Abstract
Early identification of younger, non-demented adults at elevated risk for Alzheimer’s disease (AD) is crucial because the pathological process begins decades before dementia onset. Toward that end, we showed that an AD polygenic risk score (PRS) could identify mild cognitive impairment (MCI) in adults who were only in their 50s. Participants were 1176 white, non-Hispanic community-dwelling men of European ancestry in the Vietnam Era Twin Study of Aging (VETSA): 7% with amnestic MCI (aMCI); 4% with non-amnestic MCI (naMCI). Mean age was 56 years, with 89% <60 years old. Diagnosis was based on the Jak-Bondi actuarial/neuropsychological approach. We tested six P-value thresholds (0.05–0.50) for single nucleotide polymorphisms included in the ADPRS. After controlling for non-independence of twins and non-MCI factors that can affect cognition, higher PRSs were associated with significantly greater odds of having aMCI than being cognitively normal (odds ratios (ORs) =1.36–1.43 for thresholds P < 0.20–0.50). The highest OR for the upper vs. lower quartile of the ADPRS distribution was 3.22. ORs remained significant after accounting for APOE-related SNPs from the ADPRS or directly genotyped APOE. Diabetes was associated with significantly increased odds of having naMCI (ORs =3.10–3.41 for thresholds P < 0.05–0.50), consistent with naMCI having more vascular/inflammation components than aMCI. Analysis of sensitivity, specificity, and negative and positive predictive values supported some potential of ADPRSs for selecting participants in clinical trials aimed at early intervention. With participants 15+ years younger than most MCI samples, these findings are promising with regard to efforts to more effectively treat or slow AD progression.
Introduction
Because the pathological process in Alzheimer’s disease (AD) begins long before onset of clinical dementia, early identification of at-risk individuals is of paramount importance [1–3]. Toward that end, we examined risk prediction in midlife adults with respect to mild cognitive impairment (MCI), a transitional phase in the progression from normal cognition to AD [4, 5]. Genome-wide association studies (GWAS) from the International Genomics of Alzheimer’s Project (IGAP) database have identified at least 20 AD susceptibility loci at genome-wide significance level (P < 5 × 10−8) in older adults of European ancestry [6]. Most [7–10], but not all [11], samples of older controls and AD cases have also shown evidence of a significant polygenic component of AD based on an AD polygenic risk score (ADPRS) derived from the IGAP data. Other studies have shown that higher ADPRSs were associated with greater abnormalities in endophenotypes such as cortical thickness, and hippocampal volume [9, 11–15]. Results from two studies were mixed for beta-amyloid and tau levels [9, 16]. In adults in their 20s, results were also somewhat mixed for hippocampal volume and memory [9, 17]. We are aware of two out of three studies showing associations of ADPRSs with MCI [9, 11, 18].
These results were largely based on cohorts with an average age in the mid-70s, were generally stronger when more single nucleotide polymorphisms (SNPs) were included in the PRS, and usually held up after excluding APOE. Given the critical importance of early identification, further examination of MCI—particularly in relatively younger, non-demented adults—is warranted [1–3]. Here we focused on adults who were cognitively (CN) or at increased risk for AD because they had MCI. We tested whether this ADPRS was associated with increased odds of having MCI in men in the Vietnam Era Twin Study of Aging (VETSA) [19, 20], almost 90% of whom were only in their 50s.
We used the well-validated, Jak-Bondi actuarial/neuropsychological diagnostic approach [21–30]. We have argued that a more extensive test battery is essential for detecting impairment in middle-aged, compared with older, adults [23, 31]. Thus, employing the Jak-Bondi approach with our test battery could be particularly useful because our sample was 16–22 years younger than prior samples examining MCI and ADPRSs [9, 11]. Over 20% of healthy older adults have one impaired score in 2 different cognitive domains, but <5% have ≥2 impaired within a domain [32]. By defining impairment as ≥2 tests impaired within a cognitive domain, the Jak-Bondi approach improves the balance between sensitivity and specificity [21–23]. Compared with clinical diagnosis of Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants, Jak-Bondi diagnoses resulted in higher proportions converting to AD, lower proportions reverting to normal, and higher proportions with abnormal beta-amyloid, abnormal tau, and at least one APOE-ε4 allele (the major AD risk allele) [21]. Elevated ADPRSs in our MCI group would provide further validation of this diagnostic approach.
Materials and methods
Participants
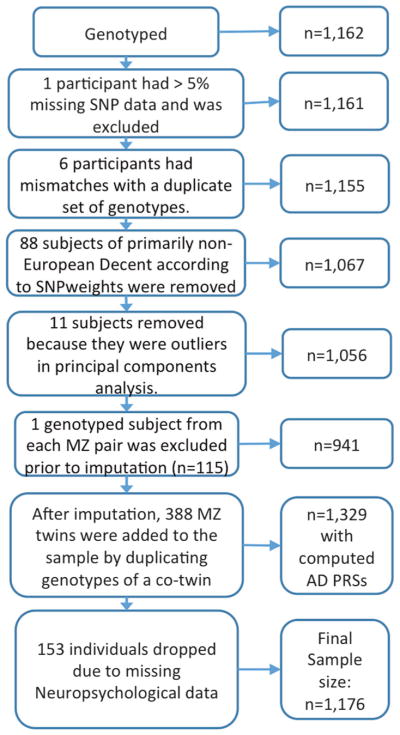
There were 1329 men in the Vietnam Era Twin Study of Aging (VETSA) [19, 20] who were determined to be of white, non-Hispanic European ancestry. As described elsewhere, we then excluded those with missing data that would preclude a possible MCI diagnosis, and with conditions that could cause cognitive deficits unrelated to MCI including seizure disorder, multiple sclerosis, stroke, HIV/AIDS, schizophrenia, substance dependence, brain cancer, or dementia [23]. The final sample comprised 1176 participants (Fig. 1).
Fig. 1.
Flow chart of participant selection
Sample characteristics are shown in Table 1. VETSA constitutes a national sample comparable to American men in their age range with respect to health and lifestyle characteristics [33]. All were in some branch of military service sometime between 1965 and 1975. Nearly 80% report no combat exposure. VETSA participants had to be 51–59 years old at the time of recruitment in wave 1, and both twins in a pair had to be willing to participate [19, 20]. Here we included wave 1 and new wave 2 participants, so that all were undergoing their initial assessment. In sum, VETSA constitutes a reasonably representative sample of community-dwelling men in their age range who were not selected for any characteristic other than age.
Table 1.
Sample characteristics
| Cognitively normal (N = 1050) Mean |
Amnestic MCI (N = 83) Mean |
Non-amnestic MCI (N = 43) Mean |
Significance test | |
|---|---|---|---|---|
| Age | 56.8 (3.3) | 57.2 (3.5) | 56.9 (3.3) | F = 0.81 (2, 537), P = 0.44 |
| Education | 13.9 (2.2) | 13.4 (2.2) | 12.7 (1.3) | F = 4.75 (2, 537), P < 0.01 |
| Age 20 general cognitive ability | 62.2 (21.8) | 65.7 (20.7) | 59.2 (20.8) | F = 4.31 (2, 537), P = 0.01 |
| Depressive symptoms | 7.6 (7.5) | 8.8 (8.6) | 10.0 (10.6) | F = 1.99 (2, 537), P = 0.14 |
| % | % | % | ||
| Hypertension | 56.6% | 66.3% | 58.1% | χ2 = 1.48 (2), P = 0.48 |
| Diabetes | 10.6% | 12.1% | 25.6% | χ2 = 9.40 (2), P < 0.01 |
| Prior head injury | 34.6% | 34.9% | 36.5% | χ2 = 0.45 (2), P = 0.80 |
Participants chose to go to the University of California, San Diego or Boston University where they underwent identical protocols. Brothers almost always chose the same study site. A small number who could or would not travel to the study sites were tested in their hometowns. The study was approved by the Institutional Review Boards of participating institutions, and all participants gave written informed consent.
Measures
Neuropsychological function
The test battery was designed to capture the breadth and depth of cognitive function without having ceiling effects in a middle-aged sample. Table 2 shows 18 measures covering 6 cognitive domains, as described elsewhere [23, 34].
Table 2.
Neuropsychological tests and scores used to define MCI
| Cognitive domain | Tests and measures | No. of measures |
|---|---|---|
| Episodic memory | CVLT-II: sum of trials 1–5; delayed free recall (composite)a WMS-III: Logical memory immediate, delayed free recall (composite)a,b WMS-III: visual reproduction immediate and delayed free recall (composite)a |
3 |
| Executive function | DKEFS Trails: switching DKEFS fluency: category switching Stroop: color-word, interference WASI: matrix reasoning |
4 |
| Attention/working memory | WMS-III: Digit span WMS-III: spatial span WMS-III: letter-number sequencing DKEFS Trails: cancellations |
4 |
| Verbal/language | DKEFS: letter fluency DKEFS: category fluency |
2 |
| Visual-spatial | Gottschaldt hidden figures Card rotation WMS-III: visual reproduction copy |
3 |
| Processing speed | DKEFS Trails: number sequencing, letter sequencing (composite)a Stroop: word condition, color condition (composite)a |
2 |
CVLT-II California Verbal Learning Test-Version II, WMS-III Wechsler Memory Scale-Version III, DKEFS Delis–Kaplan Executive Function System, WASI Wechsler Abbreviated Scale of Intelligence.
Composite refers to the mean of two measures.
Standard WMS-III instructions call for reading the second logical memory story a second time, but it was read only once in our administration
Health/medical measures
We included health/medical measures as covariates in order to know if associations with MCI were truly accounted for by the ADPRS. Depressive symptoms were assessed with the Center for Epidemiological Studies Depression Scale. Hypertension was based on measured blood pressure ≥140 systolic or 90 diastolic, having been told by a physician that the participant had hypertension, or use of medication for hypertension. Blood pressure was measured with an automated cuff: twice in the morning and twice in the afternoon, with each pair of readings separated by 1 min. Medication information was obtained by a standard “brown bag” method. Diabetes was assessed if a participant reported being told by a physician that he had diabetes or if he was taking medication for diabetes. Type 1 diabetes would have ruled out entry into the military. History of head injury was based on a question asking if the person ever had a serious head injury with loss of consciousness or confusion during their lifetime. This constitutes a very liberal threshold for head injury.
Definition of MCI
MCI is generally defined as impairment in one or more cognitive domains, but with functioning generally preserved [4]. In an early article on MCI, Petersen explained that, in the absence of prior cognitive test data, subjective cognitive concern provided a way to infer change from previous levels to avoid labeling people with lifelong static cognitive deficits as having MCI [23, 35]. VETSA is, however, one of a very few studies with actual prior cognitive test data from a period in life that would be unconfounded by aging-related changes. At an average age of 20 years, VETSA participants took the Armed Forces Qualification Test (AFQT) just prior to their induction into the military. The AFQT is an index of general cognitive ability that is correlated about .85 with Wechsler IQ in the VETSA and other studies, and had a 42-year test-retest correlation of 0.73 in VETSA [36, 37]. At VETSA wave 1, 495 individuals had 12 years of education; however, in this subsample with no educational variability, there was a normal distribution of AFQT scores. This illustrates the advantage of this direct measure of “premorbid” cognitive ability over a proxy such as education.
To account for change from “premorbid” levels, we adjusted neuropsychological scores for age 20 AFQT scores. These adjusted scores were then defined as impaired if they were >1.5 SDs below age- and education-adjusted normative means. This criterion is stricter than the more commonly used Jak-Bondi cut-off of >1SD below normative means [21–23] because we had a community-based cohort, almost all of whom were in their 50s, which is much younger than MCI samples that typically average 15–20 years older. Thresholds for defining impairment often need to be different depending on expected base rates, and base rates will differ as a function of factors such as community vs. clinic samples or sample age [38, 39].
As in prior studies using the Jak-Bondi actuarial/neuropsychological approach, impairment in a cognitive domain was defined as having at least two tests that met the criterion for impairment [21, 22]. With this criterion, out of a total of 1176, the diagnoses were as follows: 1050 CN (89%); 83 aMCI (7%); and 43 non-amnestic MIC (naMCI; 4%). Of those with aMCI, 69 had single-domain aMCI and 14 had multiple-domain aMCI. Of those with naMCI 37 had single-domain naMCI and 6 had multiple-domain naMCI. 11% of the VETSA sample was diagnosed with MCI, 7% with aMCI.
Genotyping methods
Other than a small number of MZ pairs that were included as a quality control check, genome-wide genotyping was conducted on individual DZ and unpaired twins and one randomly selected twin from each MZ pair.
Whole-genome genetic variation was assessed at deCODE Genetics (Reykjavík, Iceland). DNA samples were whole-genome amplified, fragmented, precipitated and resuspended prior to hybridization on Illumina HumanOmniExpress-24 v1.0A beadchips for 20 h at 48 °C according to the manufacturer’s protocol (Illumina, San Diego, CA). After hybridization, a single-base extension followed by a multi-layered staining process was performed. Beadchips were imaged using the Illumina iScan System and analyzed with Illumina GenomeStudio v2011.1 software containing Genotyping v1.9.4 module. A GenomeStudio project was created with a custom genotyping cluster file, and average call rate was 0.996. GenomeStudio final report files for 1162 participants were generated and supplied to VETSA investigators for cleaning and analysis.
SNP cleaning and imputation
Cleaning and quality control of genome-wide genotype data were performed using PLINK v1.9 [40]. One participant with more than 5% missing data was excluded. SNPs with more than 5% missing data or SNPs with Hardy-Weinberg equilibrium P-values < 10−6 were excluded. Relationships and zygosity (MZ vs. DZ) were confirmed by estimating the proportion of the autosomal genome shared identical by decent using PLINK’s genome procedure. Relationships based on genome-wide genotype data were concordant with the previously determined relationships derived from the microsatellite markers. As expected, analysis of X-chromosome heterozygosity indicated that all samples were from male participants.
Self-reported ancestry was confirmed using both SNPweights [41] and a principal components (PCs) analysis performed in PLINK v1.9 in conjunction with 1000 Genomes Phase 3 reference data [42]. Participants of self-reported European ancestry or with primarily (>50%) European ancestry according to SNPweights (n = 1067) were analyzed with PCs computed based on a linkage-disequilibrium (LD)-pruned set of 100,000 common (minor allele frequency (MAF) > 0.05) SNPs merged with the 1000 Genomes reference data. Weights for PCs were computed based on 1000 Genomes data and then applied to the VETSA sample. Three participants >6 SDs away from the 1000 genomes European-decent population (EUR) mean on either of the first 2 PCs were excluded. PCs were then recomputed within the remaining putatively white non-Hispanic (WNH) participants, this time without 1000 Genomes reference data. Weights for PCs were computed based on a set of unrelated participants (1 individual per twin pair) and then applied to the whole sample. Within this set, an additional 8 participants were >6 SDs away from the WNH mean on the first 10 PCs and were excluded, leaving 1056 genetically confirmed WNH participants: 115 MZ twin pairs, 264 DZ twin pairs, and 298 individuals whose twin was not genotyped.
Most of the twins whose co-twin was not genotyped were from MZ pairs for which only one twin was selected for genotyping. All of these remaining participants had a >89% European ancestry as estimated by SNPweights. PCs for use as covariates to control for population substructure were recomputed among this WNH set, again based on 100,000 randomly chosen common (MAF > 5%) markers with weights determined in a set of unrelated participants. Imputation was performed using MiniMac [43, 44] computed at the Michigan Imputation Server (https://imputationserver.sph.umich.edu). The 1000 genomes phase 3 EUR data were used as a haplotype reference panel. Owing to the concerns about potential distortion in the haplotype-phasing step of imputation, only one randomly chosen participant per genotyped MZ twin pair was submitted for imputation, and that participant’s resulting imputed data were applied to his MZ co-twin. See Supplementary text for more details.
The final WNH dataset included 1329 participants, including 388 MZ twin pairs, 264 DZ pairs, and 25 unpaired twins.
ADPRS calculation
ADPRSs were computed using summary data from the AD GWAS as presented in Lambert et al. [6]. Individual SNP effect estimates and P-values were downloaded from http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php. The ADPRS is a weighted average of VETSA sample additive imputed SNP dosages with the log-odds ratios (ORs) for each SNP estimated in the GWAS used as the weights. Rare SNPs (MAF < 1%) and SNPs with poor imputation quality (R2 < 0.5) were excluded from ADPRS calculation. The remaining SNPs were trimmed for LD using PLINK’s clumping procedure (r2 threshold of 0.2 in a 500 kb window) based on LD patterns in the 1000 Genomes EUR cohort. ADPRSs were computed by PLINK v1.9 using six different P-value thresholds: P < 0.05, 0.10, 0.20, 0.30, 0.40, and 0.50 (see Supplementary text and Supplementary Figure 1 for details on the choice of these thresholds).
To determine whether or not observed ADPRS associations were being driven by the APOE locus or were independent of APOE, a second version of the ADPRS was computed that excluded the region of LD surrounding the APOE gene (44,409,039–46,412,650 bp according to GRch37/Feb 2009). The number of SNPs included in ADPRS calculation and in the “No APOE” ADPRS for each P-value threshold is presented in Supplementary Table 1. We also examined the influence of APOE-ε4 and APOE-ε2 measured by direct genotyping (method described previously [45]).
Statistical analysis
We performed mixed effects logistic regression analyses to examine the ability of standardized ADPRSs to predict group membership using SAS PROC GLIMMIX [46]. The unit of analysis was the individual, and the non-independence within twin pairs was accounted for in these mixed models. In order to obtain more accurate parameter estimates, we used the METHOD = QUAD statement. This statement performs maximum likelihood estimation by adaptive Gauss–Hermite quadrature.
All analyses adjusted for the first three PCs to account for any cryptic population substructure [47–49]. We also adjusted for the following factors that may affect cognitive function: age; depressive symptoms; hypertension; type 2 diabetes; and history of head injury. We tested six P-value thresholds (0.05–0.50) for SNPs included in the ADPRS. Analyses included Type III (unique) effects of predictor variables.
The ADPRSs computed at the different levels were very highly correlated (pairwise rs = 0.81–0.99), making Bonferroni correction for six different tests highly conservative. We used the Li and Ji method [50] to estimate the effective number of tests = 2. Hence, we used a P-value cut-off of 0.025 two-tailed for multiple-testing significance.
To test for incremental value of the ADPRS, we compared model-fit as we added predictors. We then calculated sensitivity, specificity, positive predictive values (PPV), and negative predictive values (NPV) for the best-fitting model with the ROCR package in R v3.3.2 [51]. We focused on two optimal cut-points: (1) the probability at which the sum of sensitivity and specificity were weighted most equally; and (2) the probability that minimizes cost of misclassifications (PPV and NPV weighted most equally). These were also the probabilities at which both pairs were highest. The area under the curve (AUC) and receiver operating characteristic (ROC) curve plot was calculated with the R package pROC. The 95% confidence intervals (CI) were calculated for the AUC using a stratified bootstrap with 2000 replicates.
Results
As seen in Table 1, the CN, aMCI, and naMCI groups did not differ with respect to age, depressive symptoms, hypertension, or history of head injury. There were very small but significant differences in young adult/premorbid general cognitive ability (AFQT), and in education. The naMCI group had 0.7 years less education than the aMCI and 1.2 years less than the CN group. The mean AFQT score for the VETSA cohort was above the average of 50th percentile because individuals who scored below the 10th percentile were not admitted for military service. The highest mean percentile score (65.7) was in the aMCI group, followed by the CN group (62.2), and then the naMCI group (59.2). These are approximately equivalent to IQs ranging from 106 to 111 across the three groups. Current AFQT scores were uncorrelated with the ADPRS (rs = −0.040 to −0.064, Ps = 0.222 to 0.064 at different thresholds). The proportion of individuals with type 2 diabetes also differed significantly among the three groups. The prevalence was similar in the CN and aMCI groups (10.6% and 12.1%, respectively). It was more than double those levels in the naMCI group, with 25.6% of this group having diabetes.
After multiple-testing correction, higher ADPRSs were associated with significantly greater odds of being in the aMCI group than in the CN group (ORs estimated for 1SD increase in ADPRS: range = 1.363–1.427 for ADPRS thresholds of P < 0.20–0.50) (Table 3a). Results were essentially unchanged in a model with the first 10 PCs (Supplementary Table 2). For the most significant threshold, P < 0.50, the odds of having aMCI were 3.215 times greater (95% CI: 1.392; 7.424) for those in the upper vs. the lower quartile of the ADPRS distribution (Table 3b). ORs for naMCI vs. CN groups were only slightly lower than those for aMCI, but they were nonsignificant (Table 3c), probably due to lower power given the smaller size of the naMCI group. ORs were significant but slightly lower for the combined MCI groups vs. the CN group (not shown).
Table 3.
Associations of Alzheimer’s disease polygenic risk scores with MCI
| GWAS threshold | Odds ratios (95% confidence intervals) | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| ADPRS including APOE-region SNPs | ADPRS excluding APOE-region SNPs | |||||||
|
|
|
|||||||
| a. | b. | c. | d. | |||||
| aMCI vs CN: full sample | P | aMCI vs CN: upper vs lower PRS quartile | P | naMCI vs CN: full sample | P | aMCI vs CN: full sample | P | |
| P < 0.05 | 1.152 (0.886; 1.498) | 0.2903 | 1.376 (0.652; 2.903) | 0.4014 | 1.169 (0.839; 1.630) | 0.3560 | 1.126 (0.862; 1.471) | 0.3831 |
| P < 0.10 | 1.258 (0.966; 1.639) | 0.0884 | 2.397 (1.102; 5.218) | 0.0276* | 1.232 (0.885; 1.715) | 0.2166 | 1.236 (0.945; 1.616) | 0.1212 |
| P < 0.20 | 1.363 (1.041; 1.784) | 0.0246 | 2.971 (1.325; 6.665) | 0.0083 | 1.320 (0.946; 1.840) | 0.1019 | 1.350 (1.026; 1.776) | 0.0319* |
| P < 0.30 | 1.405 (1.074; 1.837) | 0.0131 | 2.483 (1.134; 5.435) | 0.0230 | 1.399 (0.996; 1.966) | 0.0530 | 1.397 (1.063; 1.835) | 0.0165 |
| P < 0.40 | 1.420 (1.085; 1.859) | 0.0107 | 2.463 (1.113; 5.450) | 0.0262* | 1.392 (0.992; 1.954) | 0.0555 | 1.413 (1.074; 1.858) | 0.0135 |
| P < 0.50 | 1.427 (1.090; 1.869) | 0.0099 | 3.215 (1.392; 7.424) | 0.0063 | 1.341 (0.957; 1.878) | 0.0877 | 1.419 (1.079; 1.868) | 0.0125 |
Covariates include the first three principal components from genome-wide genotyping data; age; depressive symptoms; hypertension; type 2 diabetes; ischemic heart disease; and history of head injury. All analyses were adjusted for clustering of twin data. Significant odds ratios at corrected P-value of 0.025 based on Li and Ji method are shown in bold font (see Supplementary text for details)
Indicates significant odds ratio at P < 0.05.
ADPRS Alzheimer’s disease polygenic risk score, SNP single nucleotide polymorphism, aMCI amnestic mild cognitive impairment, CN cognitively normal, naMCI non-amnestic mild cognitive impairment
After excluding SNPs within the APOE-region, the ADPRS still significantly differentiated aMCI and CN groups (ORs estimated for 1SD increase in ADPRS: range = 1.397–1.419 for ADPRS thresholds of P < 0.30–0.50) (Table 3d). In models including genotyped APOE-ε4 and APOE-ε2, the highest magnitude ORs for APOE-ε4 and APOE-ε2 were 1.437 and 0.515, respectively, but none of these were statistically significant. After adjusting for APOE-ε4 and APOE-ε2, the ADPRS remained significantly associated with increased odds of being in the aMCI group (range = 1.387–1.408 for thresholds of P < 0.30–0.50) (Table 4). In contrast to aMCI, diabetes was associated with significantly increased odds of having naMCI (ORs estimated for 1SD increase in ADPRS: range = 3.103–3.341 for ADPRS thresholds of P < 0.05–0.50) (Table 5). None of the other covariates was associated with increased odds of either aMCI or naMCI. An additional ADPRS comprising only the 19 AD risk genes (excluding APOE) previously identified at genome-wide significance level [6] was not correlated with our ADPRS, nor was it associated with increased odds of MCI (Supplementary Table 3).
Table 4.
Associations of direct genotyped APOE-ε4 and APOE-ε2 and Alzheimer’s disease polygenic risk with aMCI compared with cognitively normal participants
| GWAS threshold | Odds ratios (95% confidence intervals) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| APOE-ε4 | P | APOE-ε2 | P | ADPRS | P | |
| P < 0.05 | 1.393 (0.748; 2.593) | 0.2951 | 0.522 (0.221; 1.237) | 0.1395 | 1.171 (0.895; 1.533) | 0.2494 |
| P < 0.10 | 1.393 (0.753; 2.577) | 0.2898 | 0.524 (0.223; 1.240) | 0.1414 | 1.235 (0.942; 1.619) | 0.1261 |
| P < 0.20 | 1.411 (0.761; 2.616) | 0.2738 | 0.527 (0.223; 1.245) | 0.1437 | 1.327 (1.008; 1.748) | 0.0437* |
| P < 0.30 | 1.431 (0.773; 2.649) | 0.2540 | 0.522 (0.221; 1.231) | 0.1373 | 1.387 (1.053; 1.828) | 0.0200 |
| P < 0.40 | 1.437 (0.776; 2.662) | 0.2484 | 0.518 (0.220; 1.223) | 0.1333 | 1.398 (1.060; 1.842) | 0.0177 |
| P < 0.50 | 1.431 (0.772; 2.850) | 0.2541 | 0.515 (0.218; 1.218) | 0.1304 | 1.408 (1.067; 1.859) | 0.0159 |
Covariates include the first three principal components from genome-wide genotyping data; age; depressive symptoms; hypertension; type 2 diabetes; ischemic heart disease; and history of head injury. All analyses were adjusted for clustering of twin data. Significant odds ratios at corrected P-value of 0.025 based on Li and Ji method are shown in bold font
Indicates significant odds ratio at P < 0.05.
aMCI amnestic mild cognitive impairment
Table 5.
Associations of type 2 diabetes with MCI in analyses with the ADPRS including APOE-region SNPs
| GWAS threshold | aMCI vs cognitively normal | naMCI vs cognitively normal | ||
|---|---|---|---|---|
|
|
|
|||
| OR (95% CI) | P | OR (95% CI) | P | |
| P < 0.05 | 1.046 (0.464; 2.358) | 0.9128 | 3.103 (1.386; 6.947) | 0.0060 |
| P < 0.10 | 1.094 (0.488; 2.453) | 0.8274 | 3.181 (1.415; 7.154) | 0.0052 |
| P < 0.20 | 1.084 (0.480; 2.448) | 0.8458 | 3.284 (1.441; 7.480) | 0.0047 |
| P < 0.30 | 1.084 (0.481; 2.442) | 0.8456 | 3.341 (1.462; 7.634) | 0.0043 |
| P < 0.40 | 1.091 (0.484; 2.459) | 0.8328 | 3.313 (1.453; 7.358) | 0.0045 |
| P < 0.50 | 1.086 (0.481; 2.448) | 0.8426 | 3.273 (1.437; 7.452) | 0.0048 |
All analyses were adjusted for clustering of twin data. Significant odds ratios at corrected P-value of 0.025 based on Li and Ji method are shown in bold font
MCI mild cognitive impairment, ADPRS Alzheimer’s disease polygenic risk score, SNP single nucleotide polymorphism, aMCI amnestic MCI, naMCI non-amnestic MCI, GWAS genome-wide association study
The best-fitting model (Table 6, Model 4) shows that the ADPRS provided a significant improvement in prediction beyond age, risk factors for AD (covariates), and APOE genotype. For the best-fitting model, the AUC was 0.955 (95% CI: 0.93; 0.97) (Supplementary Figure 2). When the cut-off was selected for high and balanced sensitivity (0.910) and specificity (0.886), NPV was high (0.992) and PPV was modest (0.376). When cut-offs resulted in high and balanced NPV (0.953) and PPV (0.737), specificity was very high (0.990), and sensitivity was modest (0.359).
Table 6.
Model-fit comparisons for predictors of aMCI vs. CN
| Model | −2LL | LRT | ΔDF | P |
|---|---|---|---|---|
| 1 Age | 560.03 | |||
| 2 Age + covariates | 559.19 | 4.84 | 7 | 0.6795 |
| 3 Age + covariates + APOE-ε4 + APOE-ε2 | 552.49 | 2.70 | 2 | 0.2592 |
| 4 Age + covariates + APOE-ε4 + APOE-ε2 + ADPRS | 546.41 | 6.08 | 1 | 0.0137 |
All analyses were adjusted for clustering of twin data. Each model is compared to the preceding model. A P-value < 0.05 indicates a significant improvement in model-fit. Best-fitting model is shown in bold font.
aMCI amnestic mild cognitive impairment, CN cognitively normal, −2LL −2 log-likelihood, LRT likelihood ratio test, ΔDF change in degrees of freedom, ADPRS Alzheimer’s disease polygenic risk score
Discussion
We demonstrated the ability of ADPRSs to differentiate individuals with MCI from those who are CN in a community-dwelling sample of men, 89% of whom were only in their 50s. With the average age of this sample being 16–22 years younger than other studies of MCI to date, our results hold promise for early identification of increased risk for AD. Consistent with results in older adults [9, 18], the full ADPRS differentiated groups far better than the 19 genome-wide significant SNPs. Also, as described in the introduction, our use of the Jak-Bondi approach [21] might be more sensitive for diagnosing MCI than traditional approaches.
The ADPRS did significantly improve model-fit when added to a model with age, covariates, and APOE genotype. The best-fitting model had a very high AUC, but AUCs have been shown to be misleading and are problematic as comparative measures across studies [52]. In the most comparable study, the Rotterdam Study of older adults, the prevalence of aMCI was 4% and the AUC was 0.592 (sensitivity, specificity, NPV, and PPV were not presented). Here again, cross-study differences could be due to different ADPRSs and/or different approaches to diagnosis. Despite our high AUC, we focus on more informative indices. A threshold with high and balanced sensitive and specificity resulted in high NPV and modest PPV, which may be useful for screening individuals to be referred for further evaluation because people classified as controls would be highly likely to be true controls even though aMCI classifications would include increased false positives. A threshold with high and balanced NPV and PPV resulted in high specificity and modest sensitivity. Thus, aMCI cases would be missed but false positives for aMCI would be reduced substantially. This threshold may be useful for clinical trials by providing greater certainty that cases are truly cases.
With a male-only sample, we cannot know how our results might generalize to women. Women are at higher lifetime risk of AD than men, the APOE-ε4 allele confers greater risk in women, and postmortem studies indicate female excesses of neuritic plaques and neurofibrillary tangles and greater cognitive deficit per unit of brain amyloid [53]. Differentiating MCI and CN groups may thus be more difficult in men, yet we were able to do so in this relatively young cohort. APOE effects are somewhat weaker in men than in women, and may not yet be prominent in the relatively young age range of our cohort [54, 55]. However, as in most other studies of older adults [7–10, 13–15], the ADPRS retained predictive value after controlling for or removing APOE.
Male excesses of cerebrovascular pathologies have been associated with AD [53]. Diabetes increases risk for all-cause dementia, but that relationship may be stronger for vascular dementia than for AD [56]. Here diabetes—a cardiovascular-related condition—was associated with increased risk of naMCI, consistent with naMCI having more of a vascular component than aMCI. Although the ORs for the ADPRS did not suggest that aMCI is substantially more AD-related than naMCI, the different diabetes results do support the value of separately analyzing these MCI subtypes. Also, the most common deficits in the naMCI group were executive-attention deficits. Thus, our results are also consistent with findings that memory deficits are typically the prominent cognitive deficit in AD; yet there is also a significant proportion of AD patients who most prominently manifest a dysexecutive syndrome [57].
In conclusion, we view the young age of the sample as critically important. The results provide proof of principle for the value of ADPRSs to aid in identifying genetically at-risk individuals with MCI as early as the sixth decade of life, and they provide additional validation of the actuarial-neuropsychological approach to diagnosing MCI. The findings thus support the idea that ADPRSs can aid in predicting who will develop AD many years before they manifest any symptoms, although it must be acknowledged that we do not know which participants will, in fact, develop AD. It does, however, seem likely that the findings represent an AD-related phenomenon and not simply general cognitive ability because our MCI diagnosis included adjustment for age 20 AFQT scores and-- consistent with prior research [58]--the ADPRS and AFQT were not significantly correlated. ADPRSs may also be a useful additional tool for targeting individuals for clinical trials and individuals requiring clinical intervention at an earlier stage than is currently thought to be feasible. Such efforts at early identification and intervention are likely to be key factors for more effectively treating or slowing the progression of AD.
Supplementary Material
Acknowledgments
This work was supported by National Institute on Aging R01 AG018386, AG022381, AG022982, AG050595 (W.S.K.), R01 AG018384 (M.J.L.), R03 AG046413 (C.E.F), and K08 AG047903 (M.S.P), and the VA San Diego Center of Excellence for Stress and Mental Health. The content is the responsibility of the authors and does not necessarily represent official views of the NIA, NIH, or VA. The Cooperative Studies Program of the U.S. Department of Veterans Affairs provided financial support for development and maintenance of the Vietnam Era Twin Registry. We would also like to acknowledge the continued cooperation and participation of the members of the VET Registry and their families.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41380-018-0030-8) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest Dr. Dale is a Founder of and holds equity in CorTechs Labs, Inc, and serves on its Scientific Advisory Board. He is a member of the Scientific Advisory Board of Human Longevity, Inc. and receives funding through research agreements with General Electric Healthcare and Medtronic, Inc. The terms of these arrangements have been reviewed and approved by UCSD in accordance with its conflict of interest policies. The remaining authors declare that they have no conflict of interest.
References
- 1.Golde TE, Schneider LS, Koo EH. Anti-abeta therapeutics in Alzheimer’s disease: the need for a paradigm shift. Neuron. 2011;69:203–13. doi: 10.1016/j.neuron.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–92. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperling RA, Jack CR, Jr, Aisen PS. Testing the right target and right drug at the right stage. Sci Transl Med. 2011;3:111cm133. doi: 10.1126/scitranslmed.3002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011;7:270–9. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen RC, Morris JC. Mild cognitive impairment as a clinical entity and treatment target. Arch Neurol. 2005;62:1160–3. doi: 10.1001/archneur.62.7.1160. [DOI] [PubMed] [Google Scholar]
- 6.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–84. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer’s disease. Ann Neurol. 2017;82:311–4. doi: 10.1002/ana.24999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mormino EC, Sperling RA, Holmes AJ, Buckner RL, De Jager PL, Smoller JW, et al. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology. 2016;87:481–8. doi: 10.1212/WNL.0000000000002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tosto G, Bird TD, Tsuang D, Bennett DA, Boeve BF, Cruchaga C, et al. Polygenic risk scores in familial Alzheimer disease. Neurology. 2017;88:1180–6. doi: 10.1212/WNL.0000000000003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lupton MK, Strike L, Hansell NK, Wen W, Mather KA, Arm-strong NJ, et al. The effect of increased genetic risk for Alzheimer’s disease on hippocampal and amygdala volume. Neurobiol Aging. 2016;40:68–77. doi: 10.1016/j.neurobiolaging.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison TM, Mahmood Z, Lau EP, Karacozoff AM, Burggren AC, Small GW, et al. An Alzheimer’s disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro. 2016:3. doi: 10.1523/ENEURO.0098-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012;22:2653–61. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marioni RE, Campbell A, Hagenaars SP, Nagy R, Amador C, Hayward C, et al. Genetic stratification to identify risk groups for Alzheimer’s disease. J Alzheimers Dis. 2017;57:275–83. doi: 10.3233/JAD-161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan G, Adams HH, Bis JC, Weinstein G, Yu L, Toglhofer AM, et al. Association of Alzheimer’s disease GWAS loci with MRI markers of brain aging. Neurobiol Aging. 2015;36:1765e1767–1716. doi: 10.1016/j.neurobiolaging.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz SA, Boots EA, Darst BF, Zetterberg H, Blennow K, Edwards DF, et al. Cardiorespiratory fitness alters the influence of a polygenic risk score on biomarkers of AD. Neurology. 2017;88:1650–8. doi: 10.1212/WNL.0000000000003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foley SF, Tansey KE, Caseras X, Lancaster T, Bracht T, Parker G, et al. Multimodal brain imaging reveals structural differences in Alzheimer’s disease polygenic risk carriers: a study in healthy young adults. Biol Psychiatry. 2017;81:154–61. doi: 10.1016/j.biopsych.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams HHH, deBruijn RFAG, Hofman A, Uitterlinden AG, van Duijn CM, Vernooij MW, et al. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia. Alzheimers Dement. 2015;11:1277–85. doi: 10.1016/j.jalz.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Kremen WS, Franz CE, Lyons MJ. VETSA: the Vietnam era twin study of aging. Twin Res Hum Genet. 2013;16:399–402. doi: 10.1017/thg.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremen WS, Thompson-Brenner H, Leung YJ, Grant MD, Franz CE, Eisen SA, et al. Genes, environment, and time: the Vietnam era twin study of aging (VETSA) Twin Res Hum Genet. 2006;9:1009–22. doi: 10.1375/183242706779462750. [DOI] [PubMed] [Google Scholar]
- 21.Bondi MW, Edmonds EC, Jak AJ, Clark LR, Delano-Wood L, McDonald CR, et al. Neuropsychological criteria for mild cognitive impairment improves diagnostic precision, biomarker associations, and progression rates. J Alzheimers Dis. 2014;42:275–89. doi: 10.3233/JAD-140276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jak AJ, Bondi MW, Delano-Wood L, Wierenga C, Corey-Bloom J, Salmon DP, et al. Quantification of five neuropsychological approaches to defining mild cognitive impairment. Am J Geriatr Psychiatry. 2009;17:368–75. doi: 10.1097/JGP.0b013e31819431d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kremen WS, Jak AJ, Panizzon MS, Spoon KM, Franz CE, Thompson WK, et al. Early identification and heritability of mild cognitive impairment. Int J Epidemiol. 2014;43:600–10. doi: 10.1093/ije/dyt242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edmonds EC, Delano-Wood L, Clark LR, Jak AJ, Nation DA, McDonald CR, et al. Susceptibility of the conventional criteria for mild cognitive impairment to false-positive diagnostic errors. Alzheimers Dement. 2014;11:415–24. doi: 10.1016/j.jalz.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edmonds EC, Delano-Wood L, Galasko DR, Salmon DP, Bondi MW Alzheimer’s Disease Neuroimaging Initiative. Subtle cognitive decline and biomarker staging in preclinical Alzheimer’s disease. J Alzheimers Dis. 2015;47:231–42. doi: 10.3233/JAD-150128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edmonds EC, Delano-Wood L, Jak AJ, Galasko DR, Salmon DP, Bondi MW, et al. “Missed” mild cognitive impairment: High false-negative error rate based on conventional diagnostic criteria. J Alzheimers Dis. 2016;52:685–91. doi: 10.3233/JAD-150986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delano-Wood L, Bondi MW, Sacco J, Abeles N, Jak AJ, Libon DJ, et al. Heterogeneity in mild cognitive impairment: differences in neuropsychological profile and associated white matter lesion pathology. J Int Neuropsychol Soc. 2009;15:906–14. doi: 10.1017/S1355617709990257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jak AJ, Urban S, McCauley A, Bangen KJ, Delano-Wood L, Corey-Bloom J, et al. Profile of hippocampal volumes and stroke risk varies by neuropsychological definition of mild cognitive impairment. J Int Neuropsychol Soc. 2009;15:890–7. doi: 10.1017/S1355617709090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wierenga CE, Clark LR, Dev SI, Shin DD, Jurick SM, Rissman RA, et al. Interaction of age and APOE genotype on cerebral blood flow at rest. J Alzheimer’s Dis. 2013;34:921–35. doi: 10.3233/JAD-121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wierenga CE, Dev SI, Shin DD, Clark LR, Bangen KJ, Jak AJ, et al. Effect of mild cognitive impairment and APOE genotype on resting cerebral blood flow and its association with cognition. J Cereb Blood Flow Metab. 2012;32:1589–99. doi: 10.1038/jcbfm.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremen WS, Moore CS, Franz CE, Panizzon MS, Lyons MJ. Cognition in middle adulthood. In: Finkel D, Reynolds CA, editors. Behavior genetics of cognition across the lifespan. New York: Springer; 2013. pp. 105–34. [Google Scholar]
- 32.Palmer BW, Boone KB, Lesser IM, Wohl MA. Base rates of “impaired” neuropsychological test performance among healthy older adults. Arch Clin Neuropsychol. 1998;13:503–11. [PubMed] [Google Scholar]
- 33.Schoeneborn CA, Heyman KM. Health characteristics of adults aged 55 years and over: United States, 2004–7. Natl Health Stat Report. 2009;16:1–31. [PubMed] [Google Scholar]
- 34.Granholm EL, Panizzon MS, Elman JA, Jak AJ, Hauger RL, Bondi MW, et al. Pupillary responses as a biomarker of early risk for Alzheimer’s disease. J Alzheimer’s Dis. 2017;56:1419–28. doi: 10.3233/JAD-161078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–94. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- 36.Lyons MJ, York TP, Franz CE, Grant MD, Eaves LJ, Jacobson KC, et al. Genes determine stability and the environment determines change in cognitive ability during 35 years of adulthood. Psychol Sci. 2009;20:1146–52. doi: 10.1111/j.1467-9280.2009.02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyons MJ, Panizzon MS, Liu W, McKenzie R, Bluestone NJ, Grant MD, et al. A longitudinal twin study of general cognitive ability over four decades. Dev Psychol. 2017;53:1170–7. doi: 10.1037/dev0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Denny KG, Harvey D, Farias ST, Mungas D, DeCarli C, et al. Progression from normal cognition to mild cognitive impairment in a diverse clinic-based and community-based elderly cohort. Alzheimers Dement. 2016;13:399–405. doi: 10.1016/j.jalz.2016.07.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meehl P, Rosen A. Antecedent probability and the efficiency of psychometric signs, patterns, or cutting scores. Psychol Bull. 1955;52:194–216. doi: 10.1037/h0048070. [DOI] [PubMed] [Google Scholar]
- 40.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:1–16. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CY, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics. 2013;29:1399–406. doi: 10.1093/bioinformatics/btt144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz MR, Lyons MJ, Franz CE, Grant MD, Boake C, Jacobson KC, et al. Apolipoprotein E genotype and memory in the sixth decade of life. Neurology. 2008;70:1771–7. doi: 10.1212/01.wnl.0000286941.74372.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.SAS Institute Inc. SAS OnlineDoc 9.4. Carey, NC: SAS Institute; 2013. [Google Scholar]
- 47.Sadeh N, Spielberg JM, Logue MW, Wolf EJ, Smith AK, Lusk J, et al. SKA2 methylation predicts reduced cortical thickness in prefrontal cortex. Mol Psychiatry. 2016;21:299. doi: 10.1038/mp.2016.10. [DOI] [PubMed] [Google Scholar]
- 48.Sadeh N, Wolf EJ, Logue MW, Lusk J, Hayes JP, McGlinchey RE, et al. Polygenic risk for externalizing psychopathology and executive dysfunction in trauma-exposed veterans. Clin Psychol Sci. 2016;4:545–58. doi: 10.1177/2167702615613310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf EJ, Logue MW, Hayes JP, Sadeh N, Schichman SA, Stone A, et al. Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–62. doi: 10.1016/j.psyneuen.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 51.R Development Core Team. R: a Language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 52.Lobo JM, Jiménez-Valverde A, Real R. AUC: A misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008;17:145–51. [Google Scholar]
- 53.Finch CE, Shams S. Apolipoprotein E and sex bias in cerebrovascular aging of men and mice. Trends Neurosci. 2016;39:625–37. doi: 10.1016/j.tins.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blacker D, Haines JL, Rodes L, Terwedow H, Go RC, Harrell LE, et al. ApoE-4 and age at onset of Alzheimer’s disease: The NIMH genetics initiative. Neurology. 1997;48:139–47. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 55.Jack CR, Jr, Wiste HJ, Weigand SD, Knopman DS, Vemuri P, Mielke MM, et al. Age, sex, and APOE ε4 effects on memory, brain structure, and β-Amyloid across the adult life span. JAMA Neurol. 2015;72:511–9. doi: 10.1001/jamaneurol.2014.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katon W, Pedersen HS, Ribe AR, Fenger-Gron M, Davydow D, Waldorff FB, et al. Effect of depression and diabetes mellitus on the risk for dementia: a national population-based cohort study. JAMA Psychiatry. 2015;72:612–9. doi: 10.1001/jamapsychiatry.2015.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mez J, Mukherjee S, Thornton T, Fardo DW, Trittschuh E, Sutti S, et al. The executive prominent/memory prominent spectrum in Alzheimer’s disease is highly heritable. Neurobiol Aging. 2016;41:115–21. doi: 10.1016/j.neurobiolaging.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris SE, Davies G, Luciano M, Payton A, Fox HC, Haggarty P, et al. Polygenic risk for Alzheimer’s disease is not associated with cognitive ability or cognitive aging in non-demented older people. J Alzheimers Dis. 2014;39:565–74. doi: 10.3233/JAD-131058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.