Abstract
One of the well-known floral abnormalities in flowering plants is the double-flower phenotype, which corresponds to flowers that develop extra petals, sometimes even containing entire flowers within flowers. Because of their highly priced ornamental value, spontaneous double-flower variants have been found and selected for in a wide range of ornamental species. Previously, double flower formation in roses was associated with a restriction of AGAMOUS expression domain toward the centre of the meristem, leading to extra petals. Here, we characterized the genomic region containing the mutation associated with the switch from simple to double flowers in the rose. An APETALA2-like gene (RcAP2L), a member of the Target Of EAT-type (TOE-type) subfamily, lies within this interval. In the double flower rose, two alleles of RcAP2L are present, one of which harbours a transposable element inserted into intron 8. This insertion leads to the creation of a miR172 resistant RcAP2L variant. Analyses of the presence of this variant in a set of simple and double flower roses demonstrate a correlation between the presence of this allele and the double flower phenotype. These data suggest a role of this miR172 resistant RcAP2L variant in regulating RcAGAMOUS expression and double flower formation in Rosa sp.
Introduction
Roses are widely used as garden ornamental plants and cut flowers worldwide. A number of their agricultural and decorative traits specify their commercial value1 and have been selected during domestication. Examples of these important traits are recurrent flowering, double flowers, petal colour and fragrance2. Double flower refers to a characteristic of modern roses giving blooms with an increased number of petals that can vary from 10 to more than 200 petals per flower, whereas wild-type simple flowers are composed of 5 petals. This characteristic is tightly associated with flower development and organ identity patterning, as it results from homeotic conversion of stamens into petals3. However, the underlying molecular mechanisms are not fully understood in roses, or in other non-model species.
In the past three decades, most of the genetic and molecular networks controlling floral development have been extensively studied in model species such as Arabidopsis thaliana and Antirrhinum majus. These studies led to the establishment of the ABCE model of flower development4,5. In this model, the combinatorial actions of four classes of homeotic genes (A, B, C and E) determine flower organ identity. Briefly, from the outer to the inner whorl of the floral meristem, the A-class genes (APETALA1, AP1; APETALA2, AP2) alone determine sepal formation; the A-class genes together with the B-class genes (PISTILLATA, PI; APETALA3, AP3) determine petal fate, the C-class gene (AGAMOUS, AG) associated with the B-class genes specify stamen formation, and finally the C-class gene determines carpel fate. E-class genes are necessary for all floral organ identity. A-class genes have also an antagonistic role toward the expression of the C-class gene AG, and vice versa. This leads to the expression of the A-class genes in the sepal and petal whorls and of AG in the stamen and carpel whorls. In Arabidopsis, AG loss-of-function leads to over-accumulation of A-class genes in the third whorl and homeotic conversion of stamens into petals6. Similarly, over-accumulation of AP2 protein leads to a reduced expression of AG in the third whorl and a similar homeotic conversion of stamens into petals7. This conceptual framework for floral organ identity patterning is broadly valid for flowering plant species that have been studied8,9. However, during evolution, some genes underwent duplication and neo- or sub-functionalization, leading to small differences in their regulatory interactions. For example, the canonical C-function, performed by AG in Arabidopsis, is carried out by PLENA in Antirrhinum, that is orthologous to the Arabidopsis SHATTERPROOF genes (SHP)10. In Petunia, the restriction of the C-class gene expression needs mainly the actions of the microRNA BLIND, but involves a gene from the euAP2 family, PhBEN11,12. This diversity of the canonical ABCE functions, together with the absence of comprehensive genome data giving access to all members of each gene family, hampered the identification of the key genes determining floral organ identity in non-model species, such as in rose. Recently, efforts have been made to identify canonical rose A-, B-, C- and E-class gene orthologues, but we are still far from understanding their exact role in rose floral phenotype patterning3,13–19.
Previously, we demonstrated that a downregulation and a restricted expression domain of the rose orthologue of AGAMOUS (RcAG) correlates with an increase in petal number in domesticated roses3. This was later confirmed by transient RcAG downregulation using Virus Induced Gene Silencing20. Similar associations between AG expression and double flowers formation were shown in other species such as Ranunculids, Cyclamen, Japanese gentian and Prunus21–24. Yet, the molecular mechanism by which the restriction of the expression of RcAG occurs remains unknown. Indeed, the rose RcAG gene does not co-segregate with the major locus (Df) located on Linkage Group 3 that has been shown to control the switch from the simple flower to the double flower phenotype25,26. In roses, a yet unknown gene located in the Df locus and acting upstream of RcAG must be the determinant for double flower formation3.
In order to identify the genetic determinant of the double flower phenotype, we localized and analysed the sequence of the double flower interval using the recent high-quality Rosa chinensis ‘Old Blush’ genome assemblies27. The first corresponds to the homozygous rose assembly27,28 consisting of seven assembled pseudomolecules and representing a haplotype of the rose genome. The second assembly corresponds to the heterozygous Rosa chinensis ‘Old Blush’ consisting of 15,937 scaffolds, and provides access to the two haplotypes of the genome and information on alleles. Among the candidate genes in the interval, we identified a gene belonging to the euAP2 family, of which certain members are known to repress AG expression in many species6,12,29,30. We show that in double flower roses this gene is present as two different alleles, one of which harbours a transposable element insertion that is never found in simple flower roses. This insertion leads to a truncated RcAP2L version that lacks the miR172 binding site, meaning it is no longer negatively regulated by this microRNA. The data provide a basis for a mechanism by which double flowers are formed and open new perspectives to dissect in detail the underlying molecular and biochemical mechanisms in roses and likely in other species.
Results
Localization of the genetic interval associated with the double flower phenotype
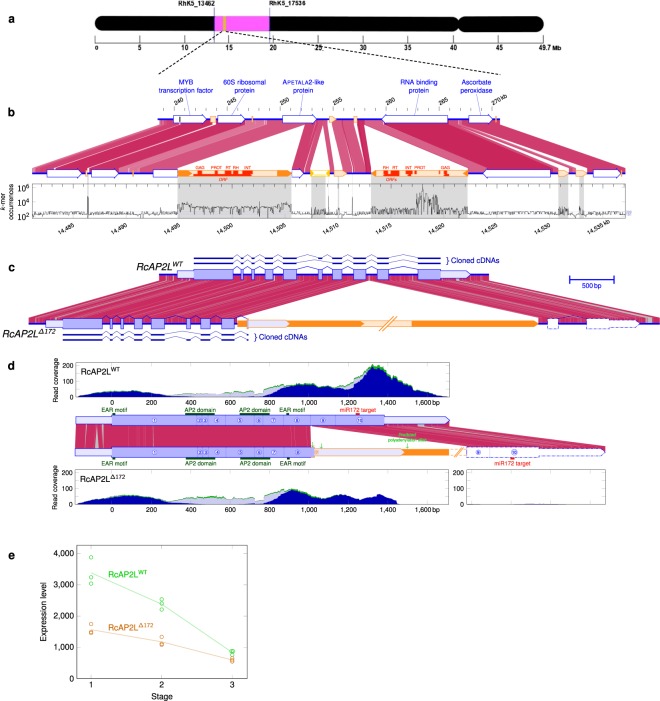
In roses, the double flower phenotype is associated with a dominant mutation in the yet unknown Df (DOUBLE FLOWER) locus. This locus was previously shown to map on LG325,26. We used the high-quality genome assembly (RcHm)27,28 to identify flanking markers that define the mapping interval containing Df. Flanking markers were retrieved from the previously reported genetic maps26,31,32 and mapped on the rose genome sequence27 and those that had unique match allowed to mark out an interval of 6.2 Mb on Chromosome 3 at coordinates 13,535,933 to 19,743,495 (Fig. 1a). Genes within this interval were then retrieved using the gene annotation of the reference rose genome27,28 (Supplementary Table 1). The assembled interval on Chromosome 3 contained 631 annotated genes (Supplementary Table 1). Alleles for each gene were then retrieved using the genome assembly of the heterozygous genome (RcHt)27. Previous studies showed that a modified expression pattern of RcAG was associated with double flower formation in rose3. RcAG maps on Chromosome 5 of the rose genome, thus corroborating previous data indicating that RcAG is not the Df gene3. Among the 631 annotated genes that lie within the double flower mapping interval, no gene showed similarities to RcAG gene. These data suggest that the gene responsible for double flower formation could be an upstream regulator of RcAG.
Figure 1.
RcAP2L lies within the double flower interval in the rose. (a) Graphical representation of chromosome 3 showing the localization of the double flower interval and its flanking markers from the genetic map. (b) Detailed comparison of the two haplotypes containing RcAP2L and neighbouring sequences. The upper haplotype contains the wild-type allele of RcAP2L. The lower haplotype contains RcAP2L∆172 with TE insertion. Blue and orange box-arrows indicate annotated genes and transposable elements, respectively. Sequence similarities between the two haplotypes are shown with purple shades. The darker is the purple colour intensity, the highest is the Blastn identity (ranging between 100 to 90%). The plot shows k-mer occurrences along the region: corresponds to the number of times each word of 47 bp in length is found in the 375 Gb set of reads used to assemble the heterozygous genome27. High occurrence counts indicate repeated sequences (grey shading). (c) Intron/exon structure of the two alleles of RcAP2L, confirmed by cDNA sequencing. Exons are shown as blue boxes. Light blue indicate 5′ and 3′ UTR. cDNA sequences confirmed by RT-PCR are shown. The transposable element inserted in RcAP2L∆172 is shown in orange. Purple lines show identical sequence and grey lines indicate single nucleotide polymorphisms and short INDELs (overall identity: 93,4%). (d) Structure of mature mRNAs from RcAP2L alleles. The three area plots show RNA-seq read coverage along mRNAs. Dark blue: reads matching specifically one of the two alleles. Light blue: reads that could come from any of the two RcAP2L alleles. Green: reads also matching at other loci in rose genome. Expression data confirm the structure of the two alleles, including the correct splicing of exon 9′ of RcAP2L∆172 originating from the inserted TE, and the appearance of a new polyadenylation site at position 1,500 bp. The region of RcAP2L∆172 homologous to exons 9 and 10 exhibits a negligible expression level (highest value ≤ 2). (e) Expression of wild-type allele RcAP2LWT (green) and RcAP2L∆172 allele (orange) early on in flower formation. Stage 1: sepal initiation, stage 2: petal initiation, stage 3: stamen initiation. Data were extracted from RNA-seq analyses. Y-axis is labelled in FPKM.
A mutant allele of an AP2-like gene lies within Double Flower interval
To narrow down the number of Df gene candidates, we searched within the assembled double flower interval for genes that share homologies with those known to regulate AG expression in Arabidopsis and that are present at heterozygous state in the double flower rose R. chinensis ‘Old Blush’. Indeed, previous genetic segregation analyses involving “Old Blush” or other rose cultivars as parents showed that the double flower trait is controlled by a dominant allele at heterozygous state25,33–35. Interestingly, one candidate gene had high sequence similarity to APETALA2 (AP2). In Arabidopsis, AP2 was shown to negatively regulate the expression of AG in the sepal and petal whorls, restricting its expression to the stamen and carpel whorls6,36. The identified rose AP2-like gene (RcAP2L, RcHm3g0468481; Fig. 1c) contains 10 exons and 9 introns, and encodes for a 460 amino-acid protein. Analysis of the predicted RcAP2L protein showed the presence of two AP2 DNA-binding domains, indicating a similar structure to the Arabidopsis A-class gene AP237. Additionally, a miR172 binding site and two EAR motifs (Ethylene-responsive element binding factor-associated amphiphilic repression) were also found in this gene. These three features are characteristic of the euAP2 family members38,39.
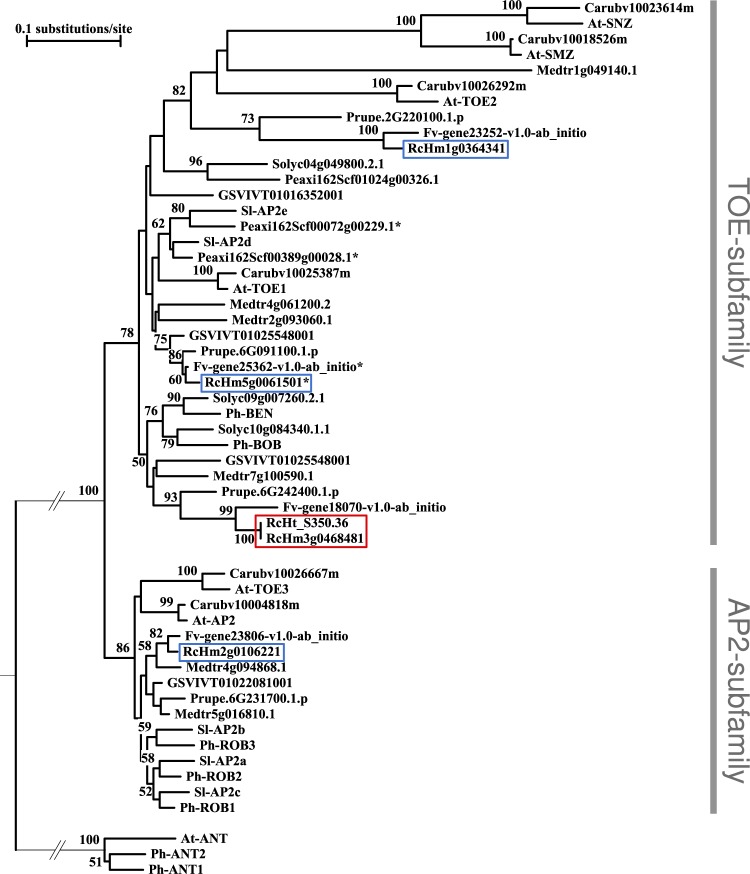
BLASTP of the Arabidopsis AP2 protein on rose and strawberry predicted protein sequences identified four potential members of the euAP2 family in each of the species (Fig. 2). Protein sequence alignments and phylogenetic analyses using the AP2 domains of euAP2 genes from Petunia hybrida, Solanum lycopersicum, Arabidopsis thaliana, Capsella rubella, Medicago truncatula, Vitis vinifera and Prunus persica showed that each of the four rose predicted proteins groups with a single and unique strawberry and Prunus predicted protein, supporting their orthologous relationship and the quality of the tree (Fig. 2). Bootstrap values highly support the presence of a single rose member of the AP2-type subfamily (RcHm2g0106221). The remaining three rose euAP2 members, including RcAP2L, likely belong to the Target Of EAT-type (TOE-type) subfamily (Fig. 2). The rose TOE-type subfamily contains a single homolog for AtTOE1 (RcHm5g0061501) and a gene (RcHm1g0364341) that groups in a branch with Arabidopsis TOE2, SMZ (SCHLAFMUTZE) and SNZ (SCHNARCHZAPFEN). The higher divergence of this last branch from the rest of the tree is likely due to the presence of a non-functional second AP2 DNA-binding domain, that could have accumulated more mutations and putatively acquired a new function12,39.
Figure 2.
RcAP2L belongs to the euAP2 family and the TOE-subfamily. Neighbor-Joining tree based on the aligned AP2 DNA binding domains of the euAP2 members from Rosa chinensis (RcHm and RcHt)27, Fragaria vesca (Fv), Petunia hybrida and P. axillaris (Ph and Peaxil, respectively), Solanum lycopersicum (Solyc or Sl), Arabidopsis thaliana (At), Capsella rubella (Carub), Medicago truncatula (Medtr), Vitis vinifera (GSVIVT) and Prunus persica (Prupe). The tree was rooted with AINTEGUMENTA (ANT) and tested with 2,000 bootstraps. EuAP2 members clearly group together with a bootstrap of 100. Among this family, the AP2- and TOE-subfamilies are also well separated (bootstrap of 86 and 78 respectively). Each distinct gene of close species generally groups together confirming the reliability of the tree. RcAP2L is part of the TOE-subfamily (observed for both the truncated RcHm3g0468481 and the wild-type RcHt_S350.36 version).
Phylogenetic analyses, using AP2 domains, revealed no direct orthologue of RcAP2L (RcHm3g0468481) in Arabidopsis genome. Interestingly, RcAP2L appears to group with the Petunia PhBEN and PhBOB gene. PhBEN was reported to repress the expression of the C-function genes in the perianth, and together with PhBOB, it is required for organ growth in the second whorl12.
Gene sequence analyses, using the assembled heterozygous genome of ‘Old Blush’, revealed that in the double flower of R. chinensis ‘Old Blush’, RcAP2L is present as two different alleles. The first allele, located on scaffold RcHt_S35027, corresponds to the wild-type sequence of RcAP2L (RcAP2LWT). A second allele, located on two assembled scaffolds (RcHt_S3277 and RcHt_S1251), contains an additional sequence of 10,790 bp inserted in its 8th intron (genome coordinates 14,494,849 – 14,505,638; Fig. 1b).
The inserted sequence is repeated in the rose genome and corresponds to a transposable element (TE) belonging to the Gypsy LTR retrotransposon family (Fig. 1b). Sequence alignment also showed that the two LTRs of the inserted TE are 100% identical on their whole length, indicating a recent insertion40. The inserted TE contains an open reading frame of 5,535 pb and DANTE software predicted the presence of at least 5 retroviral sequences coding for the structural protein GAG, a protease, a reverse transcriptase, a H-Ribonuclease and an integrase. We found 17 complete copies from this TE family in the rose genome, and 66 solo-LTRs, making it moderately repeated.
The TE insertion in RcAP2L creates a new splicing acceptor site that is predicted to lead to a fusion of the 8th exon of RcAP2L with a sequence from the 5′ LTR from the TE. This new splicing creates a premature STOP codon and the loss of the 9th and the 10th exons, which causes the formation of a truncated protein composed of 342 amino acids, and the loss of the miR172 binding site (Fig. 1d). This allele was consequently named RcAP2L∆172.
We mapped RNA-seq reads on the predicted transcripts to validate the mRNA structures; as a few SNPs and INDELs exist between the sequences of the two alleles (Fig. 1d), we were able to distinguish reads coming from each. The RNA-seq coverage drastically decreased at one of the predicted polyadenylation sites of RcAP2L∆172 identified by PASPA software41, indicating that the corresponding mRNA existed and was properly spliced, and thus must be stable. The RNA-seq mapping also showed that exons 9 and 10 of the mutated allele, located after the TE insertion, are not expressed, indicating that the mRNA from this allele no longer have a miR172 binding site. Sequencing of cDNA prepared from RNA extracted from R. chinensis ‘Old Blush’ confirmed the predicted intron/exon structures but also indicated a potential alternative splicing with the loss of the 6th exon (Fig. 1c).
Expression analyses showed that both alleles RcAP2LWT and RcAP2L∆172 are expressed during flower formation (Fig. 1e). The expression of both alleles is high in flower primordia at stages 1 and 2 (sepal and petal initiation, respectively), and their expression starts to significantly decrease at stage 3 (stamen initiation), thus consistent with a role in perianth formation.
The presence of the RcAP2L∆172 allele correlates with double flower formation in Chinese and modern roses
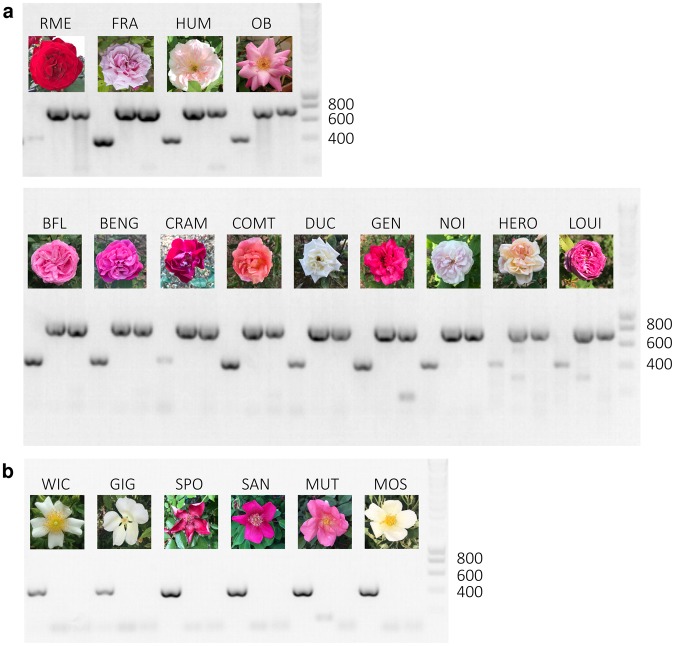
To further address the correlation between the presence of the RcAP2L∆172 allele and double flower formation, we investigated its presence in the available genomic data from five other rose genotypes that exhibit either double flowers (R. odorata ‘Hume’s Blush’, R. x hybrida ‘La France’) or simple flowers (R. chinensis ‘Sanguinea’, R. chinensis ‘Spontanea’ and R. wichurana)27. The insertion of the TE in intron 8 of RcAP2L was investigated by the presence of reads overlapping both 5′ and 3′ junctions, while its absence was confirmed by reads overlapping the intact position on the wild type RcAP2L gene (Table 1). For example, a mean of 6.9 reads per 108 reads and 9.2 reads per 108 reads were shown to overlap respectively the 3′ and 5′ TE junctions in R. odorata ‘Hume’s Blush’ and 8.4 reads per 108 reads were overlapping the wild type position of the gene (Table 1), indicating that this genotype had one of each allele. Conversely, R. wichurana had no read overlapping the TE junctions and 8.6 reads per 108 reads overlapping the intact position, indicating that this genotype only has the wild type allele at homozygous state. This analysis indicated that all double flower roses of the panel harbour both the wild-type RcAP2LWT allele and the truncated RcAP2L∆172 allele. Conversely, simple flower roses harbour only the RcAP2LWT allele and never the RcAP2L∆172. Together, these data corroborate the observation in ‘Old Blush’ and show the existence of a correlation between the presence of RcAP2L∆172 and double flower formation.
Table 1.
Identification of RcAP2L alleles present in 7 resequenced genotypes.
| Genotype | Flower multiplicity | Ploidy level | Overlapping reads per 108 reads | ||
|---|---|---|---|---|---|
WT
|
5′ of TE
|
3′ of TE
|
|||
| R. chinensis ‘Old Blush’ | Double flower | 2x | 5.7 | 7.2 | 7.4 |
| R. chinensis homozygous genome | NA | 2x | 0 | 22.2 | 16.7 |
| R. odorata ‘Hume’s Blush’ | Double flower | 2x | 8.4 | 9.2 | 6.9 |
| R. x hybrida ‘La France’ | Double flower | 3x | 5.1 | 5.1 | 8.0 |
| R. chinensis ‘Sanguinea’ | Simple flower | 2x | 22.5 | 0 | 0 |
| R. chinensis ‘Spontanea’ | Simple flower | 2x | 13.2 | 0 | 0 |
| R. wichurana | Simple flower | 2x | 8.6 | 0 | 0 |
Number of 100 bp genomic reads overlapping intra-gene (denoting the presence of a wild-type RcAP2L allele) or gene-TE (mutated allele, RcAP2L∆172) junctions, for genotypes with double or simple flowers. The read counts were normalized according to the read library size, and expressed as reads per 100 million reads. Ploidy level: According to Raymond et al.27.
To further confirm our hypothesis, we investigated the presence of RcAP2L∆172 in a set of modern rose cultivars (Supplementary Table S2). The presence or absence of RcAP2L∆172 was investigated by PCR amplification of the TE insertion junctions using DNA extracted from 6 rose plants exhibiting simple flowers and from 13 rose plants exhibiting double flowers (Supplementary Table 2; Fig. 3). DNA fragments overlapping both left and right borders of the transposon were detected in all these double flower roses (Fig. 3a), while no similar DNA fragment could be detected in the simple flower roses (Fig. 3b). Our data show a correlation between the double flower phenotype and the presence of the transposable element insertion, thus providing another argument in favour of the role of RcAP2L∆172 during double flower formation. It should be noted that none of the analysed double flower cultivars were homozygous for the truncated allele and all had wild type and truncated alleles.
Figure 3.
The transposable element insertion is only observed in the analysed double flower roses. PCR to detect the TE were performed on genomic DNA from different rose cultivars exhibiting simple (b) or double flowers deriving from R. chinensis (a). The lower band (419 bp) corresponds to the combination of primers that amplifies the wild type allele, while the two higher ones (770 bp and 754 bp) correspond to amplification of the left and right borders of the transposable element insertion, respectively.
Discussion
In roses, the formation of double flowers is associated with a shift of the A/C boundary and a restriction of RcAG expression domain toward the centre of the meristem which in turn leads to a conversion of stamens into petals3. Genetic mapping identified the major locus Df as involved in the control of rose double flower formation25. However, RcAG does not lie within the double flower interval suggesting that a yet unknown upstream regulator of RcAG must be the determinant of double flower formation.
In this study, we identified a gene of the euAP2 family, RcAP2L, that localizes within the double flower interval. We identified a truncated allele version of RcAP2L (RcAP2L∆172) whose presence correlates with double flower formation in Chinese roses and modern roses, such as R. hybrida ‘La France’, that have Chinese rose cultivars as ancestors. Such an allelic form is absent in all simple flower roses. A TE insertion in the 8th intron of RcAP2L leads to the loss of the miR172 target site. We demonstrate that the position of the TE insertion is conserved among different double flower rose varieties that have a parent from the Chinense section. These data indicate that this allele must have been inherited from a single common Chinese ancestor and spread among its double flower modern descendants due to the positive human selection during rose domestication.
Phylogenetic analysis indicates that RcAP2L is a member of the TOE-subfamily. In Arabidopsis, most studies focused on AP2 and only a few reports addressed the role of other euAP2 family members such as TOE1, TOE2 and TOE3. Arabidopsis AP2 is known to restrict AG expression to the third and fourth whorls (stamens and carpels, respectively). Knockout of AP2 results in ectopic expression of AG in the sepal and petal whorls, which is associated with a conversion of sepals to carpeloid structures and loss of petals6. Recently, ChIP-qPCR experiments in Arabidopsis showed that both AP2 and TOE3 bind to AG second intron (containing transcription cis-regulatory elements) to decrease its expression level29,42. These published data support our findings on RcAP2L as a pertinent candidate for double flower determination, likely by regulating the expression pattern of RcAG.
In Arabidopsis, TOE1 overexpression induces late flowering while its loss-of-function leads to early flowering with no apparent flower phenotype variations43. Conversely to Arabidopsis, in some species certain TOE-subfamily genes have a flower patterning function. In Petunia, PhBEN was shown to inhibit the C-function in the perianth primordium, thus consistent with a function similar to that of the Arabidopsis AP2. It is clear that TOE genes have evolved to perform different functions in different species, and in roses, their misexpression is likely at the origin of the appearance of the double flower abnormality.
EuAP2 family members are characterized by the presence of a miR172 binding site that is important for their post-transcriptional regulation. It has been reported that overexpression of miR172 induces early flowering as well as floral defects similar to the ones observed in ap2 mutants43. During flower formation, miR172 is highly expressed in whorls 3 and 4 and targets AP2 transcripts to prevent its expression in the centre of the meristem, where the stamen- and carpel-identity gene AG is expressed30. However, in Arabidopsis it has been reported that when AP2 lacks the miR172 binding site (as for the RcAP2L∆172), its expression is maintained in the centre of the meristem, which leads to continuous downregulation of AG expression30,44. Such downregulation of AG expression results into the formation of flowers with an increased number of petals or stamens and a loss of floral determinacy7,44, a phenotype resembling that of ag loss-of-function mutant45. Similarly, AG expression is reduced when a miR172-resistant TOE3 is expressed in Arabidopsis flowers29, indicating that other members of the euAP2 family can also have antagonistic role on the expression of AG.
In a recent study, Han et al.46, reported that the down-regulation of the rose AP2 orthologue leads to the reduction of petal number46. However, the RcAP2 gene (corresponding to RcHm2g010622127) studied by Han et al.46, is located on chromosome 2 and not on chromosome 3, where the interval containing the double flower mutation lies.
In double flower roses, the RcAP2L∆172 truncated allele lost its miR172 binding site but still contained both AP2 DNA binding domains and EAR domains (Ethylene-responsive element binding factor-associated amphiphilic repression), thus consistent with an RcAP2L gain of function hypothesis and the dominant character of the Df gene. Indeed, Arabidopsis AP2 is known to interact with TOPLESS via its EAR domain to recruit the histone deacetylase HDA19 to its DNA binding sites including the AG second intron47. It has been demonstrated that a fusion between the AP2 DNA binding domains and TOPLESS, with the addition of an artificial miR172 binding site, is sufficient to complement the Arabidopsis ap2 phenotype, indicating that the main floral function of AP2 is established via its TOPLESS interaction and recruitment to DNA target sites. Further investigation also revealed that TOE1, TOE2 and TOE3 interact with TOPLESS48, indicating a potentially conserved mechanism among the whole euAP2 family. Therefore, it is likely that in roses, the expression of the miR172 resistant allele (RcAP2L∆172) is responsible for the observed restricted expression of RcAG toward the centre of the flower, which in turn leads to the formation of flowers with an increased number of petals.
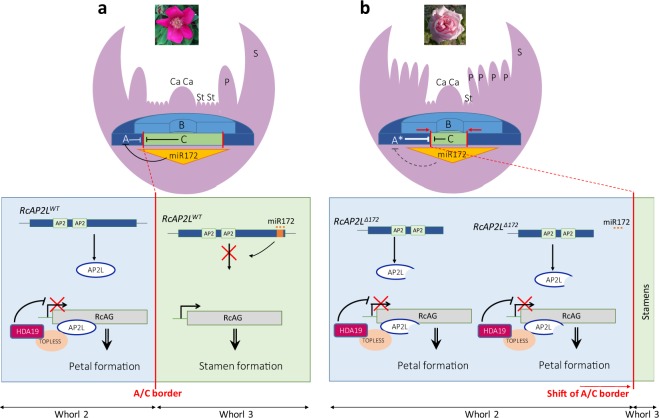
Our data, taken together with published data in Arabidopsis and other plants, suggest the following model. In simple flower roses, RcAP2L and RcAP2 proteins are produced only in the first two whorls, where they can inhibit RcAG transcription. This induces sepal and petal organ identity determination and development in whorls 1 and 2, respectively. In the third and fourth whorls, the accumulation of miR172 interferes with euAP2 mRNA accumulation, which in turn results in the expression of RcAG that mediates stamen and carpel organ identity determination and development (Fig. 4). However, in a double flower such as ‘Old Blush’, the presence of the miR172 insensitive variant RcAP2L∆172 leads to a prolonged accumulation of RcAP2L protein toward the centre of the meristem and consequently prolonged downregulation of RcAG expression (Fig. 4). This causes a restriction of RcAG expression toward the centre of the flower, as described previously3. As a consequence, the homeotic conversion of stamens into petals leads to the formation of a double flower.
Figure 4.
Model showing how a miR172-resistant euAP2 could lead to double flower formation. (a) In wild-type flowers, euAP2 are expressed in the first and second whorls where they can inhibit RcAG expression likely by recruiting cofactors and histone modifiers, such as TOPLESS and HDA19. Sepals and petals are consequently formed. In the 3rd whorl, miR172 is expressed and inhibits euAP2 proteins production, releasing the inhibition of RcAG. RcAG will then determine stamens and carpels identity and formation. (b) In double flowers, the truncated version RcAP2L∆172 mRNA (following TE insertion) is insensitive to miR172 inhibition. RcAP2L∆172 expression is maintained in the meristem and down-regulates RcAG expression. This leads to the sliding of the A/C border toward the centre of the meristem and the formation of more petals, producing the so-called double-flowers. The wild-type RcAP2L (not shown on the Figure) is degraded. Ca, carpels; St, stamens; P, petals; S, sepals. A, A-class genes (RcAP1, euAP2); A*, A-class genes including a miR172-resistant euAP2 such as RcAP2L∆172; B, B-class genes; C, C-class gene (RcAG).
Our model is supported by previous work on kiwifruit, where a downregulation of miR172 and an up-regulation of AP2 were observed in flower buds from the “Pukekohe dwarf” kiwifruit double flower cultivars, but not in the “Hayward” and “Chieftain” simple flower cultivars49.
Our data taken together with that in the literature, strengthen the conclusion that misregulation of the miR172/AP2 loop is likely the cause of the double flower phenotype in many species. To address more in deepth the molecular mechanisms that link the miR172 insensitive allele of RcAP2L to the double flower formation, a futur experiment would be to overexpress it in simple flower roses.
The fact that many double flower roses still develop carpels suggests that the accumulation of the miR172 insensitive variant RcAP2L∆172 affects RcAG expression only in the third whorl, but not in the fourth whorl. This indicates that the rose may have evolved differently from Arabidopsis where miR172 insensitive variant of AP2 affect both whorls 3 and 4. It will be interesting to address the molecular mechanisms of such difference and whether such mechanism is applicable to other species with double flowers.
Material and Methods
Plant material
Double flower rose cultivars Rosa chinensis ‘Old Blush’, R. odorata ‘Hume’s Blush’, R. x hybrida ‘La France’, R. x hybrida ‘Rouge Meilland’, R. x hybrida ‘Bébé Fleuri’, R. x hybrida ‘Bengale d’Automne’, R. x hybrida ‘Cramoisi Supérieur’, R. x hybrida ‘Comtesse de Cayla’, R. x hybrida ‘Ducher’, R. x hybrida ‘General Shablikine’, R. x hybrida ‘Blush Noisette’, R. x hybrida ‘Herodiade’ and R. x hybrida ‘Louise d’Arzens’, and simple flower cultivars R. chinensis ‘Spontanea’, R. chinensis ‘Sanguinea’, R. chinensis ‘Mutabilis’, R. wichurana, R. gigantea and R. moschata were field grown at the Lyon-Botanical-Garden and/or in environmentally controlled greenhouse conditions at the Ecole Normale Supérieure of Lyon with 16 h/8 h day/night periods and 25 °C/18 °C day/night temperatures.
Staging rose flower development
Flower development stages were distinguished and dissected under a binocular microscope as previously defined by Dubois et al.14. Stages 1 to 3 correspond to development stages when sepal, petal and stamen primordia arise, respectively. During stage 4, carpels are produced in the centre of the meristem, which will then sink below at stage 5.
DNA extraction and genotyping PCR
Young leaves or axillary buds were collected and ground in PVP and homogenization buffer (Tris pH8 15 mM, EDTA 2 mM, NaCl 20 mM, KCl 20 mM, β-mercaptoethanol 0,1%, Triton 0,5%). DNA extraction was performed using the DNeasy kit (Qiagen).
DNA fragments were amplified using the GoTaq Polymerase Chain Reaction according to the manufacturer’s recommendation (Promega). An initial denaturing step was carried at 95 °C for 5 min. Fifteen cycles of touch-down PCR were then performed 95 °C for 30 s, 65 °C (with a decrease of 1 °C per cycle) for 30 s, 72 °C for 1 min 30 s. This was followed by 30 cycles of standard PCR with the following cycle 95 °C for 30 s, 50 °C for 30s and 72 °C for 1 min 30 s. A final elongation step was performed for 10 min at 72 °C.
RNA purification and cDNA sequencing
Total RNA was prepared from floral meristems at different developmental stages (1 to 3) using the Spectrum plant total RNA kit (Sigma) and TURBO DNA-freeTM AM 1907 (Ambion), mainly as previously described3. Contaminating DNA was removed using the DNA-freeTM kit following the manufacturer’s recommendations (Ambion). One microgram of total RNA was then used in a reverse transcription assay. cDNAs were PCR amplified, cloned and sequenced using primers designed to specifically target RcAP2L or RcAP2L∆172 (Supplementary Table 3).
Characterisation of the double flower interval
The high quality rose genome from Rosa chinensis ‘Old Blush’ was recently published27 in the form of two complementary assemblies. The first one was obtained from PacBio long read sequencing using a homozygous rose material derived from the heterozygous Rosa chinensis ‘Old Blush’27,28 and consists of 7 assembled pseudomolecules representing a haplotype of the rose genome. The genome of the heterozygous Rosa chinensis ‘Old Blush’ (Illumina sequencing) consists of 15,937 scaffolds and provides access to the two haplotypes of the genome.
Flanking markers of double flower interval31 were mapped on the Rosa chinensis homozygous reference genome27 using the following parameters: evalue < 10−6, lengthHSP > 40, percentage identity >97%. Markers that had unique match were kept and used to define the corresponding physical region on the rose genome sequence. Genes within this interval were analyzed using Blast and Pfam web interface50,51.
Analysis of the presence of the TE element in RcAP2L was also performed using the available genome sequences of rose cultivars27. Single reads from resequenced genomes of the different rose cultivars27 were trimmed using cutadapt52 and custom Perl scripts. They were cut to an homogeneous length of 100 bp and aligned on the reference rose genome using bwa software53 allowing up to two mismatches on the whole length of the read (end-to-end alignment). Reads overlapping genomic positions of interest over at least 15 bp on each side were counted. Read counts were normalized on the library size for each genotype. psRNATarget webserver interface was used to detect miR targets54.
Haplotype identification and comparison
Sequence analysis was performed using the high-quality genome assembly of homozygous R. chinensis ‘Old Blush’27. The two distinct haplotypes within the double flower interval were retrieved from the heterozygous genome assembly27. Blastn55 and gene synteny were used to confirm alleles sequences. The water program from Emboss suite56 was used to obtain optimal end-to-end alignments between allelic regions and identify polymorphisms.
Gene sequence analysis
Gene models were recovered from the rose reference genome sequence annotation27. Splicing site predictions and untranslated region (UTR) boundaries were manually adjusted based on cloned cDNA sequences and RNA-seq data. Putative functions for genes flanking RcAP2L were inferred from Arabidopsis best blast hit. Unknown protein domains were identified using InterProScan software version 5.27.-66.057, Pfam database version 31.050 and manual annotation. miR172 putative binding sites were predicted using a local instance of WMD3 software (Ossowski Stephan, Fitz Joffrey, Schwab Rebecca, Riester Markus and Weigel Detlef, personal communication).
Transposable element annotation
To identify repeated regions, the genomic sequence of RcAP2L neighbourhood was cut into 47 bp overlapping k-mers, and the number of occurrences of each k-mer was counted in the 375 Gb-dataset of genomic reads used to assemble the Rosa chinensis heterozygous rose genome sequence27. These occurrence counts were plotted along the sequence (Fig. 1b) and compared to the mean occurrence counts for homozygous and heterozygous regions. Automatic transposable element (TE) annotations from the rose genome27 were used as a starting point, and manually curated.
The boundaries of the two long terminal repeats (LTRs) were accurately identified using a graphical dotplot program58 and LTR sequences were compared using bl2seq alignment55. Open reading frames were predicted in the TE internal region using Pfam software50 and protein domains were annotated by similarity search using DANTE (http://repeatexplorer.org/).
Using the LTR sequence as a Blastn query (parameters: M = 6 N = −7 Q = 8 R = 8; e-value ≤ 10−20, match length ≥600 bp) and the internal part sequence as tBlastx query (parameters: BLOSUM80 Q = 9 R = 3; e-value ≤ 10−15, overall coverage of query ≥800 bp), we looked for LTR pairs flanking a putative internal part, to detect complete copies of TEs from the same family. Using more stringent criteria (e-value ≤ 10−80 and match length ≥900 bp), we also identified solo-LTRs from the same family59.
Expression analysis
Paired-end RNA-seq data from young flower buds at stage 1, 2 and 3 were previously described27.
Pairs of reads putatively originating from RcAP2LWT and RcAP2L∆172 alleles were selected using Tophat version 2.1.160 with relaxed parameters (“–read-realign-edit-dist 0–b2-very-sensitive –max-intron-length 25000” and insert size and insert size SD estimated beforehand on the whole predicted transcriptome for each library). These read pairs were remapped on the whole genome with Tophat allowing up to 5 multimatches and secondary alignments. Based on the number of matches and the alignment scores, read pairs were sorted into four categories: (i) specific to RcAP2LWT, (ii) specific to RcAP2L∆172, (iii) coming indiscriminately from RcAP2LWT or RcAP2L∆172, and (iv) coming indiscriminately from RcAP2LWT, RcAP2L∆172 or other loci in the genome (hereafter called non-specific read pairs). Read pairs that Tophat could match on the extracted genomic sequences of RcAP2L alleles, but not on the whole genome, were put in category (iv). This case is expected for reads originating from repeated sequences. The read pairs from each category were mapped on the predicted transcripts using Bowtie2 version 2.3.4.161. Coverage at each position was computed using Samtools version 1.562. Normalization was done using the library sizes (custom Perl scripts), before adding up the coverage values from all libraries. Reads from categories (iii) and (iv) were spread between the two alleles according to the ratio (i)/(ii), computed on a sliding window of width 241 bp. After ensuring that sequence polymorphism between RcAP2LWT and RcAP2L∆172 transcripts was sufficient to estimate independently their expression level, we used Tophat version 2.1.160 on the annotated Rosa chinensis heterozygous genome27, with corrected annotations for RcAP2L alleles, and we normalized read counts using DESeq. 2 version 1.2.063.
Phylogenetic analysis
EuAP2 family members were identified by using the Arabidopsis thaliana AP2 protein as a Blast query against rose and strawberry predicted proteins27. Sequences were aligned using ClustalW64 and BioEdit software65. Where applicable, gene annotation was corrected manually.
Neighbor-Joining tree based on the aligned AP2 DNA binding domains of the euAP2 members from Rosa chinensis (RcHm and RcHt)27, Fragaria vesca (Fv), Petunia12, Solanum lycopersicum, Arabidopsis thaliana, Capsella rubella, Medicago truncatula, Vitis vinifera and Prunus persica. Sequences were downloaded from the Phytozome website (https://phytozome.jgi.doe.gov/pz/portal.html). The aligned regions containing the two AP2 domains (Supplemental Data File 1) were selected for phylogenetic analysis. Neighbor-Joining tree was computed with Treecon software66 using the following parameters (1) Distance estimation options: Tajima and Nei67; Distance calculations; insertions and deletion not taken into account; Alignment positions: all; Bootstrap analysis: yes, 2000 samples. (2) Infer tree topology options: Neighbor-joining; Bootstrap analysis: yes. (3) Root unrooted trees options: outgroup option: single sequence (forced); bootstrap analysis: yes. Tree was rooted using the Arabidopsis ANT protein.
Electronic supplementary material
Acknowledgements
We thank Judit Szécsi (ENS de Lyon) and Philippe Vergne (ENS de Lyon) for useful discussions and for critical reading of the manuscript. We thank Alexis Lacroix, Patrice Bolland and Justin Berger (ENS de Lyon), and the “Lyon Botanical Garden-France” for providing plant material. We gratefully acknowledge support from the Pôle Scientifique de Modélisation Numérique of the ENS de Lyon for the computing resources. We thank Loïs Taulelle (PSMN) for his help with computing clusters and Emmanuel Quemener (Centre Blaise Pascal, ENS de Lyon) for setting up ad-hoc servers when needed. This work was supported by funds from the French National Institute of Agronomic Research (INRA), from the Ecole Normale Supérieure-Lyon-France, and from French National Research Agency program DODO (ANR-16CE20-0024-03).
Author Contributions
L.F., D.R., X.F., J.J., O.R. performed experiments. M.B., L.F., J.J., O.R., A.B. analysed data. M.Ve., A.D., J.J., L.F. performed bioinformatics analyses. M.Va., L.F. performed phylogenetic analyses. M.B. designed and supervised the study. M.B., L.F., J.J., O.R. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-30918-4.
References
- 1.Cairns, T. Modern roses XI, The word Encyclopidia of roses, (Academic Press, San Diego, California, 2003).
- 2.Bendahmane M, Dubois A, Raymond O, Bris ML. Genetics and genomics of flower initiation and development in roses. J Exp Bot. 2013;64:847–57. doi: 10.1093/jxb/ers387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois A, et al. Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PLoS One. 2010;5:e9288. doi: 10.1371/journal.pone.0009288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irish VF, Litt A. Flower development and evolution: gene duplication, diversification and redeployment. Current Opinion in Genetics & Development. 2005;15:454–460. doi: 10.1016/j.gde.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet. 2005;6:688–98. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- 6.Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–5. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irish V. The ABC model of floral development. Curr Biol. 2017;27:R887–R890. doi: 10.1016/j.cub.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 9.Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot. 2007;100:603–19. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-R. [DOI] [PubMed] [Google Scholar]
- 11.Cartolano M, et al. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat Genet. 2007;39:901–5. doi: 10.1038/ng2056. [DOI] [PubMed] [Google Scholar]
- 12.Morel P, et al. Divergence of the Floral A-Function between an Asterid and a Rosid Species. Plant Cell. 2017;29:1605–1621. doi: 10.1105/tpc.17.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois A, et al. Transcriptome database resource and gene expression atlas for the rose. BMC Genomics. 2012;13:638–648. doi: 10.1186/1471-2164-13-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubois A, et al. Genomic approach to study floral development genes in Rosa sp. PLoS One. 2011;6:e28455. doi: 10.1371/journal.pone.0028455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hibino Y, Kitahara K, Hirai S, Matsumoto S. Structural and functional analysis of rose class B MADS-box genes ‘MASAKO BP, euB3, and B3: Paleo-type AP3 homologue ‘MASAKO B3’ association with petal development. Plant Science. 2006;170:778–785. doi: 10.1016/j.plantsci.2005.11.010. [DOI] [Google Scholar]
- 16.Kitahara K, Hibino Y, Aida R, Matsumoto S. Ectopic expression of the rose AGAMOUS-like MADS-box genes ‘MASAKO C1 and D1’ causes similar homeotic transformation of sepal and petal in Arabidopsis and sepal in Torenia. Plant Science. 2004;166:1245–1252. doi: 10.1016/j.plantsci.2003.12.040. [DOI] [Google Scholar]
- 17.Kitahara K, Hirai S, Fukui H, Matsumoto S. Rose MADS-box genes ‘MASAKO BP and B3’ homologous to class B floral identity genes. Plant Science. 2001;161:549–557. doi: 10.1016/S0168-9452(01)00442-3. [DOI] [PubMed] [Google Scholar]
- 18.Kitahara K, Matsumoto S. Rose MADS-box genes ‘MASAKO C1 and D1’ homologous to class C floral identity genes. Plant Science. 2000;151:121–134. doi: 10.1016/S0168-9452(99)00206-X. [DOI] [PubMed] [Google Scholar]
- 19.Mibus H, Heckl D, Serek M. loning and Characterization of Three APETALA1/FRUITFULL-like Genes in Different Flower Types of Rosa × hybrida L. J Plant Growth Regul. 2011;30:272. doi: 10.1007/s00344-010-9190-8. [DOI] [Google Scholar]
- 20.Ma N, et al. Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida) BMC Plant Biol. 2015;15:237. doi: 10.1186/s12870-015-0623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galimba KD, et al. Loss of deeply conserved C-class floral homeotic gene function and C- and E-class protein interaction in a double-flowered ranunculid mutant. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2267–E2275. doi: 10.1073/pnas.1203686109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Zhang D, Liu D, Li F, Lu H. Exon skipping of AGAMOUS homolog PrseAG in developing double flowers of Prunus lannesiana (Rosaceae) Plant Cell Reports. 2012;32:227–237. doi: 10.1007/s00299-012-1357-2. [DOI] [PubMed] [Google Scholar]
- 23.Nakatsuka T, et al. Isolation and characterization of the C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC plant biology. 2015;15:182. doi: 10.1186/s12870-015-0569-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka Y, et al. Multi-petal cyclamen flowers produced by AGAMOUS chimeric repressor expression. Scientific Reports. 2013;3:2641. doi: 10.1038/srep02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debener T, Mattiesch L. Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theoretical and Applied Genetics. 1999;99:891–899. doi: 10.1007/s001220051310. [DOI] [Google Scholar]
- 26.Spiller M, et al. Towards a unified genetic map for diploid roses. Theor Appl Genet. 2011;122:489–500. doi: 10.1007/s00122-010-1463-x. [DOI] [PubMed] [Google Scholar]
- 27.Raymond O, et al. The Rosa genome provides new insights into the domestication of modern roses. Nature Genetics. 2018;50:772–777. doi: 10.1038/s41588-018-0110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergne, P. et al. Production of homozygous rose line derived from heterozygous genotype. (2018).
- 29.Jung J-H, Lee S, Yun J, Lee M, Park C-M. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Science. 2014;215–216:29–38. doi: 10.1016/j.plantsci.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Wollmann H, Mica E, Todesco M, Long JA, Weigel D. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development. 2010;137:3633–42. doi: 10.1242/dev.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourke PM, et al. Partial preferential chromosome pairing is genotype dependent in tetraploid rose. Plant J. 2017;90:330–343. doi: 10.1111/tpj.13496. [DOI] [PubMed] [Google Scholar]
- 32.Koning-Boucoiran CF, et al. Using RNA-Seq to assemble a rose transcriptome with more than 13,000 full-length expressed genes and to develop the WagRhSNP 68 k Axiom SNP array for rose (Rosa L.) Front Plant Sci. 2015;6:249. doi: 10.3389/fpls.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shupert, D. A., Byrne, D. H. & Brent Pemberton, H. Inheritance of Flower Traits, Leaflet Number and Prickles in Roses. 751 edn 331-335 (International Society for Horticultural Science (ISHS), Leuven, Belgium, 2007).
- 34.Debener, T. Genetics: Inheritance of characteristics. in Encyclopedia of rose science. (eds Roberts, A.V., Debener, T. & Gudin, S.) 286–292 (Elsevier, Oxford, 2003).
- 35.Crespel L, et al. Mapping of qualitative and quantitative phenotypic traits in Rosa using AFLP markers. Theor Appl Genet. 2002;105:1207–1214. doi: 10.1007/s00122-002-1102-2. [DOI] [PubMed] [Google Scholar]
- 36.Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- 37.Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. The Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Soltis PS, Wall K, Soltis DE. Phylogeny and domain evolution in the APETALA2-like gene family. Mol Biol Evol. 2006;23:107–20. doi: 10.1093/molbev/msj014. [DOI] [PubMed] [Google Scholar]
- 39.Wang, P. et al. Expansion and Functional Divergence of AP2 Group Genes in Spermatophytes Determined by Molecular Evolution and Arabidopsis MutantAnalysis. Frontiers in Plant Science 7(2016). [DOI] [PMC free article] [PubMed]
- 40.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nature Genetics. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 41.Ji G, et al. PASPA: a web server for mRNA poly(A) site predictions in plants and algae. Bioinformatics. 2015;31:1671–3. doi: 10.1093/bioinformatics/btv004. [DOI] [PubMed] [Google Scholar]
- 42.Yant L, et al. Orchestration of the Floral Transition and Floral Development in Arabidopsis by the Bifunctional Transcription Factor APETALA2[W][OA] The Plant Cell. 2010;22:2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–41. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao L, Kim Y, Dinh TT, Chen X. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 2007;51:840–9. doi: 10.1111/j.1365-313X.2007.03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han Y, et al. An APETALA2 Homolog, RcAP2, Regulates the Number of Rose Petals Derived From Stamens and Response to Temperature Fluctuations. Front Plant Sci. 2018;9:481. doi: 10.3389/fpls.2018.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012;139:4180–4190. doi: 10.1242/dev.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS Interactome: A Framework for Gene Repression in Arabidopsis1[W][OA] Plant Physiology. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varkonyi-Gasic E, Lough RH, Moss SMA, Wu R, Hellens RP. Kiwifruit floral gene APETALA2 is alternatively spliced and accumulates in aberrant indeterminate flowers in the absence of miR172. Plant Molecular Biology. 2012;78:417–429. doi: 10.1007/s11103-012-9877-2. [DOI] [PubMed] [Google Scholar]
- 50.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.NCBI Resource Coordinators. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 45, D12–D17 (2017). [DOI] [PMC free article] [PubMed]
- 52.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research. 2011;39:W155–159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 56.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 57.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic acids research. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- 59.Vitte C, Panaud O. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol Biol Evol. 2003;20:528–40. doi: 10.1093/molbev/msg055. [DOI] [PubMed] [Google Scholar]
- 60.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 66.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Computer applications in the biosciences: CABIOS. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 67.Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–85. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.