Abstract
The formation and life-long growth of the ocular lens depends on the continuous differentiation of lens epithelial cells into lens fiber cells. To achieve their mature structure and transparent function, newly formed lens fiber cells undergo a series of cellular remodeling events including the complete elimination of cellular organelles to form the lens organelle-free zone (OFZ). To date, the mechanisms and requirements for organelle elimination by lens fiber cells remain to be fully elucidated. In previous studies, we detected the presence of mitochondria contained within autophagolysosomes throughout human and chick lenses suggesting that proteins targeting mitochondria for degradation by mitophagy could be required for the elimination of mitochondria during OFZ formation. Consistently, high-throughput RNA sequencing of microdissected embryonic chick lenses revealed that expression of a protein that targets mitochondria for elimination during erythrocyte formation, called BCL2 interacting protein 3-like protein (BNIP3L/NIX), peaks in the region of lens where organelle elimination occurs. To examine the potential role for BNIP3L in the elimination of mitochondria during lens fiber cell remodeling, we analyzed the expression pattern of BNIP3L in newborn mouse lenses, the effect of its deletion on organelle elimination and its co-localization with lens organelles. We demonstrate that the expression pattern of BNIP3L in the mouse lens is consistent with it playing an important role in the elimination of mitochondria during lens fiber cell organelle elimination. Importantly, we demonstrate that deletion of BNIP3L results in retention of mitochondria during lens fiber cell remodeling, and, surprisingly, that deletion of BNIP3L also results in the retention of endoplasmic reticulum and Golgi apparatus but not nuclei. Finally, we show that BNIP3L localizes to the endoplasmic reticulum and Golgi apparatus of wild-type newborn mouse lenses and is contained within mitochondria, endoplasmic reticulum and Golgi apparatus isolated from adult mouse liver. These data identify BNIP3L as a novel requirement for the elimination of mitochondria, endoplasmic reticulum and Golgi apparatus during lens fiber cell remodeling and they suggest a novel function for BNIP3L in the regulation of endoplasmic reticulum and Golgi apparatus populations in the lens and non-lens tissues.
Keywords: Lens, Fiber cells, Cataract, Development
Introduction
The adult vertebrate eye lens is composed of an anterior surface layer of cuboidal epithelial cells that overlie a core of elongated and organelle-free lens fiber that make up the core and bulk of the eye lens. During embryogenesis, and throughout adult life, lens epithelial cells continuously differentiate to form nascent lens fiber cells that must complete a series of cellular remodeling steps to achieve their mature elongated structure and transparent function (Bassnett et al., 2011; Menko, 2002; Wride, 2011). A hallmark remodeling step of the lens fiber cell maturation program is the complete elimination of cellular organelles to form the lens organelle free zone (OFZ) (Bassnett, 1992; Bassnett, 1995; Bassnett and Beebe, 1992; Bassnett and Mataic, 1997; Chaffee et al., 2014). Elimination of organelles during OFZ formation appears to be critical for lens transparency since failure to remove organelles during OFZ formation is associated with cataract formation (Lyu et al., 2016; Menko, 2002; Nishimoto et al., 2003; Pendergrass et al., 2005; Wride, 2011) that, despite advances in cataract surgery, remains a leading cause of world-wide visual disability (Kupfer, 2000). Identification of the mechanisms that regulate the elimination of organelles during the remodeling of lens fiber cells is therefore critical towards understanding the requirements for adult ocular lens structure and transparency.
Although a recent study identified that regulation of mTORC1 signaling results in accelerated elimination of lens fiber cell organelles by the autophagy machinery (Basu et al., 2014) and other important studies have identified critical requirements for CDK1 (Chaffee et al., 2014) and DNase IIβ (De Maria and Bassnett, 2007; Nakahara et al., 2007; Nishimoto et al., 2003) in the specific elimination of nuclei during OFZ formation, the proteins that target specific organelles for elimination during OFZ formation remain to be identified. Our previous studies detected the presence of mitochondria contained within autophagolysosomes throughout human and chick lenses undergoing organelle elimination suggesting that proteins that specifically target mitochondria for elimination by mitophagy could be required for the elimination of mitochondria during OFZ formation (Costello et al., 2013). To identify proteins that could target mitochondria and potentially other organelles for elimination during OFZ formation, we employed high-throughput RNA sequencing to analyze the gene expression profiles of micro dissected regions of embryonic day 13 (E13) chick lenses corresponding to sequential stages of lens fiber cell differentiation. This analysis revealed that the expression levels of multiple genes linked with mitochondria degradation spiked in nascent lens fiber cells just prior to the elimination of mitochondria and other organelles during OFZ formation (Chauss et al., 2014).
A particularly striking expression pattern was detected for BCL2 interacting protein 3-like protein (BNIP3L/NIX) whose transcript levels steadily increase in lens fiber cells as organelle-elimination progresses and whose protein levels spike in the micro-region of the lens where nascent lens fiber cells are just beginning to eliminate their mitochondria and other organelles (Chauss et al., 2014). BNIP3L is a BH3-only member of the Bcl-2 family, whose expression is induced by specific physiological or pathological conditions in multiple cell types (Diwan et al., 2009). BNIP3L is also up-regulated during reticulocyte differentiation (Aerbajinai et al., 2003; Ding et al., 2010) and extensive studies have demonstrated that BNIP3L functions to remove mitochondria during reticulocyte differentiation through recruitment of autophagosomes using its LC3 interacting region (LIR) (Novak et al., 2010; Sandoval et al., 2008; Schweers et al., 2007; Zhang and Ney, 2009). Consistently, fully mature erythrocytes in BNIP3L knockout mice retain mitochondria resulting in dysfunctional erythropoiesis and anemia (Diwan et al., 2007; Sandoval et al., 2008). In the heart, BNIP3L expression is upregulated in cardiac hypertrophy and contributes to programmed loss of cardiac myocytes leading to heart failure (Diwan et al., 2008). However, differentiating lens fiber cell do not undergo apoptosis suggesting that the BNIP3L does not function as a pro-apoptotic molecule in the lens. Consistently, Bellot et al. (2009) proposed that under hypoxic conditions, BNIP3L is critical for hypoxia-induced autophagy and cell survival (Bellot et al., 2009). These studies on BNIP3L in heart, brain, cancer cell lines and erythrocytes suggest that it has a condition-specific function for each tissue.
In this report, we sought to establish the potential role for BNIP3L in the targeted elimination of mitochondria during lens OFZ formation. We first established the spatial expression patterns of BNIP3L in wild-type mouse lenses and discovered that BNIP3L protein levels appear to spike in the equatorial or transition zone of the lens consistent with BNIP3L expression patterns in the chick lens. We then examined the the presence of mitochondria in the centers of newborn and day 14 (P14) BNIP3L-knockout and same-strain wild-type mouse lenses. Surprisingly, we found that deletion of BNIP3L results in retention of not only mitochondria, but also, endoplasmic reticulum (ER) and Golgi apparatus (GA) in the cores of the P1 and P14 BNIP3L knockout lenses. By contrast, elimination of nuclei during OFZ formation was unaffected by BNIP3L-deletion. Our data also show that BNIP3L localizes to the ER and GA of wild-type newborn mouse lenses and that BNIP3L is contained within mitochondria, ER and GA isolated from adult mouse liver. These data identify BNIP3L as a novel requirement for the elimination of mitochondria, endoplasmic reticulum and Golgi apparatus during lens fiber cell remodeling and they suggest a novel function for BNIP3L in the regulation of endoplasmic reticulum and Golgi apparatus populations in the lens and non-lens tissues.
Material and Methods
Animal procedures
Animal husbandry and experiments were conducted in accordance with the approved protocol of the Institutional Animal Care and Use Committee at Albert Einstein College of Medicine and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. The BNIP3L/Nix mice (Diwan et al., 2007) were kindly provided by Prof. Gerald W. Dorn, II (Washington University, St. Louis). BNIP3L mice were maintained on a mixed 129/C57BL/6 genetic background. Both male and female animals were used in this study. Mice were reared in a barrier-controlled facility (20°C; 12-h light/dark cycle) with ad libitum access to food and water. Heterozygous animals were bred and pups euthanized by decapitation. Lenses were removed under a dissecting microscope and immediately flash frozen and stored at −80°C for protein isolation. For sectioning, heads (P1) or whole eyes (P14) were fixed overnight in 4% PFA and subsequently cryoprotected in 15% sucrose. Tissues were embedded in Optimal Cutting Temperature (OCT) tissue freezing medium (Triangle Biomedical Sciences, Durham, NC) for cryo- sectioning. For genotyping two sets of primers were used (Primer 1: 5′-CAC AGC ATT GCC ACC CCT GCA GAG-3′, Primer 2: 5′-GCT GCA GAT GCC GGG CCT GAG CAA-3′, and primer 3: 5′-ACC TGC CCA TGC TCC AGA GCA GGC-3′). Primers 1 and 2 give a 375 bp product indicating the presence of a WT allele and Primers 1 and 3 give a 210 bp product indicating the presence of a KO allele.
Immunohistochemistry
Mid-sagittal lens sections were prepared from OCT embedded mouse lenses by cryosectioning (20-μm thick sections) on a Leica Crysostat. Sections were permeabilized using 0.25% Triton X-100 (catalog no. T1001, Anatrace, Santa Clara, CA) in PBS for 10 min at room temperature, followed by three washes in PBS. Nonspecific antibody binding was blocked using 2.5% horse serum (catalog no. S-2012, Vector Laboratories, Burlingame, CA) for 1 h at room temperature. Primary antibodies were diluted in 1% bovine serum albumin, 0.25% Triton X-100 in PBS, and the sections were incubated overnight at 4 °C. Sections were washed three times in PBS and incubated with fluorescently conjugated anti-mouse and/or anti-rabbit secondary antibodies (1:2000, Alexa-Fluor 488 or 555, catalog nos. A11001, A11008, A21422, and A21428, Invitrogen) diluted in 1% BSA, 0.25% Triton X-100, PBS, for 1 h at room temperature. Primary antibodies used were: TOMM20 (catalog no. sc17764, Santa Cruz Biotechnology); Grp78/Bip (catalog no. G8918, Sigma); FTCD (Catalog no. G2404, Sigma-Aldrich); Calregulin (sc166837, Santa Cruz), BNIP3L (catalog no. ADI-905-185-100, Enzo Scientific), or mouse BNIP3L (catalog no. sc166332, Santa Cruz Biotechnology). Images of mouse lens sections were obtained using a Zeiss LSM700 confocal microscope.
Analysis of BNIP3L gene expression in wild-type mouse lenses
BNIP3L transcript levels were evaluated in RNA isolated from 30–50 pooled P1 wild-type mouse lens epithelial or fiber cell preparations. BNIP3L transcript levels were measured by semi-quantitative RT-PCR using the SuperScript® III one-step RT-PCR system with Platinum Taq polymerase (Invitrogen) according to the manufacturer’s instructions. Total RNA was purified from epi or fiber using TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. 100 ng of total RNA was assayed following isolation from epi or fiber. BNIP3L transcripts were amplified for 25 PCR cycles with a 60 °C annealing temperature and the primer sequences: Forward primer – GGAGCGGAGTCCAGGTTTGT and reverse primer – CAACTCCACCCAGGAACTGTTGA. BNIP3L protein levels in epi and fiber protein were also evaluated by western blot analysis (described below) using a BNIP3L-specific antibody (Cat no: ADI-905-185-100, Enzo Life Sciences Farmingdale, NY).
Protein isolation and Western blots analysis
Total lens protein was prepared by homogenizing lenses in Nonidet P-40 lysis buffer supplemented with a protease inhibitor cocktail (catalog no. P8340, Sigma) followed by centrifugation at 10,000 rpm for 5 minutes. Levels of organelle markers were analyzed using SDS-PAGE and Western blotting. Briefly, equal amounts of protein were electrophoresed in 4–20% pre cast Mini-Protean TGX™ gradient gels (Bio-Rad) gels in the presence of running buffer containing 10% SDS using a Bio-Rad Mini Protean II system. Proteins were electroblotted onto polyvinylidine difluoride membranes (Tran-Blot® Turbo™ Transfer Packs, Bio-Rad) using the Bio-Rad Tran-Blot® Turbo™ transfer system following the manufacturer’s instructions. Membranes were blocked in either 5% nonfat dried milk in TBS-T or 5% BSA in TBS-T or Rockland block (catalog no. MB-070) for 1 hour at room temperature, followed by incubation with a primary antibody diluted in appropriate blocking solution overnight at 4 °C. After three washes in TBS-T, membranes were incubated with the secondary HRP-conjugated antibody for 1 hour. Following three washes with TBS-T and one wash in PBS, membranes were incubated for 5 minutes in Enhanced ChemiLuminescence™ Prime western blotting detection reagent (ECL; GE Lifesciences). Western blots were visualized using a LI-COR Odyssey (LI-COR Biosciences, Lincoln, NE). Densitometry analysis was performed using the Image Studio software on the LI-COR system. Western blots and bar graphs shown are triplicate technical repeats of pooled protein prepared from P1 or P14 lenses. Three separate pools were examined and the triplicate repeat for one set of pooled lenses for each time point is displayed. Primary antibodies used were: TOMM20 (catalog no. SC- 11415, Santa Cruz Biotechnology); Erp72 (catalog no. D70D12, Cell Signaling Technology); FTCD (Catalog no. G2404, Sigma-Aldrich); BNIP3 (Catalog no. AB2999, Milipore); BFSP2/CP49 (Catalog no. 346–360, Sigma-Aldrich); BFSP1/Filensin (Catalog no. 70R-4876, Fitzgerald antibodies). Equal loading was determined by stripping blots and probing for standard equal loading markers GAPDH (catalog no. SC-25778, Santa Cruz Biotechnology), β-Actin (catalog no. A5441, Sigma) or α-Tubulin (Catalog no. ab18251, Abcam).
Isolation of mitochondria, endoplasmic reticulum and Golgi apparatus from mouse liver
Mouse liver was removed from equal numbers of 3 week old male and female C57BL/6J mice following euthanization by decapitation. Mitochondria were isolated from freshly obtained mouse liver using a Mitochondrial isolation kit (Abcam, catalog no.ab110168) following the manufacturer’s instructions. Briefly, 0.53 g of liver tissue was washed in 1.5 ml of wash buffer supplied in the kit. The tissue was homogenized in a Dounce homogenizer in 2 mls of isolation buffer, following homogenization equal volumes of isolation buffer were added and the preparation centrifuged at 1000 g for 10 minutes at 4 °C. Equal volumes of isolation buffer were added to the supernatant and centrifuged at 12,000 g for 15 minutes at 4 °C. The pellet was washed in 1 ml of isolation buffer and centrifuged at 12,000 g for 15 minutes at 4 °C, following a repeat wash and centrifuge, pellets were combined and re-suspended in 500 μl of isolation buffer and stored at −20 C. Golgi apparatus was isolated using a Golgi Isolation kit (Sigma, GL0010) following the manufacturer’s instructions. Briefly, 2.5 g of tissue was washed in ice cold PBS followed by 2 washes in 0.25 M sucrose. The tissue was homogenized in equal volumes of 0.25 M sucrose and centrifuged at 3000 g for 15 mins at 4 °C. The supernatant adjusted to 1.25 M sucrose and a layered sucrose gradient prepared. Gradients were centrifuged at 120,000 g for 3 hours at 4 °C. The Golgi layer was removed and stored at −20 C. Endoplasmic reticulum isolated using an ER Isolation kit (Sigma, ER0100) following the manufacturer’s instructions. Briefly, 1.81 g of freshly isolated liver was washed twice in ice cold PBS. Liver was cut into small pieces and added to 3.5X volumes of isotonic buffer in a Dounce homogenizer. Homogenized tissue was centrifuged at 1000 g for 10 minutes at 4 °C. The supernatant was centrifuged at 12,000 g for 15 minutes at 4 °C. The supernatant was then subjected precipitation in 7.5 X volume of 8 mM calcium chloride (CaCl) for 15 minutes. Following CaCl precipitation, the mix was centrifuged at 8000 g for 10 minutes at 4 °C. The pellet corresponding to rough ER enriched microsomes was removed and re-suspended in 540 μl of 1X isotonic extraction buffer and stored at −20 °C.
Results
BNIP3L levels peak in the equatorial region of the wild-type P1 mouse lens
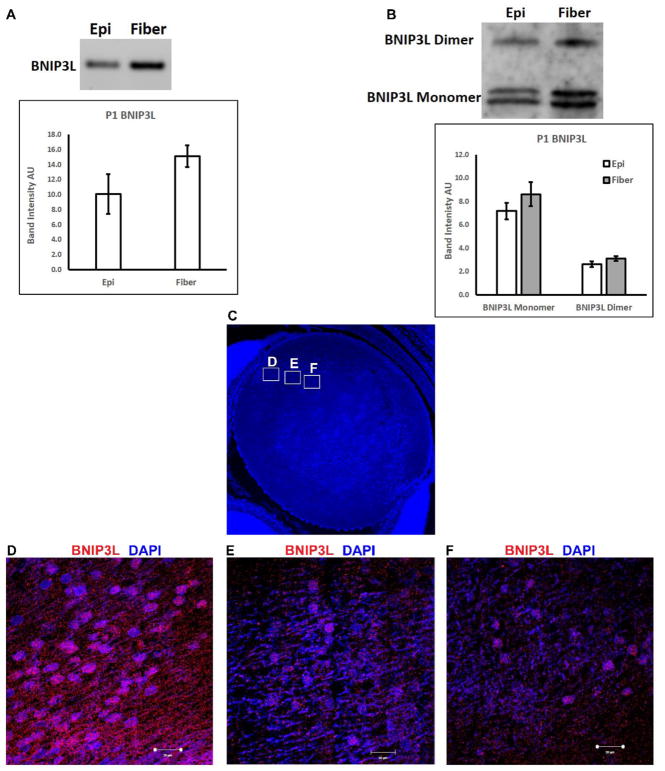
Our previous studies showed that in the embryonic chick lens BNIP3L protein levels peak in the equatorial region of the lens where mitochondria and other organelles are just beginning to be eliminated during lens OFZ formation (Chauss et al., 2014) and we detected increased levels of BNIP3L protein in human lens fiber cells relative to epithelial cells (Brennan et al., 2012). Other studies have demonstrated a requirement for BNIP3L in elimination of mitochondria during erythrocyte formation (Novak et al., 2010; Sandoval et al., 2008; Schweers et al., 2007; Zhang and Ney, 2008). Based on its spatial lens expression pattern and established function for eliminating mitochondria during erythrocyte formation, we hypothesized that BNIP3L might have a similar function for eliminating mitochondria during lens OFZ formation. To examine this possible new role for BNIP3L, we first sought to confirm that the expression pattern of BNIP3L in the newborn mouse lens paralleled the expression pattern of BNIP3L we previously established for embryonic chick and human lenses. Semi-quantitative RT-PCR using equal amounts of RNA isolated from wild-type P1 mouse lenses microdissected into lens epithelial (epi) and fiber cell (fiber) populations revealed that, consistent with both the human and chick lens studies, BNIP3L mRNA is detected at higher levels in lens fiber cells relative to lens epithelial cells (Fig. 1A). Analysis of corresponding BNIP3L protein levels by western blotting using equal amounts of protein obtained from mouse P1 lens epithelial or fiber cells also revealed increased levels of BNIP3L protein in lens fiber cells relative to lens epithelial cells (Fig. 1B). Finally, to further pinpoint the spatial distribution profile of BNIP3L in the mouse lens, fluorescent immunohistochemical staining of BNIP3L in mid-sagittal P1 mouse lens sections revealed that BNIP3L levels peak in the equatorial region or transition zone of the lens and decrease in regions of the lens where mitochondria and other organelles are being eliminated (Fig. 1C–F).
Figure 1. Spatial expression pattern of BNIP3L in wild-type P1 mouse lenses.
A. Ethidium bromide stained agarose gel showing BNIP3L transcript levels in 100 ng of RNA isolated from pooled epithelial or fiber cells (30–50 lenses) microdissected from wild-type P1 mouse lenses. Densitometric analyses of triplicate reactions. B. Immunoblot analysis of BNIP3L protein levels in 10 μg of total protein extracted from pooled epithelial or fiber cells (30–50 lenses) microdissected from wild-type P1 mouse lenses. Densitometric analyses of triplicate reactions. C. Mid-sagittal lens sections from wild-type P1 mice immunostained, for the BNIP3L (red) and co-stained with nuclear stain DAPI (blue). Low magnification images for P1 were obtained using the 10X objective, the scale bar is 200 μm. D-F High magnification images were taken from three regions of the lens to examine spatial distribution of BNIP3L in the fiber cells of the lens, images were obtained using the 40X objective, scale bar 10 μm.
Mitochondria are retained in the center of P1 and P14 BNIP3L-knockout mouse lenses
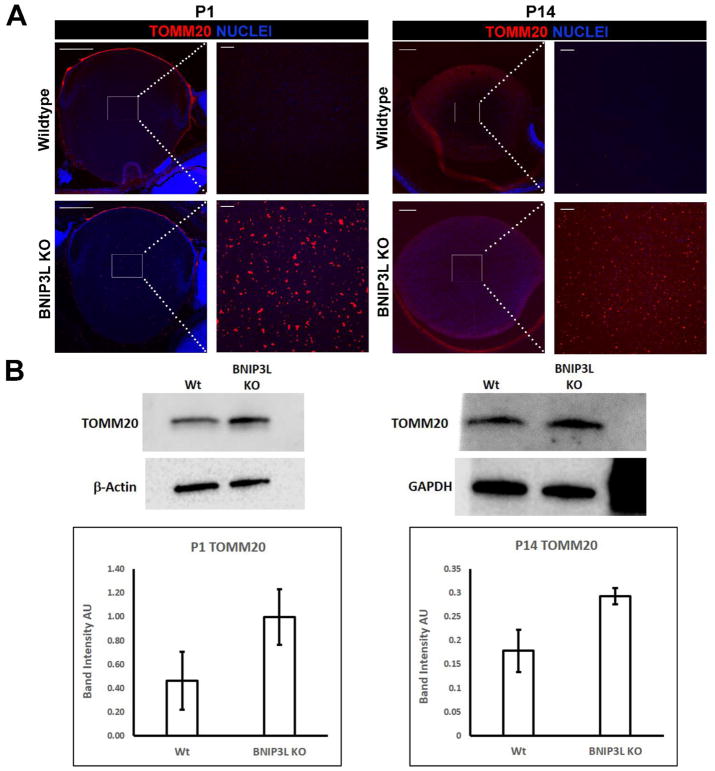
Based on the lens expression pattern of BNIP3L (Fig. 1) and its known function in the elimination of mitochondria in erythrocytes (Novak et al., 2010; Sandoval et al., 2008; Schweers et al., 2007; Zhang and Ney, 2008), we sought to establish if BNIP3L was required for elimination of mitochondria during lens OFZ formation. To establish this potential requirement for BNIP3L, we compared the mitochondrial levels between central regions of wild-type and BNIP3L−/− newborn (P1) and two week old (P14) mouse lenses by monitoring the levels of the mitochondrial marker, translocase of the outer mitochondrial membrane 20 (TOMM20). In wild-type mice, central lens fiber cells have eliminated their mitochondria and other organelles to fully form the OFZ by birth (Bassnett et al., 2011; Wride, 2011). As expected, analysis of wild-type mid-sagittal lens sections by immunohistochemistry with a TOMM20-specific antibody revealed the absence of TOMM20 in the central regions of both P1 and P14 wild-type mouse lenses suggesting the normal elimination of mitochondria during OFZ formation (Fig. 2A). By contrast, identical analysis of P1 and P14 BNIP3L−/− mouse lenses revealed the persistence of TOMM20 in the center of lens suggesting a requirement for BNIP3L in the elimination of mitochondria during lens OFZ formation (Fig. 2A). To further explore the potential role of BNIP3L in elimination of mitochondria during lens OFZ formation, western blot analysis was conducted to determine the relative levels of mitochondria between wild-type and BNIP3L−/− mouse lenses using the same TOMM20-specific antibody. The results reveal higher levels of TOMM20 in whole lens protein extracts prepared from P1 and P14 BNIP3L−/− mice relative to whole lens protein extracts prepared from age-matched wild-type mice (Fig. 2B). TOMM20 levels were 2-fold higher in the P1 BNIP3L−/− mouse lenses relative to the P1 wild-type lenses and 1.6-fold higher in the P14 BNIP3L−/− lenses relative to the P14 wild-type mouse lenses. These data collectively provide evidence that BNIP3L is required for the elimination of mitochondria during formation of the mouse lens OFZ. By contrast, the pattern of nuclear staining in the central regions of the P1 and P14 wild-type and BNIP3L−/− lenses detected by DAPI staining was similar (Fig. 2A) suggesting that elimination of nuclei during lens OFZ formation is BNIP3L-independent. These data provide evidence that BNIP3L is a novel requirement for elimination of mitochondria during lens OFZ formation.
Figure 2. Mitochondria are retained in the lenses of BNIP3L knockout mice.
A. Mid-sagittal lens sections from P1 and P14, wild-type and BNIP3L KO mice immunostained, for the mitochondrial marker TOMM20 (red) and co-stained with nuclear stain DAPI (blue). Low magnification images for P1 were obtained using the 10X objective, for P14 the 5X objective was used, the scale bar for both is 200 μm. High magnification images for both P1 and P14 were taken from the center or organelle free zone of the lens and obtained using the 40X objective, scale bar 20 μm. B. Immunoblot analysis of TOMM20 protein levels in 15 μg of total protein extract isolated from P1 and P14, wild-type and BNIP3L KO lenses. Also shown are immunoblots for β-actin (P1) and GAPDH (P14) as controls for equal protein loading. Densitometric analyses of the immunoblots was plotted as TOMM20 levels relative to its loading control. Samples were run in triplicate and displayed as mean software determined arbitrary unit (AU) ± the standard deviation.
Endoplasmic reticulum (ER) and Golgi apparatus (GA) are retained in the center of P1 and P14 BNIP3L-knockout mouse lenses
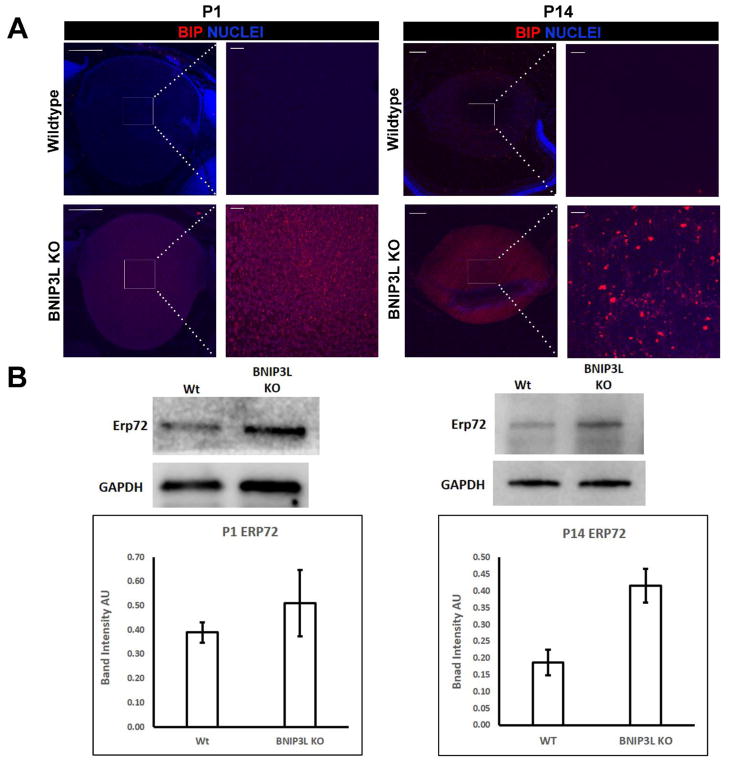
The precise timing of organelle elimination during OFZ formation indicates the possibility that a common regulator could target multiple organelles for elimination during OFZ formation. Consistent with the requirement for BNIP3L in elimination of mitochondria during OFZ formation (Fig. 2) BNIP3L has been previously localized to the mitochondria in multiple cell types (Zhang and Ney, 2011). BNIP3L has also been localized to the ER in non-lens cells (Diwan et al., 2009). Based on the BNIP3L localizing to both mitochondria and ER in non-lens cells, we hypothesized that BNIP3L might be required for elimination of ER, GA and other organelles during OFZ formation in addition to being required for mitochondria elimination (Fig. 2). To evaluate this possibility, we compared the levels of ER and GA in the centers of P1 and P14 BNIP3L−/− and wild-type mice using ER- and GA-specific protein markers. To evaluate the presence or absence of ER in the BNIP3L−/− and wild-type lenses, mid-sagittal lens sections were immunostained with Glucose-Regulated Protein, 78kDa (GRP78/BiP)-specific antibody and co-stained with nuclear stain DAPI. As expected, GRP78/BiP was absent from the central region of lenses from wild-type P1 and P14 mice confirming that the ER had been eliminated by the time of birth (Fig. 3A). Surprisingly, examination of the GRP78/BiP-immunostained lens sections from BNIP3L−/− mice revealed the persistence of GRP78/BiP staining throughout the central region of the lenses of both P1 and P14 mice (Fig. 3A). As a confirmation, western blot analysis using another ER-specific marker called Endoplasmic Reticulum Resident Protein 72 (ERP72) and protein isolates from P1 and P14 wild-type and BNIP3L−/− lenses confirmed higher levels of ERP72 in whole lens protein from P1 and P14 BNIP3L−/− mice relative to wild type mice (Fig. 3B). In the P1 BNIP3L−/− mouse lenses, ERP72 levels were 1.3 fold those detected in wild-type lenses while in the P14 mice the levels of ERP72 were 2.2 fold those detected in wild-type lenses. These data provide evidence that BNIP3L is a novel requirement for elimination of ER during lens OFZ formation.
Figure 3. Endoplasmic reticulum is retained in the lenses of BNIP3L knockout mice.
A. Mid-sagittal lens sections from P1 and P14, wild-type and BNIP3L KO mice immunostained, for the endoplasmic reticulum marker BiP/GRP78/HSPA5 (red) and co-stained with nuclear stain DAPI (blue). Low magnification images for P1 were obtained using the 10X objective, for P14 the 5X objective was used, the scale bar for both is 200 μm. High magnification images for both P1 and P14 were taken from the center or organelle free zone of the lens and obtained using the 40X objective, scale bar 20 μm. B. Immunoblot analysis of endoplasmic reticulum marker ERP72 protein levels in 15 μg of total protein extract isolated from P1 and P14, wild-type and BNIP3L KO lenses. Also shown are immunoblots for GAPDH as control for equal protein loading. Densitometric analyses of the immunoblots was plotted as ERP72 levels relative to GAPDH loading control. Samples were run in triplicate and displayed as mean software determined arbitrary unit (AU) ± the standard deviation.
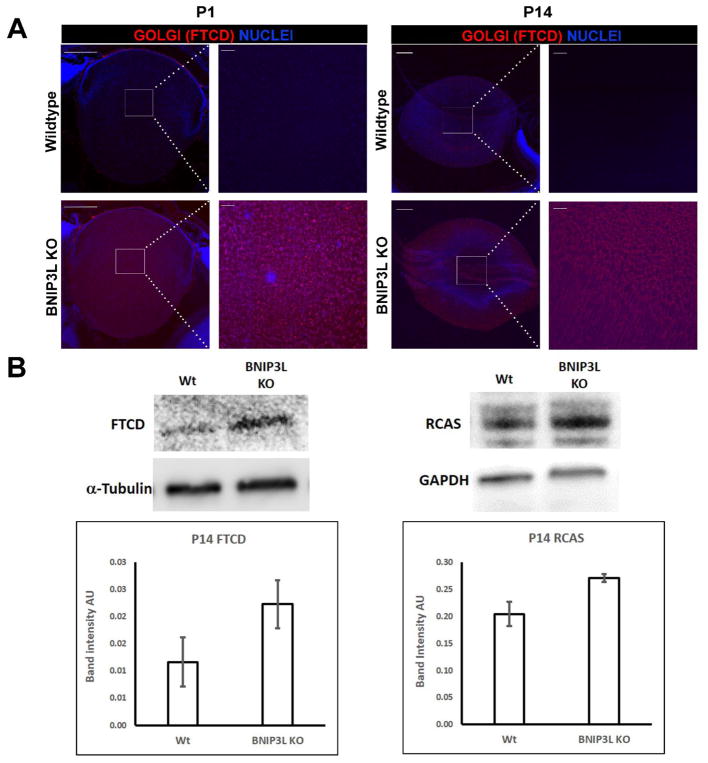
In addition to examining the role of BNIP3L in the elimination of ER during lens OFZ formation, we also explored the possible requirement for BNIP3L in the elimination of GA during lens OFZ formation by examining the levels of the GA-specific protein formimidoyltransferase cyclodeaminase (FTCD) in P1 and P14 wild-type and BNIP3L−/− midsagittal mouse lens sections. As expected, analysis of the FTCD-immunostained mid-sagittal lens sections from P1 and P14 wild-type and BNIP3L−/− mice revealed the elimination of FTCD in the centers of wild-type lenses (Fig. 4A). Surprisingly, significant levels of FTCD were detected in the central regions of both P1 and P14 BNIP3L−/− mice (Fig. 4A) suggesting a requirement for BNIP3L in the elimination of GA during lens OFZ formation. Since levels of GA were difficult to detect in protein isolates from the P1 mouse lenses, we examined the levels of two Golgi markers in protein isolates from P14 wild-type and P14 BNIP3L−/− mouse lenses (Fig. 4A). Western blot analysis of the GA-specific markers FTCD and receptor binding cancer antigen expressed on SiSo cells (RCAS1) revealed increased levels of both markers (1.9 fold and 1.3 fold respectively) in the P14 BNIP3L−/− mouse lens protein isolates relative to wild-type mouse lens protein isolates. Collectively, these data provide evidence that BNIP3L is a novel requirement for the elimination of mitochondria (Fig. 2) ER (Fig. 3) and GA (Fig. 4) during lens OFZ formation and that elimination of nuclei during OFZ formation is BNIP3L-independent.
Figure 4. Golgi apparatus is retained in the lenses of BNIP3L knockout mice.
A. Mid-sagittal lens sections from P1 and P14, wild-type and BNIP3L KO mice immunostained, for the Golgi apparatus marker FTCD (red) and co-stained with nuclear stain DAPI (blue). Low magnification images for P1 were obtained using the 10X objective, for P14 the 5X objective was used, the scale bar for both is 200 μm. High magnification images for both P1 and P14 were taken from the center or organelle free zone of the lens and obtained using the 40X objective, scale bar 20 μm. B. Immunoblot analysis of two Golgi apparatus markers FTCD and RCAS protein levels in 15 μg of total protein extract isolated from P14 wild-type and BNIP3L KO lenses. Also shown are immunoblots for Tubulin (FTCD) and GAPDH (RCAS) as controls for equal protein loading. Densitometric analyses of the immunoblots was plotted as FTCD or RCAS levels relative to their loading controls. Samples were run in triplicate and displayed as mean software determined arbitrary unit (AU) ± the standard deviation.
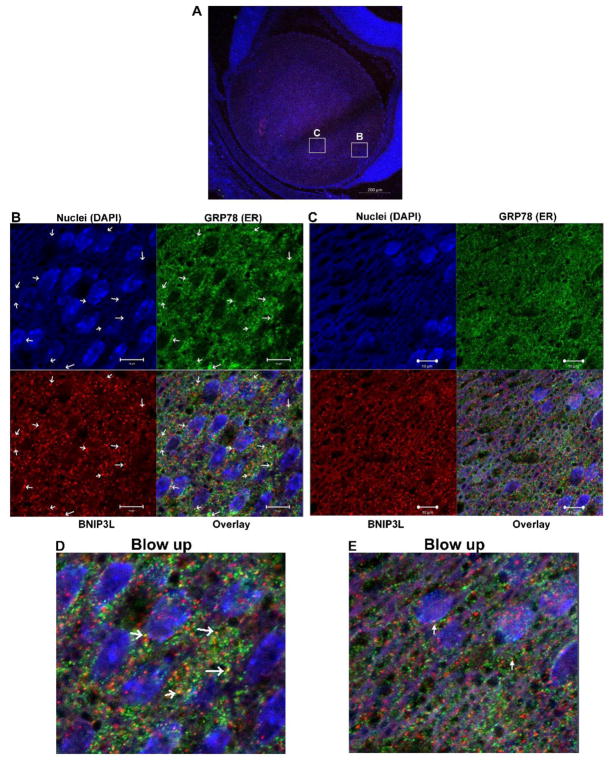
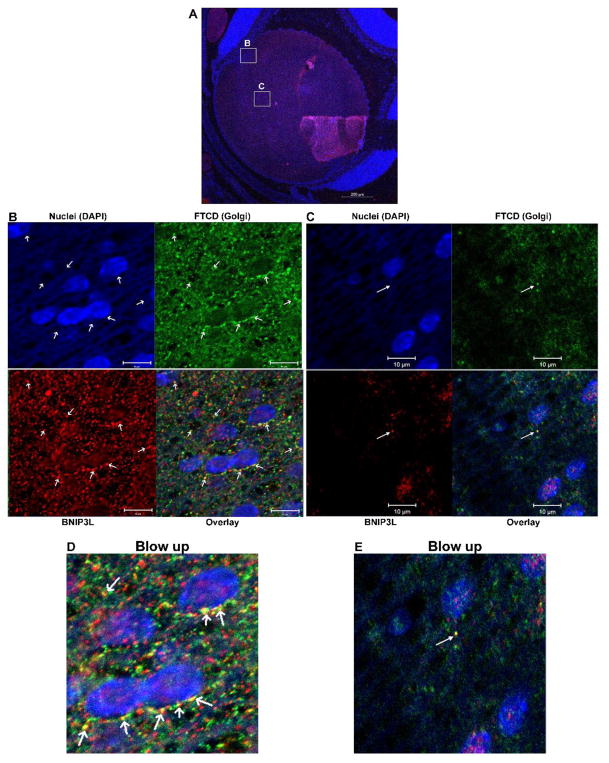
BNIP3L co-localizes with endoplasmic reticulum (ER) and Golgi apparatus (GA) in the lens
BNIP3L was first identified as an outer mitochondrial membrane protein (Aerbajinai et al., 2003) and previous work has shown that BNIP3L co-localizes with the ER in heart tissue (Diwan et al., 2009). Since this study is the first to demonstrate a requirement for BNIP3L in the degradation of ER and GA, we sought to establish if direct binding of BNIP3L to lens ER and GA is associated with the BNIP3L-dependent elimination of these organelles during lens OFZ formation. To determine if BNIP3L directly interacts with these organelles we co-localized BNIP3L with ER- and GA-specific markers in P1 wild-type mouse lenses. Mid-sagittal lens sections from P1 wild-type mice were immunostained using an antibody specific for the ER protein GRP78/BiP (Fig. 5, green) or an antibody specific for the GA marker FTCD (Fig. 6, green). Sections were co-stained with BNIP3L (red) and nuclear stain DAPI (blue). Examination of the immunostained lens sections revealed that BNIP3L co-localizes with the ER marker GRP78 (Fig. 5B) and the GA marker FTCD (Fig. 6B) in the equatorial or transition zone of the lens, as indicated by distinct yellow puncta in the overlay images. Examination of co-localization in the area closer to the OFZ (Figs. 5C and 6C) revealed decreased levels of BNIP3L and more diffuse GRP78/BiP and FTCD staining as the organelles are degraded and BNIP3L is turned over or degraded with the organelle. These data, and the data shown in Fig. 1 demonstrating peak levels of BNIP3L in the equatorial region or transition zone of the lens, suggests that BNIP3L co-localizes with ER and GA prior to elimination of these organelles, possibly to prime them for degradation. Collectively, these data provide evidence that the BNIP3L-mediated elimination of ER and GA during lens OFZ formation is associated with the binding of BNIP3L to these organelles in the equatorial or transition zone of the lens. The data also suggests that BNIP3L could coordinate the elimination of these organelles during lens OFZ formation by specifically targeting them for elimination in the early stages of fiber cell differentiation.
Figure 5. BNIP3L co-localizes with endoplasmic reticulum in the lens.
A. Mid-sagittal lens sections from P1 wild-type mice immunostained for the endoplasmic reticulum marker BiP/GRP78/HSPA5 (green), BNIP3L (red) and co-stained with nuclear stain DAPI (blue). A. Low magnification images for P1 were obtained using the 10X objective and the scale bar is 100 μm. B. High magnification image taken in the equatorial region of the lens. This image was obtained using the 40X objective and scale bar is 20 μm. C. Zoomed area of image B. (area indicated by box), arrows indicate co-localization of ER/GRP78 (green) and BNIP3L (red) resulting in distinct yellow puncta in the overlay image, scale bar 10 μm and D. A blow up of the overlay from C.
Figure 6. BNIP3L co-localizes with Golgi apparatus in the lens.
A. Mid-sagittal lens sections from P1 wild-type mice immunostained for the Golgi apparatus marker FTCD (green), BNIP3L (red) and co-stained with nuclear stain DAPI (blue). A. Low magnification images for P1 were obtained using the 10X objective and the scale bar is 100 μm. B. High magnification Image taken in the equatorial region of the lens. This image was obtained using the 40X objective and scale bar is 20 μm. C. Zoomed area of B. (area indicated by box), arrows indicate co-localization of Golgi/FTCD (green) and BNIP3L (red) resulting in distinct yellow puncta in the overlay image, scale bar 10 μm scale bar 5 μm and D. A blow up of the overlay from C.
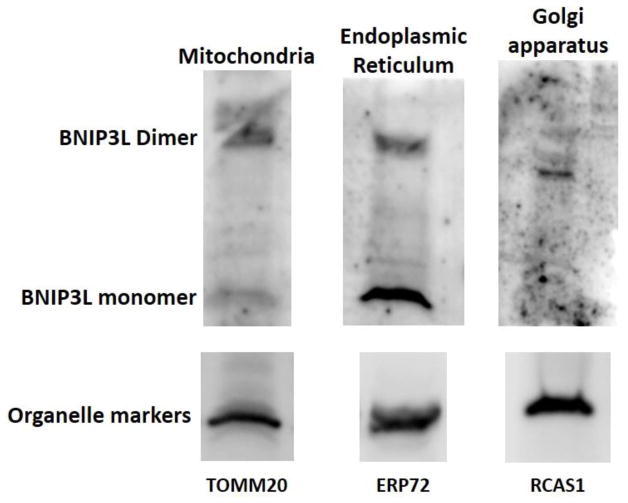
BNIP3L localizes to mitochondria, endoplasmic reticulum (ER) and Golgi apparatus (GA) of mouse liver
Since this study is the first to establish the binding of BNIP3L to GA we next sought to establish if binding of BNIP3L to these organelles might extend to non-lens tissues by evaluating the BNIP3L levels in purified mitochondria, ER and GA populations isolated from mouse liver. The liver is a good choice for these studies as it provides the large amount of tissue required to obtain sufficient amounts of mitochondria, ER and GA for purification. Western blot analysis of purified mitochondria, ER and GA fractions were probed with TOMM20-, ERP72- and RCAS1-specific antibodies as specific markers for these organelles, respectively. Immunoblotting for BNIP3L in each of these purified organelle fractions confirmed the association of BNIP3L with mitochondria, ER, and GA (Fig. 7). These data provide evidence that BNIP3L-localization to these organelles could extend beyond the lens and suggests that BNIP3L may play an important role in regulating populations of these organelles in non-lens cells.
Figure 7. BNIP3L is localized to the mitochondria, endoplasmic reticulum and Golgi apparatus in mouse liver.
Immunoblot analysis of BNIP3L protein levels in 50 μg of mitochondria, endoplasmic reticulum or Golgi apparatus isolated from wild-type mouse liver. Also shown are immunoblots for mitochondrial marker TOMM20, endoplasmic reticulum maker ERP72 and Golgi apparatus marker RCAS1 as controls for purity of organelle isolates.
Discussion
Removal of cellular organelles is a key step in the remodeling program leading to the terminal differentiation of lens fiber cells and the formation of the lens OFZ (Bassnett, 1992; Bassnett, 1995; Bassnett and Beebe, 1992; Bassnett and Mataic, 1997; Chaffee et al., 2014). Since failure to remove organelles and form the OFZ is associated with cataract formation (Menko, 2002; Nishimoto et al., 2003; Pendergrass et al., 2005; Wride, 2011), identification of the requirements and mechanisms governing the elimination of organelles during lens OFZ formation has been a long-sought goal of the lens field. Recent advances in our understanding of OFZ formation include a study that identified a key upstream pathway governing the elimination of organelles during lens OFZ formation through JNK-mediated regulation of mTORC1 signaling and activation of the autophagy machinery (Basu et al., 2014). Consistent with a critical role for the autophagy machinery in lens fiber cell function, another recent study demonstrated that mutation of the autophagosomal transport protein FYCO1 results in congenital nuclear cataract formation (Chen et al., 2011). Also consistent with an important role for the autophagy machinery in the elimination of lens organelles, another study detected increased expression of multiple key autophagy genes in lens fiber cells undergoing organelle-elimination (Brennan et al., 2012) and mitochondria contained within autophagolysosomes were detected in lens fiber cells also during organelle elimination (Costello et al., 2013).
To date, no studies have identified the specific requirements for elimination of mitochondria, ER and GA during lens OFZ formation and in the present report, we provide evidence that deletion of BNIP3L results in the retention of mitochondria, ER and GA in mouse lenses identifying BNIP3L as a novel requirement for elimination of these organelles during OFZ formation. These data also suggest a novel function for BNIP3L in the regulation of ER and GA populations in the lens that may also extend in non-lens tissues. The data are consistent with previous studies showing that BNIP3L levels spike in lens fiber cells where organelle-elimination is just being initiated (Chauss et al., 2014); that BNIP3L is induced in multiple cell-types under specific physiological or pathological conditions (Diwan et al., 2009), and that BNIP3L functions to eliminate mitochondria during reticulocyte differentiation (Novak et al., 2010; Sandoval et al., 2008; Schweers et al., 2007; Zhang and Ney, 2009). The data also suggest that nuclear elimination during lens OFZ formation is BNIP3L-independent since elimination of nuclei during OFZ formation was not impacted by BNIP3L-deletion. Conversely, inactivation of ATP-dependent chromatin remodeling enzyme Snf2h (Smarca5) disrupts nuclear degradation while mitochondria and ER are normally eliminated (He et al., 2016).
The data also demonstrate that BNIP3L co-localizes with the mitochondria, ER and GA in lens fiber cells that are actively eliminating their organelles suggesting that binding of BNIP3L to these organelles could initiate their elimination through recruitment of the autophagy and/or other degradation machinery. These results are consistent with studies identifying BNIP3L as an outer mitochondria membrane protein (Aerbajinai et al., 2003) and studies co-localizing BNIP3L to the ER in HEK293, rat ventricular myocytes and in heart tissue (Diwan et al., 2009). To our knowledge, this is the first study to localize BNIP3L to the GA or to suggest a role for BNIP3L in regulation of ER and GA degradation. Our data suggests that the localization of BNIP3L to the ER and GA extends to non-lens tissues since we detected BNIP3L in ER and GA isolated from mouse liver (Fig. 7). These data suggest a potential role for BNIP3L in regulating the populations of these organelles in non-lens tissues where BNIP3L is expressed. Consistently, BNIP3L has been shown to be induced in multiple tissues under various pathological conditions (Diwan et al., 2009). These data are the first to demonstrate binding of BNIP3L to the GA in any tissue suggesting a novel function for BNIP3L in regulating GA populations.
Although further studies are required to determine the exact mechanism(s) for BNIP3L-dependent elimination of mitochondria, ER and GA during lens OFZ formation, we believe that BNIP3L likely recruits the autophagy machinery for elimination of mitochondria, ER and GA during OFZ formation. This is consistent our previous study that identified significant numbers of autophagolysosomes containing mitochondria in lens fiber cells undergoing organelle elimination (Costello et al., 2013). BNIP3L contains a WXXL-like motif facing the cytosol that binds the autophagosome proteins MAP1LC3 (LC3B), GABARAPL1 and GABARAPL2 (GATE16) to directly recruit autophagosomes for the elimination of mitochondria in other tissues (Ashrafi and Schwarz, 2013; Randow and Youle, 2014; Youle and Narendra, 2011). Previous work has determined that BNIP3L has weak affinity for LC3B but a much higher affinity for its homologues GABARAPL1 and GATE16 (Novak et al., 2010) suggesting that these proteins may participate in the BNIP3L-mediated elimination of lens fiber cell mitochondria, ER and GA. Other studies have shown that mitophagy is primarily (but not only) due to alternative autophagy since knockdown of RAB9A and RAB9B suppressed mitophagy to a greater degree than knockdown of ATG5/7 in hypoxia treated HeLa cells (Hirota et al., 2015). Interestingly our studies on up-regulation of autophagy molecules during fiber cell remodeling revealed that both ATG5/7 and Ras-related protein Rab-9A were up-regulated in fiber cells suggesting that multiple autophagy pathways operate during organelle elimination (Brennan et al., 2012).
It is possible that other proteins in addition to BNIP3L function to facilitate the elimination of mitochondria, ER and GA during OFZ formation. Consistently, we have shown that expression of the mitophagy proteins PINK1 and Parkin, that like BNIP3L, target mitochondria for elimination persists throughout the process of mitochondria elimination during the remodeling of lens fiber cells (Chauss et al., 2014) and we have previously established that the PINK1/Parkin pathway operates to degrade damaged mitochondria in cultured lens epithelial cells (Brennan et al., 2017). PINK1/Parkin degradation of mitochondria is dependent on loss of mitochondrial membrane potential (MMP) and, consistent with a potential cooperative role for the PINK1/Parkin pathway in the BNIP3L-dependent elimination of mitochondria during lens OFZ formation, a previous study demonstrated that mitochondria in nascent lens fiber cells exhibit decreased MMP just prior to their elimination during lens fiber cell remodeling (Weber and Menko, 2005). Other studies have suggested that BNIP3L depolarizes mitochondria (Zhang and Ney, 2008) and BNIP3L-mediated mitochondrial depolarization in the lens could recruit PARKIN to target mitochondria for elimination during OFZ formation. Consistent with a possible role for the PINK1/PARKIN pathway in the BNIP3L-dependent elimination of mitochondria during lens fiber cell remodeling, another study found that following cerebral ischemia-reperfusion injury both BNIP3L and the PINK1/Parkin pathways are simultaneously activated to cooperatively eliminate damaged mitochondria (Yuan et al., 2017).
In summary, the present data provide evidence that BNIP3L is a novel requirement for the specific elimination of mitochondria, ER and GA during lens fiber cell remodeling and they suggest that regulation of ER and GA populations may be a novel function for BNIP3L that may extend to non-lens tissues. Future studies will seek to identify the pathways and mechanism required for the BNIP3L-dependent elimination of these organelles during lens OFZ formation, to identify additional proteins required for elimination of mitochondria, ER and GA during OFZ formation, and to identify the effects of retaining these organelles upon deletion of BNIP3L on lens fiber cell homeostasis and transparency.
Supplementary Material
Highlights.
Mitochondria, ER and Golgi apparatus are retained in the lenses of BNIP3L KO mice
BNIP3L localizes to the Mitochondria, ER and Golgi apparatus in the lens and liver
BNIP3L contributes toward organelle free zone formation in the developing lens.
Acknowledgments
We thank Dr. Gerald Dorn II MD for the gift of the BNIP3L-knockout mice used in these studies. We thank Jie Zhao for her help with the BNIP3L mouse colony. This work was supported by the NIH/NEI RO1 EY026478 (MK and ASM) and EY014237 (AC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aerbajinai W, Giattina M, Lee YT, Raffeld M, Miller JL. The proapoptotic factor Nix is coexpressed with Bcl-xL during terminal erythroid differentiation. Blood. 2003;102:712–717. doi: 10.1182/blood-2002-11-3324. [DOI] [PubMed] [Google Scholar]
- Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S. Mitochondrial dynamics in differentiating fiber cells of the mammalian lens. Curr Eye Res. 1992;11:1227–1232. doi: 10.3109/02713689208999548. [DOI] [PubMed] [Google Scholar]
- Bassnett S. The Fate of the Golgi Apparatus and the Endoplasmic Reticulum During Lens Fiber Cell Differentiation. Investigative Ophthalmology & Visual Science. 1995;36:1793–1803. [PubMed] [Google Scholar]
- Bassnett S, Beebe DC. Coincident Loss of Mitochondria and Nuclei During Lens Fiber Cell Differentiation. Development Dynamics. 1992;194:85–93. doi: 10.1002/aja.1001940202. [DOI] [PubMed] [Google Scholar]
- Bassnett S, Mataic D. Chromatin Degradation in Differentiating Fiber Cells of the Eye Lens. The Journal of Cell Biology. 1997;137:37–49. doi: 10.1083/jcb.137.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Shi Y, Vrensen GF. Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci. 2011;366:1250–1264. doi: 10.1098/rstb.2010.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Rajakaruna S, Reyes B, Van Bockstaele E, Menko AS. Suppression of MAPK/JNK-MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during terminal differentiation of lens fiber cells. Autophagy. 2014;10:1193–1211. doi: 10.4161/auto.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan L, Khoury J, Kantorow M. Parkin elimination of mitochondria is important for maintenance of lens epithelial cell ROS levels and survival upon oxidative stress exposure. Biochim Biophys Acta. 2017;1863:21–32. doi: 10.1016/j.bbadis.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan LA, Kantorow WL, Chauss D, McGreal R, He S, Mattucci L, Wei J, Riazuddin SA, Cvekl A, Hejtmancik JF, Kantorow M. Spatial expression patterns of autophagy genes in the eye lens and induction of autophagy in lens cells. Mol Vis. 2012;18:1773–1786. [PMC free article] [PubMed] [Google Scholar]
- Chaffee BR, Shang F, Chang ML, Clement TM, Eddy EM, Wagner BD, Nakahara M, Nagata S, Robinson ML, Taylor A. Nuclear removal during terminal lens fiber cell differentiation requires CDK1 activity: appropriating mitosis-related nuclear disassembly. Development. 2014;141:3388–3398. doi: 10.1242/dev.106005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauss D, Basu S, Rajakaruna S, Ma Z, Gau V, Anastas S, Brennan LA, Hejtmancik JF, Menko AS, Kantorow M. Differentiation state-specific mitochondrial dynamic regulatory networks are revealed by global transcriptional analysis of the developing chicken lens. G3 (Bethesda) 2014;4:1515–1527. doi: 10.1534/g3.114.012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ma Z, Jiao X, Fariss R, Kantorow WL, Kantorow M, Pras E, Frydman M, Pras E, Riazuddin S, Riazuddin SA, Hejtmancik JF. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am J Hum Genet. 2011;88:827–838. doi: 10.1016/j.ajhg.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello MJ, Brennan LA, Basu S, Chauss D, Mohamed A, Gilliland KO, Johnsen S, Menko AS, Kantorow M. Autophagy and mitophagy participate in ocular lens organelle degradation. Exp Eye Res. 2013;116:141–150. doi: 10.1016/j.exer.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maria A, Bassnett S. DNase IIbeta distribution and activity in the mouse lens. Invest Ophthalmol Vis Sci. 2007;48:5638–5646. doi: 10.1167/iovs.07-0782. [DOI] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan A, Koesters AG, Odley AM, Pushkaran S, Baines CP, Spike BT, Daria D, Jegga AG, Geiger H, Aronow BJ, Molkentin JD, Macleod KF, Kalfa TA, Dorn GW., 2nd Unrestrained erythroblast development in Nix−/− mice reveals a mechanism for apoptotic modulation of erythropoiesis. Proc Natl Acad Sci U S A. 2007;104:6794–6799. doi: 10.1073/pnas.0610666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan A, Matkovich SJ, Yuan Q, Zhao W, Yatani A, Brown JH, Molkentin JD, Kranias EG, Dorn GW., 2nd Endoplasmic reticulum-mitochondria crosstalk in NIX-mediated murine cell death. J Clin Invest. 2009;119:203–212. doi: 10.1172/JCI36445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW., 2nd Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Limi S, McGreal RS, Xie Q, Brennan LA, Kantorow WL, Kokavec J, Majumdar R, Hou H, Jr, Edelmann W, Liu W, Ashery-Padan R, Zavadil J, Kantorow M, Skoultchi AI, Stopka T, Cvekl A. Chromatin remodeling enzyme Snf2h regulates embryonic lens differentiation and denucleation. Development. 2016;143:1937–1947. doi: 10.1242/dev.135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y, Yamashita S, Kurihara Y, Jin X, Aihara M, Saigusa T, Kang D, Kanki T. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 2015;11:332–343. doi: 10.1080/15548627.2015.1023047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer C. The National Eye Institute’s low vision education program: improving quality of life. Ophthalmology. 2000;107:229–230. doi: 10.1016/s0161-6420(99)00094-9. [DOI] [PubMed] [Google Scholar]
- Lyu L, Whitcomb EA, Jiang S, Chang ML, Gu Y, Duncan MK, Cvekl A, Wang WL, Limi S, Reneker LW, Shang F, Du L, Taylor A. Unfolded-protein response-associated stabilization of p27(Cdkn1b) interferes with lens fiber cell denucleation, leading to cataract. FASEB J. 2016;30:1087–1095. doi: 10.1096/fj.15-278036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menko AS. Lens epithelial cell differentiation. Experimental Eye Research. 2002;75:485–490. doi: 10.1006/exer.2002.2057. [DOI] [PubMed] [Google Scholar]
- Nakahara M, Nagasaka A, Koike M, Uchida K, Kawane K, Uchiyama Y, Nagata S. Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 2007;274:3055–3064. doi: 10.1111/j.1742-4658.2007.05836.x. [DOI] [PubMed] [Google Scholar]
- Nishimoto S, Kawane K, Watanabe-Fukunaga R, Fukuyama H, Ohsawa Y, Uchiyama Y, Hashida N, Ohguro N, Tano Y, Morimoto T, Fukuda Y, SN Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003;424:1071–1074. doi: 10.1038/nature01895. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass W, Penn P, Possin D, Wolf N. Accumulation of DNA, nuclear and mitochondrial debris, and ROS at sites of age-related cortical cataract in mice. Invest Ophthalmol Vis Sci. 2005;46:4661–4670. doi: 10.1167/iovs.05-0808. [DOI] [PubMed] [Google Scholar]
- Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GF, Menko AS. The canonical intrinsic mitochondrial death pathway has a non-apoptotic role in signaling lens cell differentiation. J Biol Chem. 2005;280:22135–22145. doi: 10.1074/jbc.M414270200. [DOI] [PubMed] [Google Scholar]
- Wride MA. Lens fibre cell differentiation and organelle loss: many paths lead to clarity. Philos Trans R Soc Lond B Biol Sci. 2011;366:1219–1233. doi: 10.1098/rstb.2010.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zheng Y, Zhang X, Chen Y, Wu X, Wu J, Shen Z, Jiang L, Wang L, Yang W, Luo J, Qin Z, Hu W, Chen Z. BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy. 2017;13:1754–1766. doi: 10.1080/15548627.2017.1357792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ney PA. Nix induces mitochondrial autophagy in reticulcytes. Autophagy. 2008;4:354–356. doi: 10.4161/auto.5552. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ney PA. Mechanisms and biology of B-cell leukemia/lymphoma 2/adenovirus E1B interacting protein 3 and Nip-like protein X. Antioxid Redox Signal. 2011;14:1959–1969. doi: 10.1089/ars.2010.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.