Abstract
Mouse embryos undergo genome-wide methylation reprogramming by demethylation in early preimplantation development, followed by remethylation thereafter. Here we show that genome-wide reprogramming is conserved in several mammalian species and ask whether it also occurs in embryos cloned with the use of highly methylated somatic donor nuclei. Normal bovine, rat, and pig zygotes showed a demethylated paternal genome, suggesting active demethylation. In bovine embryos methylation was further reduced during cleavage up to the eight-cell stage, and this reduction in methylation was followed by de novo methylation by the 16-cell stage. In cloned one-cell embryos there was a reduction in methylation consistent with active demethylation, but no further demethylation occurred subsequently. Instead, de novo methylation and nuclear reorganization of methylation patterns resembling those of differentiated cells occurred precociously in many cloned embryos. Cloned, but not normal, morulae had highly methylated nuclei in all blastomeres that resembled those of the fibroblast donor cells. Our study shows that epigenetic reprogramming occurs aberrantly in most cloned embryos; incomplete reprogramming may contribute to the low efficiency of cloning.
Epigenetic modification of DNA by methylation in mammals occurs predominantly at CpG dinucleotides and is involved in a number of key genome functions (1, 2). These include roles in imprinting, X chromosome inactivation, genome stability, silencing of retrotransposons, and inactivation of genes in cancers. Whether methylation also has a role in regulating gene expression during development is still debated (2, 3). In the mouse, mutations in the genes for the maintenance methyltransferase, Dnmt1 (4), or the two de novo methyltransferases, Dnmt3a and -b (5), result in genome demethylation and lethality at postimplantation stages or after birth, possibly involving apoptosis (6).
Genomic methylation patterns in somatic differentiated cells are generally stable and heritable by virtue of maintenance methylation by Dnmt1 (2, 4). However, in the mouse there are at least two developmental periods in which methylation patterns are reprogrammed genome wide (7). The first occurs in primordial germ cells of both sexes and leads to rapid demethylation of imprinted genes and single copy sequences, followed by de novo methylation in male and female germ cells several days later (8). This reprogramming of methylation patterns is essential for imprinting and may be important for the erasure of acquired epigenetic modifications (7, 8).
A similar reprogramming cycle occurs in mouse preimplantation embryos. Only hours after fertilization, before DNA replication in the fertilized embryo, the paternal genome undergoes genome-wide demethylation (9, 10), which is likely to occur by active demethylation, but the mechanisms are unknown. The maternal genome in contrast does not become actively demethylated in the zygote. Instead sequential stepwise demethylation occurs during the first cleavage divisions (11–13); this demethylation is likely to be caused by exclusion from the nucleus of Dnmt1o (14). As a result of these two demethylation processes, mouse morulae are substantially undermethylated. De novo methylation is thought to occur at some stage soon after implantation (11). In contrast to the reprogramming that occurs during germ cell development, however, imprinted genes are not demethylated in preimplantation embryos or de novo methylated after implantation (15). The oocyte form of Dnmt1 has recently been identified as being important for the maintenance of imprinted gene methylation. This form is generally excluded from the nucleus in preimplantation embryos, except in eight-cell embryos. This nuclear localization for one cell cycle is apparently important for the maintenance of imprinted methylation at the eight-cell stage (14).
All information about methylation reprogramming in mammalian preimplantation development is currently limited to the mouse. However, in zebrafish (16) and in Xenopus (I. Stancheva, O. El-Maarri, J. Walter, and R. Meehan, personal communication) there is no demethylation in early embryos, thus raising the question of whether demethylation is restricted to mammals. This is a particularly important question in light of recent successes in cloning of various mammalian species (17–19). Although cloning is possible now in mammals, the rate of success of obtaining live young is very low, and it has been proposed that epigenetic reprogramming of somatic donor nuclei is important for attaining totipotency (15, 17–21). Recently a study on bovine cloned preimplantation embryos that made use of methylation analysis by bisulfite sequencing (21) found that some DNA sequences were more highly methylated in cloned versus normal morulae. Here we show that methylation reprogramming is conserved in eutherian mammals, but that reprogramming of methylated somatic donor nuclei occurs aberrantly in many cloned preimplantation embryos, thus explaining how hypermethylation arises in cloned embryos on a genome-wide scale.
Materials and Methods
Collection of Mammalian Oocytes and Embryos.
Mouse fertilized oocytes and embryos were collected from superovulated females on appropriate days for cleavage-stage embryos according to standard procedures (22). Embryos used were derived from a cross of (C57BL/6J × CBA/Ca) F1 females mated to (C57BL/6J × CBA/Ca) F1 males. The day after mating is termed day 1.
Rat fertilized oocytes were collected from naturally mated Wistar rats at ≈12 h after fertilization. Fertilized pig oocytes were obtained from artificially inseminated gilts that had been superovulated. Fertilized oocytes were collected and centrifuged to visualize the pronuclei before fixation. The zonae were removed with acidic Tyrode's solution before further processing. Bovine fertilized oocytes and embryos were obtained from in vitro matured oocytes harvested from ovaries obtained from the abattoir. Matured oocytes were fertilized in vitro and subsequently cultured before fixation.
Cloning of Bovine Embryos.
The nuclear transfer procedure was that described in ref. 23. Non-starved fetal fibroblasts were used as nuclear donor cells.
Indirect Immunofluorescence.
Fertilized oocytes and early preimplantation embryos were washed in PBS, fixed for 15 min in 4% paraformaldehyde in PBS, and permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature. For the detection of 5-methyl-cytosine, oocytes, zygotes, and embryos were treated with 2 M HCl at room temperature for 30 min and subsequently neutralized for 10 min with 100 mM Tris⋅HCl buffer (pH 8.5) after permeabilization. After extensive washing with 0.05% Tween-20 in PBS, all samples were blocked overnight at 4°C in 1% BSA/0.05% Tween-20 in PBS. Anti-5-methyl-cytosine antibodies (24) were detected by a secondary antibody coupled with either Cy3 or Texas-Red, respectively (Jackson ImmunoResearch). DNA was stained with either the intercalating dye YOYO-1 iodide (Molecular Probes) at 100 nM or 5 μg/ml 4′,6-diamidino-2-phenylindole and mounted in 50% glycerol in PBS (Sigma).
Digital Imaging Microscopy.
Observations were performed with an Olympus BX40 epifluorescence microscope. Images were recorded digitally with a high-resolution charge-coupled device camera (F-View) and analysis 3.0 image analysis software (SIS GmbH, Münster, Germany). Greyscale images were pseudocolored after capture by separate filter sets for YOYO-1, CY3/Texas Red, and 4′,6-diamidino-2-phenylindole and merged with adobe photoshop 5.0 software.
Confocal Microscopy.
Digital optical sections from preimplantation embryos (blastocysts) were recorded with a confocal laser scanning Ultraview microscope (Perkin–Elmer). For each wavelength a z series of 0.2 μm slices were scanned and exported as 8-bit tagged image file format (TIFF) files. The images were later projected with imagej 1.19z and pseudocolored with adobe photoshop 5.0.
Results
Conservation of Methylation Reprogramming.
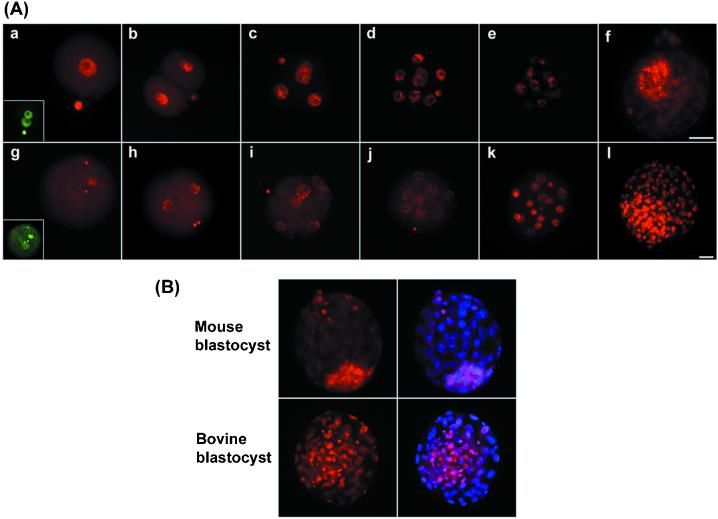
Genome-wide demethylation can be examined comprehensively with the use of immunostaining with a 5-methyl cytosine antibody (10, 13, 24). Here we used immunostaining of interphase nuclei in mammalian preimplantation embryos. Interphase nuclei were chosen because they give an overall impression of genome-wide methylation, and normal nuclear organization is retained. Normal bovine, pig, and rat zygotes showed demethylation of the larger (male) pronucleus, as in the mouse (Fig. 1). Because the sperm genome is highly methylated in all of these species (25), this finding shows that genome-wide demethylation of the paternal genome is conserved in eutherian mammals. We next examined bovine cleavage stage embryos in comparison with the mouse (Fig. 2). From the two-cell to the eight-cell stage there was a further reduction in methylation, consistent with passive demethylation occurring during DNA replication, as demonstrated in the mouse (Fig. 2). Strikingly, however, there was considerable de novo methylation in bovine embryos from the eight-cell to the 16-cell stage, which resulted in a finely granular staining pattern in many nuclei (Fig. 2 Aj and Ak, and Fig. 3Bc). This pattern resembled that of the DNA itself, suggesting that the nuclear methylation pattern reflects at least in part the distribution of interphase chromosomes in the nucleus (Fig. 3B). In contrast, mouse 16-cell embryos continued to remain demethylated, and genome-wide de novo methylation occurred approximately four cell divisions later, in inner cell mass (ICM) cells of the blastocyst (Fig. 2Af), regardless of whether in vivo developed or in vitro cultured embryos were analyzed. As a result, bovine blastocysts show considerably higher levels of methylation specifically in trophectodermal cells (Fig. 2B). Thus although the basic events of active demethylation in the zygote, and passive demethylation during cleavage divisions, followed by de novo methylation, appear to be conserved in mammals, their timing with respect to developmental events can apparently be different.
Figure 1.
Selective demethylation of the male pronucleus is conserved. Indirect immunofluorescence with the use of an antibody to 5-methyl cytosine (in red) shows that the male pronucleus is selectively demethylated immediately after fertilization, whereas the female pronucleus remains methylated, in mouse (a and b), rat (c and d), pig (e and f), and cow (g and h). DNA staining is in blue. The larger of the two pronuclei is the male, except in cows, where they are of the same size. In all cases embryos were collected and fixed before DNA replication. (Scale bar, 20 μm.)
Figure 2.
Demethylation and remethylation are conserved during preimplantation development. (A) Normal mouse (a–f) and bovine (g–l) embryos were stained for 5-methyl cytosine (red) from the zygote to the blastocyst stage. In the mouse (a–f), there is an initial loss of methylation specifically from the male pronucleus [(Inset) DNA stained to identify two pronuclei, green]. Thereafter the remaining decline in signal occurs in a stepwise fashion up to the morula stage (e). The ICM, but not the trophectoderm, has undergone de novo methylation by the blastocyst stage (f). Bovine zygotes also show loss of methylation from one pronucleus (g) followed by a further stepwise decline in methylation to the eight-cell stage (h–j). De novo methylation by the 16-cell stage results in heterogeneity with highly and moderately methylated nuclei (k) such that at the blastocyst stage (l) the ICM contains highly methylated nuclei and the trophectoderm moderately methylated ones. (B) To better define the location of the methylated nuclei images are presented with the methylation signal (red) and the merged image of the DNA (blue) superimposed on the methylation signal (pink). This superimposition of images clearly shows that in the mouse the ICM has become remethylated, but in bovine nuclei both ICM and trophectoderm are methylated.
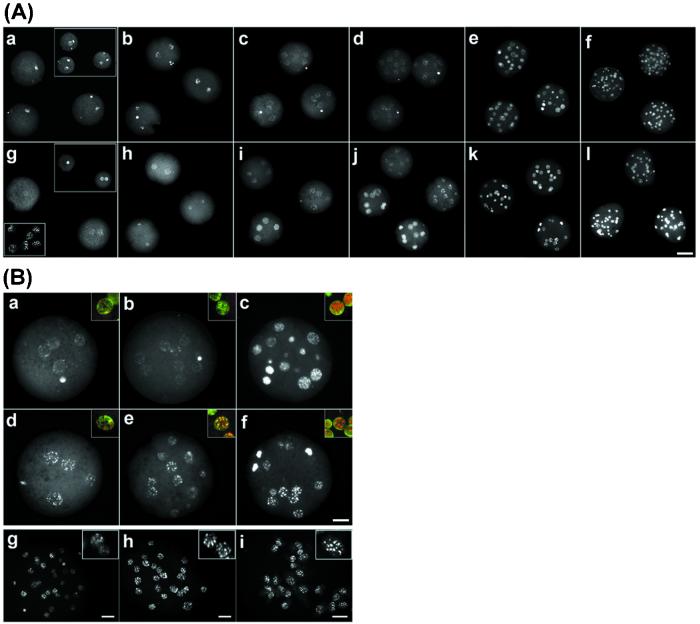
Figure 3.
Aberrant methylation patterns in cloned bovine preimplantation embryos. (Aa–Af) Anti-5-methyl cytosine immunofluorescence of normal bovine embryos. (a) Zygotes 12 h after fertilization [(Inset) staining of the DNA; n = 25, number of embryos analyzed]. (b) Two-cell embryos stain intensely (n > 10). (c and d) Four-cell embryos (n > 15; c) and eight-cell embryos (d) show reduced staining (n >10). (e) Ten- to sixteen-cell embryos undergo a dramatic increase in methylation (n >10). ( f) Morulae (>24 cells) maintain the high-intensity signal (n > 10). (g–l) Cloned embryos. (g) Cloned one-cell embryos stain faintly (n > 10). (Inset) DNA staining. (Lower Inset) Fibroblast donor nuclei. (h) Two-cell cloned embryos (n = 15). (i and j) In four- (i) and eight-cell (j) clones there are two populations of embryos with high and low staining, respectively (n = 7/14), in each group. (k) Ten to sixteen cells (n = 20). (l) Morula stage (n > 10). (Scale bar, 50 μm.) (B) Organization of methylation patterns in normal and cloned bovine embryos. (a–c) Normal bovine embryos at four- to 16-cell stage. (d–f) Cloned bovine embryos at the 4- to 16-cell stage. Note the heterogeneous de novo methylation and fine granular staining in normal 16-cell embryos (c) and the precocious de novo methylation and organization into fewer and more intense foci in cloned 4- to 16-cell embryos (d–f). (Insets) Merged images of methylation staining (red) and DNA staining (green), with yellow showing overlapping patterns. (g–i) Confocal projection of normal and cloned morula, and fibroblast donors. (g) Normal morula stains heterogeneously. (Inset) Nuclei indicate the two patterns of staining. (h) Cloned morula stains homogeneously. (i) Methylation organization in fetal fibroblast donor cells. (Scale bar, 20 μm.) All insets are enlarged ×3.
Aberrant Reprogramming in Clones.
Because the three basic methylation reprogramming events appeared to be conserved in mammalian preimplantation embryos, it was important to determine the extent to which a highly methylated somatic nucleus would be reprogrammed during cloning. Bovine fetal fibroblast nuclei were used as donors for cloned embryos (23); in previous and parallel experiments these experiments typically resulted in rates of development to blastocysts in the range of 30–50%, and in live births in 2–5% (23). The fibroblast nuclei were uniformly highly methylated but with a pattern that was concentrated in much fewer but larger and more intense foci than in normal 16-cell embryos (Fig. 3Bi). The large intense foci have been attributed to pericentromeric heterochromatin in bovine somatic cells (26). Because there are fewer foci than there are centromeres, this finding means that centromeres of different chromosomes are probably colocalized in specific areas. On introduction of fibroblast nuclei into enucleated oocytes and activation, pseudopronuclei are formed in the zygote (Fig. 3A). These stained much less intensely than the female pronucleus in the normal zygote, consistent with loss of methylation from the somatic nucleus in the reconstituted zygote (Fig. 3A). The comparison at the two-cell stage shows this point very clearly. Whereas there was substantial staining in both nuclei in the normal embryo, staining was considerably reduced in cloned two-cell embryos (Fig. 3 Ab and Ah). Because the zygotic genome is approximately half as methylated as a somatic nucleus, considerable demethylation must have occurred.
After the two-cell stage, however, cloned embryos did not appear to undergo further demethylation, in contrast to the normal ones (Fig. 3). Instead, in cloned four-cell and eight-cell embryos there was heterogeneity between individual embryos, with their methylation patterns falling into two groups. In approximately half of the embryos all nuclei stained relatively dimly; they were comparable to cloned two-cell embryos. Strikingly, however, in the other half all nuclei stained very brightly and had clearly undergone de novo methylation by the four-cell or eight-cell stage and thus were ahead of schedule (Fig. 3). This precocious de novo methylation was associated with a characteristic change in morphology of the methylation pattern. In some of the cloned four-cell embryos, the finely granular pattern that is characteristic of the normal 16-cell embryo was observed; however, in others there was a significant change in the methylation pattern of the nucleus. This altered pattern persisted in all subsequent stages evaluated; was characterized by methylation foci that were much fewer, larger, and more intense; and was reminiscent of the donor fibroblast nuclei (Fig. 3 Bd–Bf). Although we cannot exclude the possibility that these changes are brought about by demethylation of specific regions and de novo methylation of others, we feel it is more likely that the change in pattern is brought about by a different intranuclear organization of the DNA, because similar changes can be seen when we stain for DNA only (Fig. 3B).
As a result of the aberrant reprogramming events in cloned embryos, particularly of precocious de novo methylation, all nuclei in cloned morulae stained very brightly, and the pattern of staining was very similar to that of fibroblast donors (Fig. 3Bh). In contrast, normal morulae were much more heterogeneous in their levels of staining, and many nuclei stained only dimly (Fig. 3Bg).
Discussion
Our study shows that methylation reprogramming is conserved in eutherian mammals and that somatic nuclei undergo some genome-wide reprogramming events in cloned embryos, but that in most cloned embryos aspects of reprogramming are aberrant (Fig. 3). Rapid demethylation of the paternal genome in the zygote is conserved in the eutherian mammals tested. The mechanism of this presumably active demethylation process remains unknown; recent studies in the mouse that made use of an MBD2 knockout have excluded this protein as a candidate in vivo (27). However, because there is no early demethylation in Xenopus and zebrafish, organisms without imprinting, the observed conservation in mammals lends further support to the suggestion that paternal demethylation is related to imprinting (28).
Further loss of methylation during cleavage-stage divisions is consistent with passive demethylation (of predominantly the maternal genome) occurring as in the mouse. Thus Dnmt1 is presumably absent or excluded from the nucleus, which is similar to the finding in mouse embryos. However, substantial de novo methylation occurs in bovine embryos at the 8–16-cell stage. This finding is remarkable for two reasons. One is that it coincides with the major wave of transcriptional activation of the embryonic genome (29), suggesting the possibility that de novo methylation enzymes such as Dnmt3a or -b are activated at that stage. The other reason is that this stage is precisely the same one at which in the mouse Dnmt1 enters the nucleus for one cell cycle, being important at this stage for maintenance of imprinted methylation (14). The early de novo methylation in bovine embryos leads to relatively higher levels of methylation in trophectoderm cells of the blastocyt than in the mouse. However, de novo methylation does occur in mouse blastocysts but is limited to ICM cells. This finding explains why mouse embryonic stem cells are relatively highly methylated (5). Thus the first differentiation event in mammalian embryos (that of trophectoderm and ICM cells) and the resulting loss of totipotency (of ICM cells) is accompanied by genome-wide de novo methylation. Because in cattle extraembryonic tissues in the placenta are required for a much longer time (more than 270 days) than in the mouse (15 days), their higher methylation level may confer added stability on the differentiated state.
Somatic donor nuclei appear to be considerably demethylated in the recipient oocyte within hours of their introduction and activation. This demethylation may occur by the same putative active demethylation mechanism by which the paternal genome is demethylated at fertilization, which may suggest that an important trigger for demethylation is remodeling of chromatin by factors present in the oocyte cytoplasm. Presence of the chromatin factor imitation switch (ISWI) has recently been shown to be important for remodeling in cloned Xenopus embryos (30). If active demethylation occurs in the somatic nucleus in clones, this raises the question of whether differential methylation in imprinted genes will be protected against demethylation, or whether clones could have altered imprinting patterns. Mice cloned from embryonic stem cells can have altered imprinting patterns (31) because embryonic stem donor cells appear to be epigenetically unstable (32). Imprinting defects could lead to postimplantation failure of cloned embryos, which often show characteristic placental abnormalities (17–19), or to perinatal failures associated with abnormal functioning of the cardiovascular system in particular (17–19), which are also characteristic of altered imprinted gene expression.
Cloned embryos seemed to lack further passive demethylation, and a large proportion became precociously methylated de novo at the four- and eight-cell stages. Although the bovine Dnmts have not been extensively characterized, by analogy with the mouse the oocyte form of Dnmt1 may be excluded from the nucleus, whereas the somatic form of Dnmt1 will have been introduced with the somatic nucleus and may remain associated with it. This pattern of exclusion and association may result in increased levels of Dnmt1, and passive demethylation may therefore not occur in clones. De novo methylation is likely to be caused by Dnmt3a and -b; whether these enzymes are present in the oocyte cytoplasm or are made from embryonic transcripts is not known. It is striking that precocious de novo methylation occurred in cloned embryos mainly at the four-cell stage, at the same stage at which precocious transcriptional activation of somatic donor nuclei in cloned bovine embryos has been observed (33). On the other hand, many somatic tissues continue to express Dnmt3a and -b, so it is possible that transcription of these genes is not silenced in transferred somatic nuclei.
Precocious de novo methylation and precocious nuclear reorganization in cloned embryos may be independent events. On the other hand, de novo methylation in cloned embryos at the four-cell stage was accompanied by nuclear reorganization, whereas in normal embryos de novo methylation at the 16-cell stage was followed by nuclear reorganization at the morula to blastocyst stage. It is therefore possible that genome-wide methylation, nuclear reorganization, and differentiation are intricately linked and lead to stable programs of gene expression and repression (34). Perturbation of the normal timing of these events, as observed in most cloned embryos here, may lead to aberrant development, which would be consistent with the very substantial losses of cloned embryos during preimplantation and early postimplantation development (17–19). Cloned mouse fetuses had normal patterns of X chromosome inactivation, suggesting that this aspect of epigenetic reprogramming was successful in embryos that survived (35). Our results show that global epigenetic reprogramming of somatic nuclei is aberrant in most preimplantation cloned embryos; the effects on expression of individual genes, particularly imprinted ones, have yet to be addressed. Our study strongly supports the view that correct epigenetic reprogramming is necessary for successful and normal development of clones.
Acknowledgments
We thank A. Niveleau for providing the antibody against 5-methyl cytosine and P. Lipp for expert help with confocal microscopy. This work was funded by Biotechnology and Biological Sciences Research Council Grant GTH 12511.
Abbreviation
- ICM
inner cell mass
Note Added in Proof.
A complementary analysis of metaphase chromosomes in cloned embryos using the 5-methyl cytosine antibody has been published by Bourc'his et al. (36).
References
- 1.Bird A P, Wolffe A P. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 2.Bestor T H. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 3.Stancheva I, Meehan R R. Genes Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 5.Okano M, Bell D W, Haber D A, Li E. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 6.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 7.Reik W, Dean W, Walter J. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 8.Surani A. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 9.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 10.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Nature (London) 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 11.Monk M, Boubelik M, Lehnert S. Development (Cambridge, UK) 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 12.Howlett S K, Reik W. Development (Cambridge, UK) 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 13.Rougier N, Bourc'his D, Gomes D M, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Genes Dev. 1998;12:2108–2118. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell CY, Bestor T H, Ding F, Latham K E, Mertineit C, Trasler J M, Chaillet J R. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 15.Reik W, Walter J. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 16.Macleod D, Clark V H, Bird A. Nat Genet. 1999;23:139–140. doi: 10.1038/13767. [DOI] [PubMed] [Google Scholar]
- 17.Wilmut I, Young L, Campbell K H. Reprod Fertil Dev. 1998;10:639–643. doi: 10.1071/rd98047. [DOI] [PubMed] [Google Scholar]
- 18.Solter D. Nat Rev Genet. 2000;1:199–207. doi: 10.1038/35042066. [DOI] [PubMed] [Google Scholar]
- 19.Colman A. Cloning. 2000;1:185–199. doi: 10.1089/15204559950019825. [DOI] [PubMed] [Google Scholar]
- 20.Kikyo N, Wolffe A P. J Cell Sci. 2000;113:11–20. doi: 10.1242/jcs.113.1.11. [DOI] [PubMed] [Google Scholar]
- 21.Kang Y K, Koo D B, Park J S, Choi Y H, Chung A S, Lee K K, Han Y M. Nat Genet. 2001;28:173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- 22.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 23.Zakhartchenko V, Durcova-Hills G, Stojkovic M, Schernthaner W, Prelle K, Steinborn R, Muller M, Brem G, Wolf E. J Reprod Fertil. 1999;115:325–331. doi: 10.1530/jrf.0.1150325. [DOI] [PubMed] [Google Scholar]
- 24.Coffigny H, Bourgeois C, Ricoul M, Bernardino J, Vilain A, Niveleau A, Malfoy B, Dutrillaux B. Cytogenet Cell Genet. 1999;87:175–181. doi: 10.1159/000015460. [DOI] [PubMed] [Google Scholar]
- 25.Jabbari J, Caccio S, Barros J P P, Desgres J, Bernardi G. Gene. 1997;205:109–118. doi: 10.1016/s0378-1119(97)00475-7. [DOI] [PubMed] [Google Scholar]
- 26.Schnedl W, Erlanger B F, Miller O J. Hum Genet. 1976;31:21–26. doi: 10.1007/BF00270395. [DOI] [PubMed] [Google Scholar]
- 27.Santos, F., Hendrich. B., Reik, W. & Dean, W. (2001) Dev. Biol., in press. [DOI] [PubMed]
- 28.Reik W, Walter J. Nat Genet. 2001;27:255–256. doi: 10.1038/85804. [DOI] [PubMed] [Google Scholar]
- 29.Memili E, First N L. Zygote. 2000;8:87–96. doi: 10.1017/s0967199400000861. [DOI] [PubMed] [Google Scholar]
- 30.Kikyo N, Wade P A, Guschin G, Ge H G, Wolffe A P. Science. 2000;289:2360–2362. doi: 10.1126/science.289.5488.2360. [DOI] [PubMed] [Google Scholar]
- 31.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout W M, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 32.Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses J J, Reik W, Feil R. Development (Cambridge, UK) 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 33.Kanka J, Smith S D, Soloy E, Holm P, Callesen H. Mol Reprod Dev. 1999;52:253–263. doi: 10.1002/(SICI)1098-2795(199903)52:3<253::AID-MRD2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Brown K E, Baxter J, Graf D, Merkenschlager M, Fisher A G. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 35.Eggan K, Akutsu H, Hochedlinger K, Rideout W, III, Yanagimachi R, Jaenisch R. Science. 2000;290:1578–1581. doi: 10.1126/science.290.5496.1578. [DOI] [PubMed] [Google Scholar]
- 36.Bourc'his D, Le Bourhis D, Patin D, Niveleau A, Comizzoli P, Renard J, Viegas-Pequignot E. Curr Biol. 2001;11:1542–1546. doi: 10.1016/s0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]