Mfng or Lfng have a reduced response to Jagged1 in the coculture assay (2, 11). To show that this effect requires fringe β3GlcNAcT activity, the conserved DDD motif at amino acids 142–144 in Mfng was converted to DEE, a mutation known to inactivate both β3GlcNAcT activity and function in Drosophila fringe (2). Mfng.DEE was, however, not produced at 37°C, although it could be expressed equivalently to Mfng.DDD at 32°C. By contrast, a different mutation, Mfng.DDA, was stable at 37°C and expressed at similar levels to wild-type MfngDDD on the basis of AP activity in secretions and lysates. Both Mfng.DDA and Lfng.DDA (at amino acids 199–201) tagged at the C terminus with IgG1 Fc were purified on beads and assayed for GlcNAc to Fuc-O-pNp transfer. Although wild-type fringe proteins Mfng.DDD/Fc and Lfng.DDD/Fc had β3GlcNAcT activity, none was detected for the DDA fringe mutants (Table 1). When CHO cells stably expressing Mfng.DDA were tested in the coculture assay, there was no inhibition of Jagged1-induced Notch signaling (Fig. 1A). Therefore, fringe β3GlcNAcT activity is essential for fringe to modulate Jagged1-induced Notch signaling in CHO cells.
Table 1.
GlcNAc-transferase activity of mutant Mfng and Lfng
| Substrate | Fringe activity, cpm/μg of protein
|
|||
|---|---|---|---|---|
| Mfng (D142DD144) | Mfng (D142DA144) | Lfng (D199DD201) | Lfng (D199DA201) | |
| Fuc-O-pNp | 11,273 | 193 | 96,118 | 283 |
| None | 200 | 200 | 260 | 285 |
In vitro GlcNAc-transferase activity of fringe proteins are the average of duplicates.
Figure 1.
Fringe glycosyltransferase activity and fucose transfer are required for fringe modulation of Notch signaling. (A) The histogram shows the fold-activation of normalized luciferase units in control (vector), wild-type Mfng (Mfng.DDD), and mutant (Mfng.DDA) CHO and Lec1 transfectants stimulated by J1 vs. L cells (J1/L). Error bars represent SEM; n = 4. (B) The synthesis of GDP-fucose in CHO cells is blocked in Lec13 cells as a result of reduced transcription of the GDP-4,6-DH gene. (C) Immunofluorescence of FITC-pea lectin on the surface of Lec13, CHO, and Lec13 cells transiently transfected with pMirb/GDP-4,6-DH. (D) Fold-activation of normalized luciferase units in CHO, Lec13.Mfng, and Lec13.Mfng transiently transfected with pMirb/GDP-4,6-DH. Error bars represent SEM; n = 4 for each.
O-fucose that is transferred from GDP-fucose to serine/threonine residues in EGF repeats of NECD is the substrate of fringe (2). Lec13 CHO cells have low GDP-fucose levels because of a mutation that essentially abrogates expression of the GDP-4,6-DH gene (Fig. 1B; refs. 14 and 18). Correction of Lec13 cells by transfection with a cDNA encoding GDP-4,6-DH restored fucose on cell surface glycoproteins as shown by rescue of their ability to bind pea lectin (Fig. 1C). Jagged1-induced Notch signaling was also restored to wild-type levels, as was modulation of Notch signaling by fringe (Fig. 1D). Rescue was more complete than was previously obtained by adding exogenous fucose to the medium (2). Therefore, Jagged1-induced Notch signaling is enhanced by the presence of O-fucose, and both fucose and fringe β3GlcNAcT activity are essential for fringe to modulate Notch signaling.
Fringe Action Is Necessary but Not Sufficient to Inhibit Jagged1-Induced Notch Signaling.
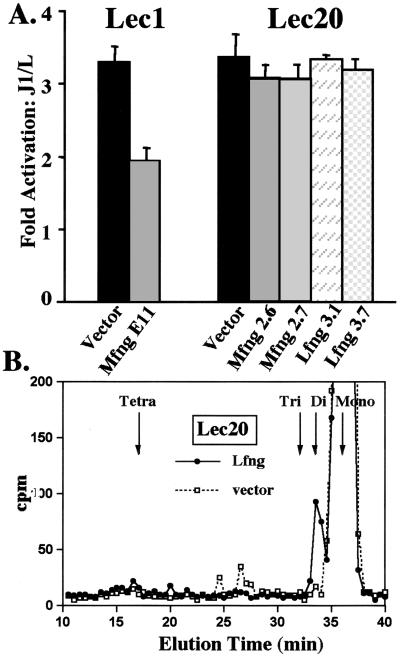
Lec1 CHO cells have defective N-glycan synthesis but normal O-glycan synthesis (22, 23). They respond to Jagged1 stimulation and exhibit inhibition of Notch signaling by Mfng similarly to parent CHO cells (2). O-fucose glycans released from the mouse Notch1 fragment EGF19–23 secreted from Lec1 CHO cells reveal that Mfng causes elongation of a proportion of O-fucose MS to the tetrasaccharide, SAα3Galβ4GlcNAcβ3Fuc (2). Therefore, the tetrasaccharide or di- or trisaccharide intermediates might be responsible for modulation of Notch signaling by fringe.
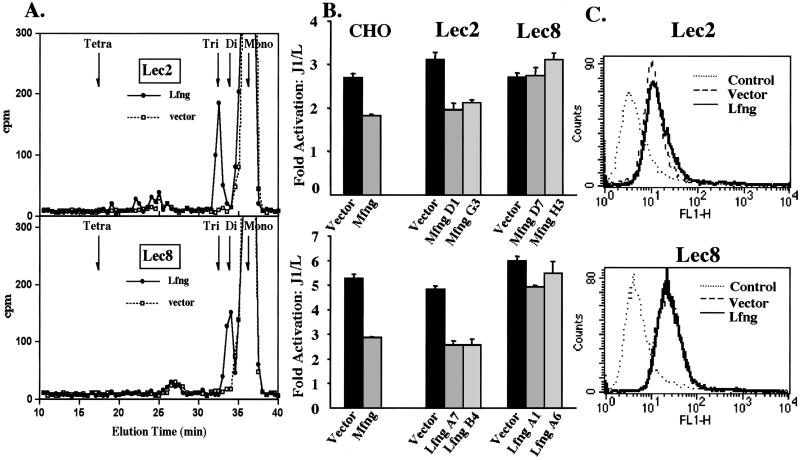
Lec2 CHO glycosylation mutants have mutations in the CMP-SA transporter that prevent CMP-SA from being translocated into the Golgi, markedly inhibiting sialylation of glycoproteins (16). Notch1 EGF19–23 secreted from Lec2 cells expressing Lfng (Lec2.Lfng) contained MS and trisaccharide O-fucose glycans (Fig. 2A). No tetrasaccharide was detected, as expected if CMP-SA was not available. A secreted Notch1 fragment containing all of the EGF repeat substrates of fringe (EGF1–36) also contained no tetrasaccharide when expressed in Lec2.Mfng cells (see Supporting Information). In the absence of Lfng (Lec2.vector), O-fucose was not elongated on EGF19–23 (Fig. 2A). When Lec2.vector cells were assayed for Jagged1-induced Notch signaling, they were as active as CHO or Lec1 cells (Fig. 2B). In addition, both Mfng and Lfng inhibited Jagged1-induced Notch signaling to a similar degree in independent Lec2 transfectants (Fig. 2B). The inhibition of Notch signaling by fringe was not the result of reduced expression of cell surface Notch1 in Lec2 transfectants (Fig. 2C). Thus, SA is not essential for either Jagged1-induced Notch signaling or for fringe to inhibit Notch signaling. However, inhibition occurs when O-fucose is elongated to the trisaccharide, Galβ4GlcNAcβ3Fuc.
Figure 2.
Galactose but not SA is required for a fringe effect. (A) Gel filtration of [3H]fucose-labeled O-glycans released by β-elimination from Notch1 EGF19–23 secreted from Lec2 or Lec8 cells expressing Lfng (filled circles) or vector (open squares). The major species (≈1,500 cpm) was O-fucose MS (fucitol). Elution positions of standard O-fucose glycans (Tetra, SAα3Galβ4GlcNAcβ3Fucitol; Tri, Galβ4GlcNAcβ3Fucitol; Di, GlcNAcβ3Fucitol; Mono, Fucitol) are indicated. (B) Fold-activation of normalized luciferase units in CHO, Lec1, Lec2, or Lec8 clones expressing vector, Mfng, or Lfng stimulated by J1 vs. L cells. Error bars represent SEM; n = 4. (C) FACS analysis with Notch1 ECD Ab. Dotted line, normal rabbit serum; dashed line, Lec2.vector or Lec8.vector; solid line, Lec2.Lfng or Lec8.Lfng.
To determine whether the terminal Gal residue on Galβ4GlcNAcβ3Fuc is essential for the fringe effect, CHO mutants defective in the addition of Gal were examined. Lec8 CHO mutants have mutations in the UDP-Gal transporter that prevent UDP-Gal from being translocated into the Golgi and inhibit galactosylation of glycoproteins (17). The results in Fig. 2B show that, in contrast to Lec2 mutants, Lec8 CHO mutants expressing Mfng or Lfng were refractory to the fringe inhibitory effect (Fig. 2B). Although Jagged1-induced Notch signaling was similar in Lec8.vector and Lec2.vector cells, neither Lec8.Lfng nor Lec8.Mfng cells exhibited inhibition of Notch signaling. Cell surface Notch1 was expressed at similar levels in Lec8.vector and Lec8.fng cells (Fig. 2C), and AP assays showed that fringe proteins were expressed equivalently. In addition, structural analyses of O-fucose glycans showed that fringe was active in Lec8.fng cells (Fig. 2A). Characterization of Notch1 EGF1–36 secreted from Lec8.Mfng cells (see Supporting Information) and Notch1 EGF19–23 secreted from Lec8.Lfng (Fig. 2A) showed that O-fucose was elongated only to the disaccharide in Lec8.fng cells, whereas Lec8.vector cells did not elongate O-fucose. As expected, Lec8.fng cells did not produce O-fucose trisaccharide or tetrasaccharide (Fig. 2A). Therefore, whereas the trisaccharide Galβ4GlcNAcβ3Fuc on Notch in Lec2.fng cells supports inhibition of Jagged1-induced Notch signaling, the disaccharide GlcNAcβ3Fuc synthesized by either Mfng or Lfng in Lec8 cells does not. To obtain a fringe effect on Notch signaling, the action of a β4GalT is required subsequent to fringe.
β4GalT-1 Is Required for Fringe to Modulate Jagged1-Induced Notch Signaling.
Although CHO cells may express all six β4GalT genes known to transfer Gal to GlcNAc in mammals (12), our wild-type CHO line, Pro−5, lacks transcripts for β4GalT-6 (12). However, Pro−5Lec1 cells express fringe-generated O-fucose tri- and tetrasaccharides (ref. 2; see Fig. 5, which is published as supporting information on the PNAS web site) and also exhibit inhibition of Notch signaling by fringe (ref. 2; Fig. 2B). Therefore, β4GalT-6 is not essential for the synthesis of tri- or tetrasaccharide O-fucose glycans. By contrast, Pro−5Lec20 CHO cells that lack both β4GalT-6 and β4GalT-1 (12) did not exhibit a fringe effect, although they responded well to Jagged1 (Fig. 3A). Like Lec8 cells, independent Lec20 clones expressing Mfng or Lfng were not reduced in Notch signaling (Fig. 3A). Cell surface expression of Notch1 was equivalent in fringe and vector expressing Lec20 cells, and all Lec20.fng cells produced similar amounts of fringe as detected by AP activity. Structural analyses of [3H]fucose-labeled O-fucose glycans on Notch1 EGF1–36 secreted from Lec20.Mfng revealed only the disaccharide (see Supporting Information), as was also observed in EGF19–23 secreted from Lec20.Lfng (Fig. 3B). Neither the O-fucose trisaccharide nor the tetrasaccharide were synthesized in Lec20.fng cells. Thus, neither β4GalT-2, β4GalT-3, β4GalT-4, nor β4GalT-5 that are expressed in Pro−5Lec20 cells (12) are able to elongate the fringe disaccharide, GlcNAcβ3Fuc.
Figure 3.
The β4GalT-1 mutant Lec20 does not exhibit a fringe effect. (A) Fold-activation of normalized luciferase activity in Lec1 and Lec20 clones stably expressing vector, Mfng, or Lfng stimulated by J1 vs. L cells. Error bars represent SEM; n = 4. (B) Gel filtration profiles of [3H]fucose-labeled O-glycans released by β-elimination from Notch1 EGF19–23 produced by Lec20.Lfng (filled circles) or Lec20.vector (open squares). Elution positions of standard O-fucose glycans are indicated.
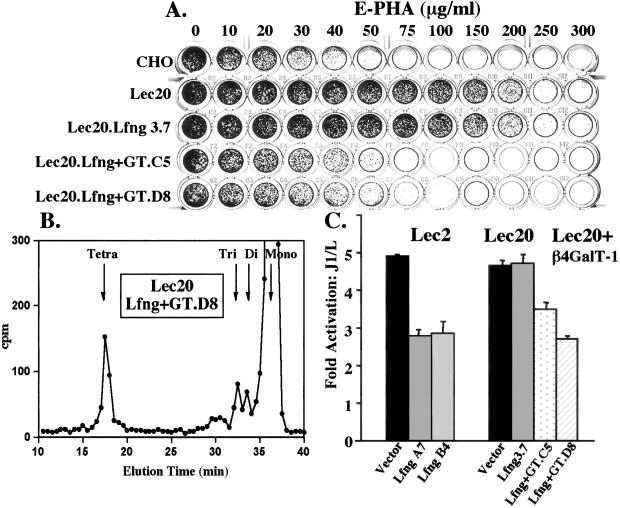
Further evidence that β4GalT-1 is required to generate the O-fucose trisaccharide was obtained by correcting the defect in Lec20 cells with a cDNA encoding bovine β4GalT-1. Lec20 cells stably expressing Lfng are resistant to the Gal-binding lectin E-phytohemagglutinin (PHA) like Lec20.vector and Lec20 cells (Fig. 4A). Transfection of a bovine β4GalT-1 cDNA almost completely rescued the E-PHA sensitivity of Lec20 cells. Assay of β4GalT activity in cell lysates with GlcNAc as acceptor showed correction of the reduced β4GalT activity in Lec20 cells (not shown). Structural analyses of [3H]fucose-labeled O-glycans released from Notch1 EGF19–23 showed that expression of bovine β4GalT-1 in Lec20.fng cells led to the synthesis of tetrasaccharide (Fig. 4B). Correction of the Lec20 β4GalT-1 defect also led to the rescue of a fringe effect on Notch signaling in Lec20 cells. Lec20.Lfng clone D8 expressing β4GalT-1 was inhibited in Notch signaling similarly to Lec2.Lfng cells (Fig. 4C). The correction of Lec20 CHO cells by a β4GalT-1 cDNA demonstrates that this particular β4GalT acts on the disaccharide product of fringe and is a novel modulator of Notch signaling.
Figure 4.
Rescue of a fringe effect with β4GalT-1. (A) Lectin sensitivity test. Cells surviving in E-phytohemagglutinin (PHA) were stained with methylene blue. (B) Gel filtration profiles of [3H]fucose-labeled O-glycans released by β-elimination from Notch1 EGF19–23 produced by Lec20 cells expressing Lfng and β4GalT-1. Elution positions of O-fucose glycan standards are indicated. (C) Fold-activation of normalized luciferase units in Lec2 or Lec20 clones expressing vector, Lfng, or Lfng + GT (β4GalT-1) stimulated by J1 vs. L cells. Error bars represent SEM; n = 4.
Discussion
The discovery that, in CHO cells, fringe β3GlcNAcT causes not only the synthesis of GlcNAcβ3Fuc disaccharide on NECD, but also tri- and tetrasaccharides (2), immediately begged the question: Is the action of fringe sufficient to cause modulation of Notch signaling? By assaying fringe function in CHO glycosylation mutants defective in the addition of Fuc (Lec13), Gal (Lec8 and Lec20), or SA (Lec2), we found that fringe inhibition of Jagged1-induced Notch signaling requires both fucose as the substrate for fringe and elongation of the fringe-generated disaccharide by a Gal residue. The results summarized in Table 2 show that O-fucose facilitates Notch signaling by Jagged1, and that Mfng or Lfng inhibition of Jagged1-induced Notch signaling requires the action not only of fringe, but also of β4GalT-1. The minimal O-fucose glycan that functions to inhibit Notch signaling is the trisaccharide, Galβ4GlcNAcβ3Fuc, the longest O-fucose glycan synthesized in Lec2 CHO cells. These conclusions identify key aspects of essential roles for O-fucose glycans in Notch signaling. They also provide important insights into fringe mechanism of action and focus experiments to be performed in organisms.
Table 2.
O-fucose glycans that facilitate or modulate Notch signaling
| Cells | Glycosylation defect | O-fucose glycans | Notch activation | Fringe effect |
|---|---|---|---|---|
| CHO (Pro−5) | β4GalT-6 | SAα3Galβ4GlcNAcβ3Fuc-O-S/T | + | + |
| Lec1 | GlcNAcT-1 | SAα3Galβ4GlcNAcβ3Fuc-O-S/T | + | + |
| Lec2 | CMP-SA transporter | Galβ4GlcNAcβ3Fuc-O-S/T | + | + |
| Lec8 | UDP-Gal transporter | GlcNAcβ3Fuc-O-S/T | + | − |
| Lec13 | GDP-4,6-dehydratase | S/T | ↓ | − |
| Lec20 | β4GalT-1, β4GalT-6 | GlcNAcβ3Fuc-O-S/T | + | − |
| Lec20 + GT | Corrected | SAα3Galβ4GlcNAcβ3Fuc-O-S/T | + | + |
A β4GalT related to β4GalT-1 may also be required for fringe modulation of Notch signaling in Drosophila. The Drosophila genome encodes three genes with strong homology to that of β4GalTs. One of them (CG11780) has several features typical of β4GalT-7 that add Gal to xylose (24). The other two, CG8536 and CG14517, are more similar to those that transfer Gal to GlcNAc (25, 26). Both encode a type II transmembrane glycoprotein that contains four conserved cysteine residues. The Drosophila genome also contains at least one gene that is homologous to a sialyltransferase (CG4871) but, in light of our findings, a sialyltransferase need not be necessary for fringe modulation of Notch signaling.
Notch signaling pathways are significantly more complex in mammals. Whereas Drosophila express a single Notch and only two ligands (Serrate and Delta), mammals express four Notch receptors, two Jagged/Serrate and three Delta ligands, and three fringe glycosyltransferases. Thus, although β4GalT-1 knockout mice (27, 28) and mice lacking Lfng (29, 30) differ in gross morphological phenotype, they are predicted to share a subset of features because of disruption of fringe modulation of Notch signaling. These phenotypes may be difficult to uncover, however, because β4GalT-1 is ubiquitously expressed and acts on many glycoproteins other than Notch. In addition, mutations in mice and humans reveal different in vivo functions for essentially all of the potentially redundant molecules in Notch signaling pathways.
In Drosophila and mammalian cells, fringe in a Notch-expressing cell alters the binding of soluble forms of ligand (3, 11). Therefore, O-fucose glycans on the EGF repeats of Notch play a direct or indirect role in ligand binding. They might induce a conformational change in NECD that alters the nature or efficiency of ligand binding. Our findings predict that the O-fucose disaccharide would not be sufficient to induce the change, whereas O-fucose trisaccharide is the minimal determinant that would suffice. Although the presence of O-fucose MS on an EGF repeat of clotting factor VII did not alter the solution conformation determined by NMR (31), it is not known whether elongated O-fucose glycans would affect the conformation of tandem EGF repeats.
It is also possible that Notch ligands bind directly to O-fucose glycans through a lectin-like recognition. Different Notch ligand/receptor pairs may form by means of recognition of specific O-fucose glycans attached to particular EGF repeats, because the substrate of fringe is a properly folded EGF repeat with O-fucose (2). In fact, there are differences among the four mammalian Notch homologues in the locations of predicted O-fucose sites, although many sites are conserved (20). Alternatively, lectins at the cell surface may bind different O-fucose glycans on Notch and thereby affect Notch–ligand interactions. For example, galectins bind terminal Gal residues and may bind Gal capped with SA (32). In this case, the tetrasaccharide could work as effectively as the trisaccharide at inhibiting Jagged1-induced Notch signaling, as indeed we observed. Future studies may determine the O-fucose glycans required for different Notch receptors and ligands to signal and the mechanism by which different O-fucose glycans on various EGF repeats of NECD induce or modulate Notch signaling.
Supplementary Material
Acknowledgments
We thank Subha Sundaram for excellent technical assistance; Thomas Vogt and Stuart Johnston for fringe constructs; Gerry Weinmaster for J1 L cells; Joel Shaper for β4GalT-1 cDNA; Robert Haltiwanger for O-fucose standards and mNotch1 EGF1–36; Francis Sullivan for GDP-4,6-dehydratase cDNA; Diane Hayward for JH-26, Philipp Scherer for HEK-293T cells; and Ken Irvine, Tom Vogt, Bob Haltiwanger, and Philipp Scherer for helpful discussions. This work was supported by National Institutes of Health Grant RO1 36434, the Mizutani Foundation (to P.S.), and by a Belfer Fellowship (to D.J.M.). Partial support was provided by Albert Einstein Cancer Center Grant PO1 13330.
Abbreviations
- EGF
epidermal growth factor
- NECD
Notch extracellular domain
- CHO
Chinese hamster ovary
- SA
sialic acid
- Mfng
manic fringe
- Lfng
lunatic fringe
- 4GalT-1
β4galactosyltransferase-1
- AP
alkaline phosphatase
- MS
monosaccharide
- GDP-4.6-DH
GDP-4.6-dehydratase
- HBSS
Hanks' balanced salt solution
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mumm J S, Kopan R. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 2.Moloney D J, Panin V M, Johnston S H, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine K D, Haltiwanger R S, Vogt T F. Nature (London) 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 3.Bruckner K, Perez L, Clausen H, Cohen S. Nature (London) 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 4.Munro S, Freeman M. Curr Biol. 2000;10:813–820. doi: 10.1016/s0960-9822(00)00578-9. [DOI] [PubMed] [Google Scholar]
- 5.Panin V M, Papayannopoulos V, Wilson R, Irvine K D. Nature (London) 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 6.Fleming R J, Gu Y, Hukriede N A. Development (Cambridge, UK) 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- 7.Irvine K D. Curr Opin Genet Dev. 1999;9:434–441. doi: 10.1016/S0959-437X(99)80066-5. [DOI] [PubMed] [Google Scholar]
- 8.Grammont M, Irvine K D. Development (Cambridge, UK) 2001;128:2243–2253. doi: 10.1242/dev.128.12.2243. [DOI] [PubMed] [Google Scholar]
- 9.Hicks C, Johnston S H, diSibio G, Collazo A, Vogt T F, Weinmaster G. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu K, Chiba S, Hosoya N, Kumano K, Saito T, Kurokawa M, Kanda Y, Hamada Y, Hirai H. Mol Cell Biol. 2000;20:6913–6922. doi: 10.1128/mcb.20.18.6913-6922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu K, Chiba S, Saito T, Kumano K, Takahashi T, Hirai H. J Biol Chem. 2001;276:25753–25758. doi: 10.1074/jbc.M103473200. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Sundaram S, Shaper N L, Raju T S, Stanley P. J Biol Chem. 2001;276:13924–13934. doi: 10.1074/jbc.M010046200. [DOI] [PubMed] [Google Scholar]
- 13.Russo R N, Shaper N L, Shaper J H. J Biol Chem. 1990;265:3324–3331. [PubMed] [Google Scholar]
- 14.Sullivan F X, Kumar R, Kriz R, Stahl M, Xu G Y, Rouse J, Chang X J, Boodhoo A, Potvin B, Cumming D A. J Biol Chem. 1998;273:8193–8202. doi: 10.1074/jbc.273.14.8193. [DOI] [PubMed] [Google Scholar]
- 15.Chaney W, Stanley P. J Biol Chem. 1986;261:10551–10557. [PubMed] [Google Scholar]
- 16.Eckhardt M, Gotza B, Gerardy-Schahn R. J Biol Chem. 1998;273:20189–20195. doi: 10.1074/jbc.273.32.20189. [DOI] [PubMed] [Google Scholar]
- 17.Oelmann S, Stanley P, Gerardy-Schahn R. J Biol Chem. 2001;28:26291–26300. doi: 10.1074/jbc.M011124200. [DOI] [PubMed] [Google Scholar]
- 18.Ohyama C, Smith P L, Angata K, Fukuda M N, Lowe J B, Fukuda M. J Biol Chem. 1998;273:14582–14587. doi: 10.1074/jbc.273.23.14582. [DOI] [PubMed] [Google Scholar]
- 19.Lindsell C E, Shawber C J, Boulter J, Weinmaster G. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 20.Moloney D J, Shair L H, Lu F M, Xia J, Locke R, Matta K L, Haltiwanger R S. J Biol Chem. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 21.Johnston S H, Rauskolb C, Wilson R, Prabhakaran B, Irvine K D, Vogt T F. Development (Cambridge, UK) 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 22.Robertson M A, Etchison J R, Robertson J S, Summers D F, Stanley P. Cell. 1978;13:515–526. doi: 10.1016/0092-8674(78)90325-2. [DOI] [PubMed] [Google Scholar]
- 23.Li E, Kornfeld S. J Biol Chem. 1978;253:6426–6431. [PubMed] [Google Scholar]
- 24.Almeida R, Levery S B, Mandel U, Kresse H, Schwientek T, Bennett E P, Clausen H. J Biol Chem. 1999;274:26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 25.Almeida R, Amado M, David L, Levery S B, Holmes E H, Merkx G, van Kessel A G, Rygaard E, Hassan H, Bennett E, Clausen H. J Biol Chem. 1997;272:31979–31991. doi: 10.1074/jbc.272.51.31979. [DOI] [PubMed] [Google Scholar]
- 26.Lo N W, Shaper J H, Pevsner J, Shaper N L. Glycobiology. 1998;8:517–526. doi: 10.1093/glycob/8.5.517. [DOI] [PubMed] [Google Scholar]
- 27.Lu Q, Hasty P, Shur B D. Dev Biol. 1997;181:257–267. doi: 10.1006/dbio.1996.8444. [DOI] [PubMed] [Google Scholar]
- 28.Asano M, Furukawa K, Kido M, Matsumoto S, Umesaki Y, Kochibe N, Iwakura Y. EMBO J. 1997;16:1850–1857. doi: 10.1093/emboj/16.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evrard Y A, Lun Y, Aulehla A, Gan L, Johnson R L. Nature (London) 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- 30.Zhang N, Gridley T. Nature (London) 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
- 31.Kao Y H, Lee G F, Wang Y, Starovasnik M A, Kelley R F, Spellman M W, Lerner L. Biochemistry. 1999;38:7097–7110. doi: 10.1021/bi990234z. [DOI] [PubMed] [Google Scholar]
- 32.Leffler H. Results Probl Cell Differ. 2001;33:57–83. doi: 10.1007/978-3-540-46410-5_4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.