Abstract
The pentose phosphate pathway (PPP) plays an important role in the synthesis of ribonucleotides and aromatic amino acids. During bioethanol production from cellulosic biomass composed mainly of d-glucose and d-xylose, the PPP is also involved in xylose metabolism by engineered Saccharomyces cerevisiae. Although the activities and thermostabilities of the four PPP enzymes (transaldolase: TAL1, transketolase: TKL1, ribose-5-phosphate ketol-isomerase: RKI1 and d-ribulose-5-phosphate 3-epimerase: RPE1) can affect the efficiency of cellulosic ethanol production at high temperatures, little is known about the suitable expression levels of these PPP genes. Here, we overexpressed PPP genes from S. cerevisiae and the thermotolerant yeast Kluyveromyces marxianus either singly or in combination in recombinant yeast strains harboring a mutant of xylose isomerase (XI) and evaluated xylose consumption and ethanol production of these yeast transformants in glucose/xylose mixed media at 36 °C. Among the PPP genes examined, we found that: (1) strains that overexpressed S. cerevisiae TKL1 exhibited the highest rate of xylose consumption relative to strains that overexpressed other PPP genes alone; (2) overexpression of RKI1 and TAL1 derived from K. marxianus with S. cerevisiae TKL1 increased the xylose consumption rate by 1.87-fold at 24 h relative to the control strain (from 0.55 to 1.03 g/L/h); (3) the strains with XI showed higher ethanol yield than strains with xylose reductase and xylitol dehydrogenase and (4) PHO13 disruption did not improve xylose assimilation under the experimental conditions. Together these results indicated that optimization of PPP activity improves xylose metabolism in genetically engineered yeast strains, which could be useful for commercial production of ethanol from cellulosic material.
Electronic supplementary material
The online version of this article (10.1186/s13568-018-0670-8) contains supplementary material, which is available to authorized users.
Keywords: Bio-ethanol, Glucose/xylose co-fermentation, Xylose isomerase, Thermostability, Saccharomyces cerevisiae, Kluyveromyces marxianus
Introduction
Second-generation technology has been developed for bioethanol production from non-edible biomass composed of cellulose and hemicellulose, which are sugar polymers of d-glucose, d-xylose and l-arabinose (Naik et al. 2010) that can be hydrolyzed by cellulase and hemicellulase (Bhattacharya et al. 2015). The yeast Saccharomyces cerevisiae readily produces ethanol from d-glucose, but not from pentose sources such as d-xylose and l-arabinose (Young et al. 2010). In 1990, introduction of two Scheffersomyces stipites (formerly known as Pichia stipites) genes, XYL1 and XYL2, encoding NADPH-dependent xylose reductase (XR) and NAD+-dependent xylitol dehydrogenase (XDH), respectively, into S. cerevisiae was shown to allow successful xylose fermentation (Kötter et al. 1990). Moreover, various genetic engineering trials with endogenous or exogenous genes have been applied to yeast transformants carrying xylose-metabolizing enzyme genes to improve ethanol production in xylose-containing media. For instance, xylose metabolism was markedly improved by deletion of the putative alkaline phosphate gene PHO13 in XLY1/XYL2 transformants (Van Vleet et al. 2008; Kim et al. 2015; Xu et al. 2016; Kobayashi et al. 2017). Overexpression of HXT7, a known xylose-permeable hexose transporter in S. cerevisiae, also improved xylose consumption (Gonçalves et al. 2014). However, critical issues remained for efficient xylose fermentation by XYL1/XYL2 transformants such as accumulation of xylitol, sedoheptulose-7-phosphate and pyruvate, and depletion of fructose-1,6-bisphosphate because of insufficient capacity of the glycolytic pathway (Kötter and Ciriacy 1993). An imbalance of cofactors can also slow xylose metabolism (Van Vleet and Jeffries 2009). Previous studies reported that overexpression of non-oxidative pentose phosphate pathway (PPP) genes did not improve the xylose fermentation rate (Walfridsson et al. 1995; Johansson and Hahn-Hägerdal 2002; Matsushika et al. 2009).
Another strategy to enhance xylose metabolism in S. cerevisiae depended on the xylose isomerase (XI) gene derived from Thermus thermophiles (Walfridsson et al. 1996). The S. cerevisiae XI transformants had a better ethanol yield than did XYL1/XYL2 since production inhibition by xylitol and cofactor imbalances was avoided (Brat et al. 2009). Meanwhile, deletion of either PHO13 (Lee et al. 2014; Bamba et al. 2016) or the aldo–keto reductase gene GRE3 (Träff et al. 2001), as well as introduction of an HXT7 mutant (Reider Apel et al. 2016) improved xylose metabolism in the XI transformants.
In addition to introduction of the xylose metabolism gene, balanced expression of PPP genes is important for bioethanol production (Kuyper et al. 2005). Many studies revealed that overexpression of four non-oxidative PPP genes, transaldolase (TAL1), transketolase (TKL1), ribose-5-phosphate ketol-isomerase (RKI1) and d-ribulose-5-phosphate 3-epimerase (RPE1) increased ethanol production in medium in which d-xylose was the sole carbon source (Kuyper et al. 2005; Karhumaa et al. 2007; Lee et al. 2014; Qi et al. 2015). However, the effect of exogeneous expression of PPP genes on the rates of xylose consumption and ethanol production, particularly in a glucose/xylose-mixed medium, remained unclear, and no studies had addressed suitable activities for PPP enzymes that would enhance xylose metabolism. For ethanol production from multiple sugar sources, the metabolic flow of each sugar through the glycolytic pathway (GP) and PPP may be intricately involved in xylose metabolism. Furthermore, fermentation at high temperatures is preferable in the simultaneous saccharification and co-fermentation (SSCF) process because the optimal temperature for the cellulase reaction is much higher than that for fermentation by most ethanologenic microorganisms (Paulova et al. 2014; Kawaguchi et al. 2016).
To explore the optimal expression levels of PPP genes needed to maximize ethanol production from cellulosic sources, here we systematically investigated the effect of overexpression of single and multiple PPP genes derived from S. cerevisiae and the thermotolerant yeast Kluyveromyces marxianus in a S. cerevisiae strain harboring the XI gene that allows glucose/xylose co-fermentation at high temperatures. We explored differences in the favorable combination of PPP genes in S. cerevisiae between our previous study using XR-XDH transformants in medium having d-xylose as the sole sugar (Kobayashi et al. 2017) and this study using the XI transformants in the glucose/xylose-mixed medium.
Materials and methods
Media, general cultivation conditions, plasmid vectors, oligonucleotides and establishment of engineered S. cerevisiae strains
Yeast were cultivated in YPD medium (20 g/L Bacto™ peptone (BD Biosciences, Franklin Lakes, NJ, USA), 10 g/L Bacto™ yeast extract (BD Biosciences) and 20 g/L d-glucose (Sigma-Aldrich, St. Louis, MO, USA)) on YPD agar plates (20 g/L Bacto™ Agar, BD Biosciences) with Geneticin® (Thermo Fisher Scientific, Waltham, MA, USA), Zeocin™ (Thermo Fisher Scientific) or Aureobasidin A (Takara Bio, Shiga, Japan) if necessary.
All yeast strains used in this study are listed in Table 1 with their genotypes. S. cerevisiae IR-2, a flocculating yeast isolated from waste water from a food-processing company in Indonesia, was used as the host strain (Kuriyama et al. 1985). A draft genome sequence of IR-2 was also available (Sahara et al. 2014, Accession Nos. BAUI01000001–BAUI01000322). Based up on the genome sequence, strain SS29 was first constructed as a reference strain. SS29 carries a deletion mutation of the HO gene, which maintains the haploid state, and in the GRE3 gene, an intrinsic xylose reductase gene in S. cerevisiae that suppresses xylitol production (Träff et al. 2001). A mutant xylose isomerase gene derived from Lachnoclostridium phytofermentans LpXI(T63I) (Brat et al. 2009, Accession No. LC390328) driven by the HSP12 promoter of S. cerevisiae was constructed in an AUR101 plasmid vector (Takara-Bio) with an additional copy of the XKS1 gene from Saccharomyces cerevisiae IR-2 strain that provided compensatory xylulokinase activity. This plasmid was linearized by StuI digestion and introduced at the AUR1 locus of SS29 by single crossover homologous recombination. Selection with Aureobasidin A established the basal strain SS82. For chromosomal integration of exogeneous PPP gene(s), recipient vectors were constructed with the neomycin-resistant gene cassette kanMX6 and several combinations of PPP genes from S. cerevisiae and/or K. marxianus DMB1, which are codon-optimized for suitable expression in S. cerevisiae, and inserted between the TDH3 promoter and CYC1 terminator of S. cerevisiae (Kobayashi et al. 2017, Accession Nos. LC390324 for KmRKI1opt, LC390325 for KmRPE1opt, LC390326 for KmTAL1 and LC390327 for ScTKL1). The DNA fragments of the PPP gene cassettes including kanMX6 were amplified with primers to yield 50 bp overhangs for deletion of GPD2 or PHO13 loci via transformation (Additional file 1: Table S1). Yeast transformation was performed using a traditional LiOAc method (Gietz and Schiestl 2007). All DNA sequences of the constructed plasmid vectors and DNA fragments were verified by Sanger sequencing (Eurofins Japan, Tokyo Japan).
Table 1.
S. cerevisiae strains used in this study
| Code | Genotype, full description | |
|---|---|---|
| IR-2 | MATa/α | (Kuriyama et al. 1985) |
| IR-2 2a-3-34A | MATα hoΔ::bleMX6 | Lab. stock |
| SS29 | IR-2 2a-3-34A gre3Δ::hphMX6 | (Seike et al. unpublished observations) |
| SS82 | SS29 AUR1-C::HSP12p-XImut-CYC1t-XKS1-PGK1p | (Seike et al. unpublished observations) |
| SS118 | SS82 gpd2Δ::kanMX6 | (Seike et al. unpublished observations) |
| YK184 | SS82 gpd2Δ::TDH3p-KmTAL1opt-CYC1t-kanMX6 | This study |
| YK185 | SS82 gpd2Δ::TDH3p-ScTKL1-CYC1t-kanMX6 | This study |
| YK183 | SS82 gpd2Δ::TDH3p-KmRKI1opt-CYC1t-kanMX6 | This study |
| YK186 | SS82 gpd2Δ::TDH3p-KmRPE1opt-CYC1t-kanMX6 | This study |
| YK223 | SS82 gpd2Δ::TDH3p-KmTAL1opt-CYC1t-TDH3p-ScTKL1-CYC1t-kanMX6 | This study |
| YK193 | SS82 gpd2Δ::TDH3p-KmRKI1opt-CYC1t-TDH3p-ScTKL1-CYC1t-kanMX6 | This study |
| YK224 | SS82 gpd2Δ::TDH3p-KmRPE1opt-CYC1t-TDH3p-ScTKL1-CYC1t-kanMX6 | This study |
| YK246 | SS82 gpd2Δ::TDH3p-KmRKI1opt-CYC1t-TDH3p-KmTAL1opt-CYC1t- TDH3p-ScTKL1-CYC1t-kanMX6 |
This study |
| YK247 | SS82 gpd2Δ::TDH3p-KmRPE1opt-CYC1t-TDH3p-KmTAL1opt-CYC1t- TDH3p-ScTKL1-CYC1t-kanMX6 |
This study |
| YK248 | SS82 gpd2Δ::TDH3p-KmRKI1opt-CYC1t-TDH3p-KmRPE1opt-CYC1t- TDH3p-KmTAL1opt-CYC1t -kanMX6 |
This study |
| YK249 | SS82 gpd2Δ::TDH3p-KmRKI1opt-CYC1t-TDH3p-KmRPE1opt-CYC1t- TDH3p-ScTKL1-CYC1t-kanMX6 |
This study |
| YK197 | SS82 gpd2Δ::TDH3p-KmRKI1opt-CYC1t-TDH3p-KmRPE1opt-CYC1t- TDH3p-KmTAL1opt-CYC1t-TDH3p-ScTKL1-CYC1t-kanMX6 |
This study |
| YK001 | IR-2 2a-3-12A AUR1-C::PGK1p-SsXR-PGK1t-PGK1p-SsXDH-PGK1t- PGK1p-ScXKS1-PGKt |
(Kobayashi et al. 2017) |
| YK002 | YK001 pho13Δ::kanMX6 | (Kobayashi et al. 2017) |
| YK115 | YK001 pho13Δ::TDH3p-ScRKI1-CYC1t-TDH3p-ScRPE1-CYC1t- TDH3p-ScTAL1-CYC1t-TDH3p-ScTKL1-CYC1t-kanMX6 |
(Kobayashi et al. 2017) |
| YK149 | SS82 pho13Δ::blaMX6 | This study |
| YK150 | SS82 pho13Δ::TDH3p-ScRKI1-CYC1t-TDH3p-ScRPE1-CYC1t- TDH3p-ScTAL1-CYC1t-TDH3p-ScTKL1-CYC1t-blaMX6 |
This study |
Fermentation examination and quantitative analyses of fermentation performance by HPLC
Recombinant yeast strains were cultivated at 30 °C in YPD media under aerobic conditions by shaking at 150 rpm for 2 days until the stationary phase was reached. The cells were recovered and washed immediately with YPDX medium (20 g/L Bacto™ peptone, 10 g/L Bacto™ yeast extract, 85 g/L d-glucose and 35 g/L d-xylose) and diluted to an OD600 of 3 in 70 mL of YPDX medium. A silicon plug with needles and a three-way stopcock for sampling and aeration was used as a cap to aliquot the culture medium in 100 mL Erlenmeyer flasks. Fermentation studies were performed at 36 °C for up to 72 h with periodic sampling under micro-aerobic conditions.
The supernatants were collected by centrifugation, and the concentrations (g/L) of residual sugars, produced polyols, organic acids and ethanol in the fermentation media were measured using an HPLC equipped with a refractive index (RI) detector (JASCO Co., Tokyo, Japan). For separation of these chemical compounds, an Aminex HPX-87 H column (Bio-Rad, CA, USA) with a guard column (Cation H Cartridges 30 × 4.6 mm, Bio-Rad) was used under the following conditions: column oven temperature of 65 °C, 5 mM H2SO4 as the mobile phase buffer, and flow rate of 0.6 mL/min. Further details are described in our previous publication (Kobayashi et al. 2017).
Results
Effects of overexpressing a single PPP gene on glucose/xylose co-fermentation
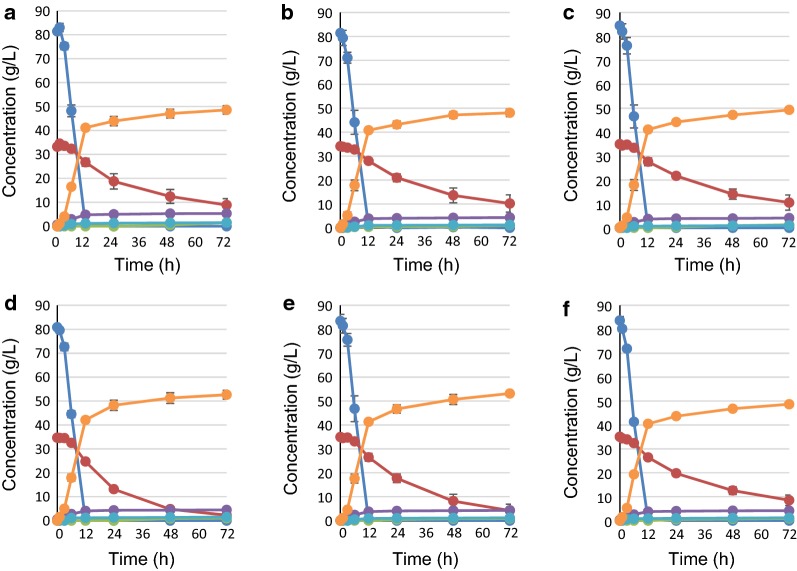
To determine the effect of overexpression of each PPP gene on glucose/xylose co-fermentation at 36 °C, batch-fermentation was first carried out under microaerobic conditions using strains YK184 (KmTAL1opt), YK185 (ScTKL1), YK183 (KmRKI1opt) and YK186 (KmRPE1opt) and the fermentation profiles and metabolic characteristics were determined (Fig. 1, Table 2, Additional file 2: Table S2). Under microaerobic conditions, the control strain SS82 consumed approximately 85 g/L of d-glucose within 12 h, by which point the d-xylose concentration had slightly decreased. Across 72 h, the ethanol concentration gradually increased to reach 48.6 g/L. The xylose consumption rate over 24 h was about 0.60 g/L/h, and the ethanol yield over 72 h was 0.41 g-ethanol/g-total sugars (Table 2). Under this co-fermentation condition, consumption of d-xylose was incomplete at 72 h; an alternative calculation for the ethanol yield for 72 h was 0.44 g-ethanol/g-consumed sugars (calculation from Additional file 2: Table S2). Notably, 1.5 g/L xylitol was detected at 72 h even though GRE3 was disrupted in SS82 (Fig. 1a, Additional file 2: Table S2). SS118, a control strain for overexpression of PPP gene(s), had a metabolic profile for co-fermentation that was similar to that of SS82 (Fig. 1b). The xylose consumption rate of YK185 expressing S. cerevisiae TKL1 was 1.66-fold higher than that of SS118 at 24 h, whereas YK183 expressing K. marxianus RKI1 was about 1.42-fold higher. Meanwhile, YK184 and YK186 showed no obvious improvement in xylose consumption or ethanol production rates (Figs. 1c–f). The amount of residual xylose for YK185 and YK183 was less than 5 g/L at 72 h. Therefore, the ethanol yields for these strains were clearly increased at 72 h (0.45 g-ethanol/g-total sugars) relative to the control strain SS118, while no change in the ethanol yields for YK184 and YK186 was observed (0.40–0.41 g-ethanol/g-total sugars). Together these results indicate that expression of TKL1 and RKI1 could contribute to increased xylose metabolism and concurrent increased ethanol yield.
Fig. 1.
Fermentation profiles of strains expressing XI and overexpressing single PPP genes. Cells were initially inoculated at OD600 = 3 and cultivated in medium containing 85 g/L glucose and 35 g/L xylose for 72 h at 36 °C. a SS82, b SS118 (GPD2Δ), c YK184 (KmTAL1opt), d YK185 (ScTKL1), e YK183 (KmRKI1opt), f YK186 (KmRPE1opt). Blue lines: glucose, Red lines: xylose, Green lines: xylitol, Purple lines: glycerol, Light blue lines: acetate, Orange lines: ethanol. Each value is an average of triplicate cultures
Table 2.
Comparison of fermentation performance
| Strain | Yeth (g/g) | Cxyl (g/L/h) | ||
|---|---|---|---|---|
| 72 h | 24 h | 48 h | 72 h | |
| SS82 | 0.41 ± 0.01 | 0.60 ± 0.08 | 0.43 ± 0.04 | 0.34 ± 0.02 |
| SS118 | 0.40 ± 0.02 | 0.55 ± 0.07 | 0.43 ± 0.06 | 0.33 ± 0.05 |
| YK184 | 0.41 ± 0.01 | 0.55 ± 0.03 | 0.44 ± 0.04 | 0.34 ± 0.04 |
| YK185 | 0.45 ± 0.03 | 0.90 ± 0.04 | 0.62 ± 0.02 | 0.45 ± 0.01 |
| YK183 | 0.45 ± 0.02 | 0.72 ± 0.06 | 0.56 ± 0.06 | 0.43 ± 0.04 |
| YK186 | 0.41 ± 0.01 | 0.64 ± 0.05 | 0.47 ± 0.04 | 0.37 ± 0.04 |
| YK223 | 0.44 ± 0.01 | 0.73 ± 0.06 | 0.58 ± 0.04 | 0.44 ± 0.02 |
| YK193 | 0.46 ± 0.01 | 0.94 ± 0.03 | 0.66 ± 0.02 | 0.46 ± 0.01 |
| YK224 | 0.44 ± 0.00 | 0.71 ± 0.04 | 0.55 ± 0.03 | 0.42 ± 0.02 |
| YK246 | 0.45 ± 0.01 | 1.03 ± 0.03 | 0.68 ± 0.01 | 0.48 ± 0.01 |
| YK247 | 0.43 ± 0.01 | 0.76 ± 0.08 | 0.58 ± 0.04 | 0.44 ± 0.01 |
| YK248 | 0.43 ± 0.00 | 0.82 ± 0.01 | 0.61 ± 0.00 | 0.44 ± 0.00 |
| YK249 | 0.44 ± 0.02 | 0.83 ± 0.02 | 0.61 ± 0.02 | 0.45 ± 0.01 |
| YK197 | 0.42 ± 0.01 | 0.41 ± 0.08 | 0.33 ± 0.06 | 0.26 ± 0.04 |
| YK001 | 0.39 ± 0.01 | 0.68 ± 0.08 | 0.44 ± 0.07 | 0.31 ± 0.05 |
| YK002 | 0.38 ± 0.00 | 0.62 ± 0.03 | 0.40 ± 0.07 | 0.28 ± 0.06 |
| YK115 | 0.35 ± 0.01 | 0.32 ± 0.13 | 0.17 ± 0.07 | 0.13 ± 0.04 |
| YK149 | 0.40 ± 0.00 | 0.61 ± 0.02 | 0.43 ± 0.01 | 0.33 ± 0.00 |
| YK150 | 0.40 ± 0.01 | 0.50 ± 0.02 | 0.36 ± 0.03 | 0.29 ± 0.03 |
Effects of overexpressing multiple PPP genes on glucose/xylose co-fermentation
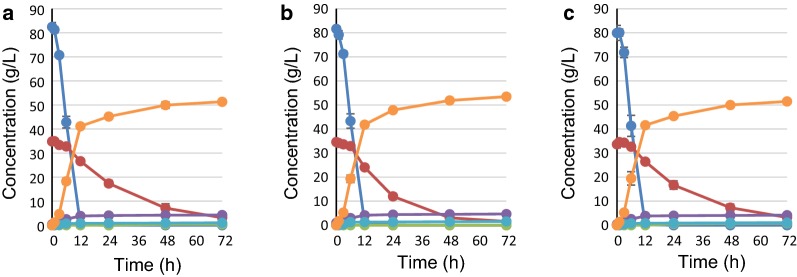
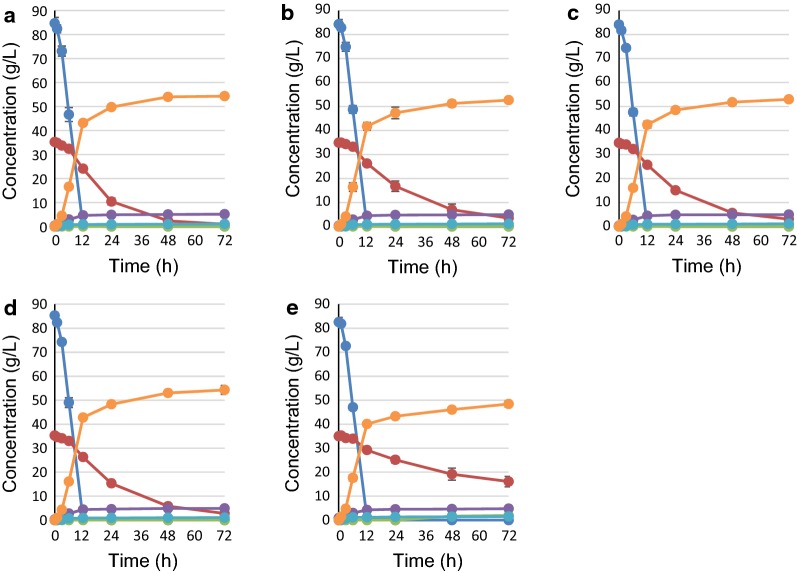
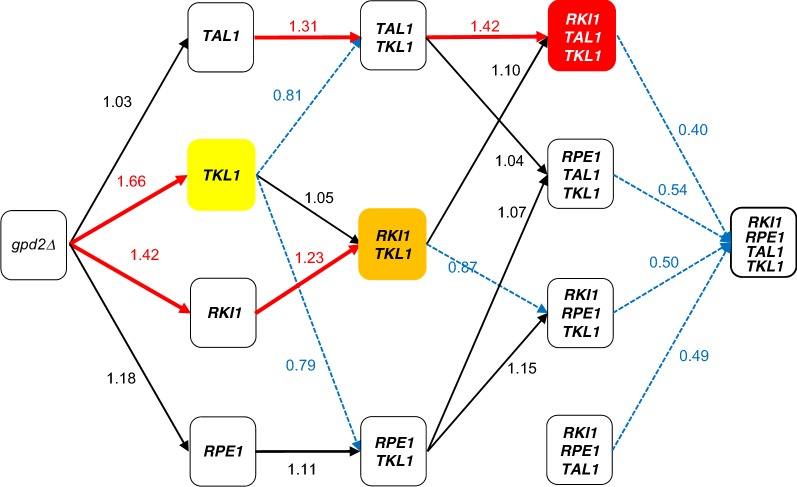
Based on the results described above, the fermentation profiles and metabolic characteristics of strains YK223 (gpd2Δ::KmTAL1opt-ScTKL1), YK193 (gpd2Δ::KmRKI1opt-ScTKL1) and YK224 (gpd2Δ::KmRPE1opt-ScTKL1) that co-expressed two PPP genes; and YK246 (gpd2Δ::KmRKI1opt- KmTAL1opt-ScTKL1), YK247 (gpd2Δ::KmRPE1opt-KmTAL1opt-ScTKL1), YK248 (gpd2Δ::KmRKI1opt-KmRPE1opt-KmTAL1opt) and YK249 (gpd2Δ::KmRKI1opt-KmRPE1opt-ScTKL1) that co-expressed three PPP genes; as well as YK197 that co-expressed all PPP genes (gpd2Δ::KmRKI1opt-KmRPE1opt-KmTAL1opt-ScTKL1) were examined on glucose/xylose co-fermentation at 36 °C (Figs. 2, 3, Table 2, Additional file 2: Table S2). The xylose consumption rates at 24 h for strains YK223 (co-express KmTAL1 and ScTKL1), YK193 (co-express KmRKI1 and ScTKL1) and YK224 (co-express KmRPE1 and ScTKL1) increased by about 1.32-, 1.71- and 1.29-fold, respectively, relative to SS118. The addition of ScTKL1 to singly expressed KmTAL1opt (YK184) and KmRKI1opt (YK183) increased xylose consumption rates at 24 h by about 1.31- and 1.23-fold, respectively, whereas no increase was seen for co-expression of ScTKL1 with KmRKI1opt (YK193). The xylose consumption rates at 24 h for strains co-expressing three PPP genes (YK246 (KmRKI1- KmTAL1- ScTKL1), YK247 (KmRPE1- KmTAL1- ScTKL1), YK248 (KmRKI1- KmRPE1- KmTAL1) and YK249 (KmRKI1- KmRPE1- ScTKL1) increased by 1.87-, 1.38-, 1.49- and 1.51-fold, respectively, compared to SS118. The xylose consumption rate at 24 h for YK246 increased by 1.42- and 1.10-fold relative to that for YK223 and YK193, respectively. Meanwhile, the xylose consumption rate at 24 h for YK247 did not increase compared to that for YK223 or YK224. The xylose consumption rate for YK249 at 24 h increased by 1.17-fold over that for YK224 but was only increased by 0.87-fold relative to YK193. Finally, the xylose consumption rate of YK197 at 24 h was 0.68-fold higher than that of SS118. These changes in the xylose consumption rate at 24 h in the presence of PPP gene overexpression are summarized in a schematic drawing (Fig. 4). YK246, which co-expressed three PPP genes (KmRKI1- KmTAL1-ScTKL1), displayed the highest xylose consumption rate at 24 h (1.03 g/L/h) among the yeast strains tested in this study, and produced an ethanol yield of 0.45 g-ethanol/g-total sugars.
Fig. 2.
Fermentation profiles of strains expressing XI and overexpressing two PPP genes. Cells were initially inoculated at OD600 = 3 and cultivated in medium containing 85 g/L glucose and 35 g/L xylose for 72 h at 36 °C. a YK222 (KmTAL1opt-ScTKL1), b YK193 (KmRKI1opt-ScTKL1), c YK223 (KmRPE1opt-ScTKL1). Blue line: glucose, Red line: xylose, Green line: xylitol, Purple line: glycerol, Light blue line: acetate, Orange line: ethanol. Each data point is an average of triplicate cultures
Fig. 3.
Fermentation profiles of strains expressing XI and combinatorial expression of multiple PPP genes. Cells were initially inoculated at OD600 = 3 and cultivated in a medium containing 85 g/L glucose and 35 g/L xylose for 72 h at 36 °C. a YK246 (KmRKI1opt- KmTAL1opt -ScTKL1), b YK247 (KmRPE1opt-KmTAL1opt-ScTKL1), c YK248 (KmRKI1opt-KmRPE1opt-KmTAL1opt), d YK249 (KmRKI1opt-KmRPE1opt-ScTKL1), e YK197 (KmRKI1opt-KmRPE1opt-KmTAL1opt-ScTKL1). Blue line: glucose, Red line: xylose, Green line: xylitol, Purple line: glycerol, Light blue line: acetate, Orange line: ethanol. Each data point is an average of triplicate cultures
Fig. 4.
Successive changes in d-xylose consumption by introduction of each PPP gene. Black line: increased by less than 1.2-fold, Blue and dashed line decreasing, Red thick line: increased by more than 1.2-fold. Colored panels: gene combination with highest consumption rate in single (Yellow), double (Orange) and triple (Red) of PPP gene(s), respectively
Comparison of XR-XDH with XI, the effect of PHO13 deletion and overexpression of all PPP genes on glucose/xylose co-fermentation
To reveal the effect of sugar composition of the fermentation media, we examined and compared the effect of PHO13 disruption and overexpression of all PPP genes on glucose/xylose co-fermentation (85 g/L d-glucose, 35 g/L d-xylose, Additional file 3: Fig. S1) and xylose fermentation (60 g/L d-xylose, Additional file 4: Fig. S2) at 36 °C using the recombinant S. cerevisiae strains YK001 (XR-XDH), YK002 (XR-XDH, pho13Δ), YK115 (XR-XDH, pho13Δ::ScRKI1-ScRPE1-ScTAL1-ScTKL1), SS82 (XI), YK149 (XI, pho13Δ) and YK150 (XI, pho13Δ:ScRKI1-ScRPE1-ScTAL1-ScTKL1). A control strain with XR-XDH driven by the TDH3 promoter (YK001) assimilated d-glucose completely within 24 h, but the concentration of residual d-xylose was about 12 g/L at 72 h. The concentration of ethanol produced from d-glucose to d-xylose reached about 45 g/L at 72 h. On the other hand, the kinetics of d-glucose and d-xylose consumption of the control strain with XI (SS82) were similar to those for YK001 (XR-XDH), but the concentration of ethanol produced at 72 h was much higher at about 48.5 g/L. The production of xylitol and glycerol by SS82 was less than that by YK001 (xylitol: 2.8-fold, glycerol: 1.5-fold). Deletion of PHO13 decreased the ethanol yields in both XR-XDH- (YK001) and XI-containing (YK149) strains at 72 h (YK002: 0.28 g/g, YK149: 0.33 g/g) on glucose/xylose co-fermentation. Furthermore, overexpression of four PPP genes in addition to the deletion of PHO13 resulted in decreased ethanol yields in both XR-XDH and XI strains at 72 h (YK115: 0.13 g/g, YK150: 0.29 g/g) compared to strains that had only the PHO13 deletion (YK002 and YK149) and control strains (YK001 and SS82). The strain expressing XI produced higher ethanol yields than did XR-XDH. Thus, PHO13 disruption and overexpression of all PPP genes in these strains with the presence of XR-XDH and XI for xylose metabolism did not improve ethanol yield or xylose consumption rate in YPDX medium at 36 °C.
Discussion
For the commercial production of bioethanol from cellulosic materials, development of microorganisms capable of highly efficient xylose fermentation is critical. To achieve this objective, many studies have demonstrated that expression of PPP genes can improve xylose metabolism (Kuyper et al. 2005; Karhumaa et al. 2007; Lee et al. 2014; Qi et al. 2015). We recently showed that the best combination of PPP gene overexpression in the S. cerevisiae haploid strain IR-2 that carries XR-XDH genes depended on the fermentation temperature in 60 g/L xylose medium (Kobayashi et al. 2017). For the SSCF process, one of the most economical fermentation processes to produce ethanol from cellulosic material at an industrial scale, the preferable temperature is higher than that for ethanologenic microorganisms. In addition, simultaneous consumption of d-glucose and d-xylose is desirable for economical ethanol production from cellulosic biomass. The concentrations of sugars in the raw material and the utilization of these sugars during the course of fermentation are also important factors for economical ethanol production, because changes in the concentrations of these sugars during glucose/xylose co-fermentation can alter gene expression levels in the microorganisms. For these reasons, it is critical to optimize expression levels of non-oxidative PPP genes under conditions that mimic those in the industrial process rather than experimental conditions such as low temperature growth on d-xylose-containing medium that was used for our previous study. However, the effects of overexpression of PPP genes on glucose/xylose co-fermentation, particularly at higher temperature, of S. cerevisiae IR-2 expressing XI were unknown.
In the present study, we systematically investigated how overexpression of PPP genes derived from S. cerevisiae and thermotolerant yeast Kluyveromyces marxianus expressed either singly or in combination by a S. cerevisiae strain harboring the XI gene affected glucose/xylose co-fermentation at 36 °C. The concentration of sugars, 85 g/L d-glucose and 35 g/L d-xylose, used in the fermentation experiments are based on typical sugarcane bagasse saccharified solutions. Among strains overexpressing a single PPP gene, TKL1 (YK185) or RKI1 (YK183) showed a dramatically increased d-xylose consumption rate at 24 h on glucose/xylose co-fermentation, whereas TAL1 (YK184) and RPE1 (YK186) did not (Fig. 1 and Table 2, also summarized in Fig. 4). Further experiments with yeast expressing multiple PPP genes clearly showed an optimal combination of PPP genes that could be used to improve xylose fermentation at high temperature: overexpression of TKL1 from S. cerevisiae with RKI1 and TAL1 from K. marxianus produced the highest rates of xylose consumption and ethanol productivity among the strains examined (YK246, Figs. 2, 3 and Table 2, also summarized in Fig. 4). This result may appear to be inconsistent with a previous report suggesting that the efficiency of xylose fermentation should be attributed, at least in part, to increased TAL1 expression in a S. cerevisiae strain expressing XI obtained by an adaptive evolution experiment (Vilela Lde et al. 2015). This study suggested that the amount of intermediary metabolites would differ between glucose/xylose co-fermentation at 36 °C and xylose fermentation alone at 30 °C. Other studies also reported that overexpression of four PPP genes could improve xylose metabolism (Kuyper et al. 2005; Qi et al. 2015). However, comparisons of our results with these previous studies is difficult due to differences in the experimental conditions, including genes for xylose metabolism, genetic background of the parental strain, fermentation temperature, inoculum dose and sugar concentrations. Thus, several points raised by our results should be addressed by future studies, including: (i) the suggestion that sedoheptulose 7-phosphate levels will be deficient in strains that do not overexpress RKI1 and TKL1 during glucose/xylose co-fermentation at high temperature because TKL1 synthesizes sedoheptulose 7-phosphate and glyceraldehyde 3-phosphate from xylose 5-phosphate and ribose 5-phosphate; and (ii) whether the synthesis of ribose 5-phosphate from ribulose 5-phosphate by RKI1 is insufficient. Overexpression of RPE1 may not be needed to increase the d-xylose consumption rate on glucose/xylose co-fermentation at a high temperature as its gene product could induce competition of enzyme reactions between xylulose 5-phosphate from xylose to ribulose 5-phosphate from glucose. In fact, only slight differences in d-xylose consumption rates at 24 h and ethanol production at 72 h were seen between YK246 and YK193 (Fig. 1 and Table 2, also summarized in Fig. 4). Thus, the present study clearly showed that overexpression of the PPP genes KmRKI1, KmTAL1 and ScTKL1 provided the best performance for xylose consumption and ethanol production on glucose/xylose co-fermentation at high temperature, indicating that optimizing xylose metabolism according to the individual and realistic experimental conditions, especially fermentation temperature, type and concentration of sugars and cell growth during fermentation, will be important for generating strains that have the best performance under these conditions.
To improve xylose metabolism, factors other than PPP may also need to be considered. Elimination of NAD-dependent glycerol 3-phosphate dehydrogenase (GPD) that has two homologous genes, GPD1 and GPD2, which are involved in glycerol metabolism by S. cerevisiae (Eriksson et al. 1995), increased ethanol yield while decreasing glycerol production (Björkqvist et al. 1997; Henningsen et al. 2015). In a preliminary examination, we verified that SS118 (XI gpd2Δ) exhibited decreased glycerol production and improved ethanol production (Seike et al. unpublished observations). As such, in the present study, each PPP gene was inserted in the GPD2 locus. As expected, glycerol production in all the strains was reduced compared with the parental strain SS82 (Additional file 2: Table S2).
On the other hand, whether deletion of PHO13, a putative alkaline phosphate gene, improves xylose fermentation is controversial. Several studies showed that deletion of PHO13 improves xylose metabolism in S. cerevisiae expressing XR-XDH. The improvement yielded by PHO13 deletion could be due to altered redox levels on d-xylose (Van Vleet et al. 2008) and xylulose-5-phosphate phosphatase (Kim et al. 2013) that induce a global transcriptional response (Kim et al. 2015). The deletion of PHO13 had a beneficial effect on fermentation with improved activity of the XI pathway, and high ethanol yield (0.45 g/g) in 40 g/L d-xylose medium at 30 °C (Lee et al. 2014). Another study demonstrated that disruption of PHO13 enhanced the d-xylose consumption rate (0.31 g/g-cell/h), ethanol productivity (0.11 g/g-cell/h) and ethanol yield (0.45 g/g) in an XI-expressing, xylose-fermenting yeast strain under oxygen-limited conditions in 50 g/L d-xylose at 30 °C (Bamba et al. 2016). In contrast to these reports, deletion of PHO13 in S. cerevisiae expressing XI sharply weakened growth on xylose plates (Shen et al. 2012) and promoted accumulation of 4-phospho-erythronate and 6-phospho-gluconate (Collard et al. 2016). Based on these earlier results, we examined and evaluated the effect of deletion of PHO13 and overexpression of all PPP genes on a glucose/xylose mixed medium or only xylose medium at 36 °C in the recombinant S. cerevisiae strains YK001 (XR-XDH), YK002 (XR-XDH pho13Δ), YK115 (XR-XDH pho13Δ:ScRKI1-ScRPE1-ScTAL1-ScTKL1), YK149 (XI pho13Δ) and YK150 (XI pho13Δ:ScRKI1-ScRPE1-ScTAL1-ScTKL1). S. cerevisiae strains expressing XR-XDH consumed 85 g/L d-glucose within 24 h but had incomplete consumption of 35 g/L d-xylose even at 72 h on glucose/xylose co-fermentation at 36 °C when cells were initially inoculated at OD600 = 3 (Additional file 3: Figure S1). Notably, no difference was observed between the parental (YK001, SS82) and PHO13Δ (YK002, YK149) strains, but xylose consumption was decreased in the strain that overexpressed all four PPP genes (YK115, YK150) on glucose/xylose co-fermentation (Additional file 3: Figure S1). Incidentally, the concentrations of ethanol production of YK001 and SS82 were about 45.1 g/L and 48.6 g/L at 72 h, respectively. Ethanol production by strains carrying XI could have been higher than those with XR-XDH due to a decrease in the by-product xylitol compared with xylose consumption. Although S. cerevisiae strains expressing XI can directly convert d-xylose to d-xylulose without a redox reaction, GRE3 encoding an intrinsic aldo-keto reductase and producing xylitol should be deleted to reduce xylitol formation (Yamanaka. 1969; Träff et al. 2001).
In xylose fermentation, however, markedly different results were obtained for strains with XR-XDH and XI. For strains harboring XR-XDH, xylose metabolism was clearly improved by deletion of PHO13 (YK002) and additional overexpression of the four PPP genes (YK115) as expected. For strains harboring XI, however, xylose consumption was decreased by the deletion of PHO13 and recovered to control levels by overexpression of the four PPP genes (Additional file 4: Figure S2, Kobayashi et al. 2017). Ismail et al. (2013) showed that the expression of several genes was up- or downregulated by more than twofold at 38 °C compared to at 30 °C in S. cerevisiae carrying XR-XDH. These results suggested that PPP could be a limiting step during ethanol production from d-xylose at high temperature when severely downregulated PPP gene expression might result in accumulation of certain intermediates that in turn affect the metabolic flux in PPP. Whatever the case, our results indicate that PHO13 deletion should not be adopted due to its negative effect on strains carrying XI.
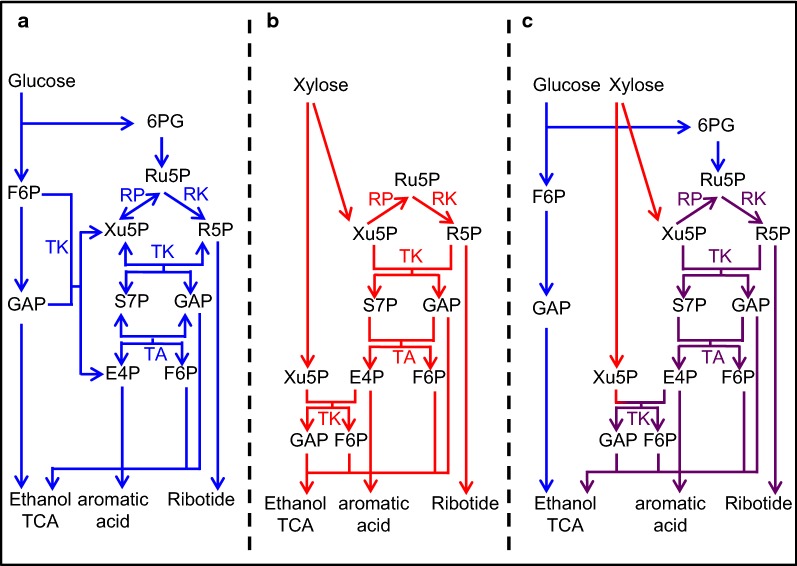
Developing an ideal strain for cellulosic ethanol production is challenging due to the complex nature of glucose/xylose co-fermentation. The entire fermentation process can be divided into three phases: (i) glucose assimilation (on catabolite repression), (ii) simultaneous assimilation of glucose and xylose (off catabolite repression), and (iii) subsequent assimilation of residual xylose. The interphase transitions can dramatically affect the expression of various metabolic genes such as hexose transporters since activities of gene promoters can be altered by extracellular concentration and uptake of sugars (Sedlak and Ho 2004). Here, we used the well-characterized strong promoter TDH3 and CYC1 terminator to control expression of PPP genes. Future studies should examine suitable promoters for the regulation of the expression of individual PPP genes to provide the necessary levels of PPP enzymes that promote harmonized xylose metabolic flux. Taken together, the activities of PPP enzymes required for ideal xylose metabolism differ according to glucose/xylose co-fermentation and for xylose fermentation at high temperature (Fig. 5).
Fig. 5.
Differences in metabolic pathways for production of ethanol and energy supply with glucose, xylose and mixed sugars fermentation. Ethanol, aromatic acids and ribotides are synthesized from glucose (blue lines), xylose (red lines), or both glucose and xylose (purple lines) on glucose fermentation (a), xylose fermentation (b), and glucose/xylose co-fermentation (c) via the glycolytic pathway and pentose phosphate pathway
Additional files
Additional file 1: Table S1. Oligonucleotide sequences used for synthesis of DNA fragments for homologous recombination.
Additional file 2: Table S2. Metabolic characteristics of yeast strains grown in YPDX medium.
Additional file 3: Figure S1. Fermentation profiles of XR-XDH-inserted and XI-inserted strains with pho13Δ on glucose/xylose co-fermentation.
Additional file 4: Figure S2. Fermentation profiles of XR-XDH-inserted and XI-inserted strains with pho13Δ on xylose containing medium.
Authors’ contributions
YK carried out all the experimental work and participated in the experimental design, as well as coordination and writing of the manuscript; TS, SO and YK participated in project conception and coordination; KEF participated in the study design and coordination and provided critical review of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by the New Energy and Industrial Technology Development Organization (NEDO).
Competing interests
The authors declare that they have are no competing interests and certify that there is no conflict of interest with any financial or non-financial organization regarding the contents of the manuscript.
Availability of data and materials
Not applicable (raw fermentation profiles were attached as supporting materials).
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
Not applicable (written in Acknowledgements).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- PPP
pentose phosphate pathway
- S. cerevisiae or Sc
Saccharomyces cerevisiae
- TAL1
transaldolase
- TKL1
transketolase
- RKI1
ribose-5-phosphate ketol-isomerase
- RPE1
d-ribulose-5-phosphate 3-epimerase
- XI
xylose isomerase
- XR or XYL1
NADPH-dependent xylose reductase
- XDH or XYL2
NAD+-dependent xylitol dehydrogenase
- PHO13
putative alkaline phosphate
- HXT7
xylose-permeable hexose transporter
- SSCF
simultaneous saccharification and co-fermentation
- K. marxianus or Km
Kluyveromyces marxianus
- GRE3
aldo–keto reductase
- Lp
Lachnoclostridium phytofermentans
- AUR1
phosphatidylinositol:ceramide phosphoinositol transferase (Aureobasidin A resistance gene)
- LiOAc
lithium acetate
- DNA
deoxyribonucleic acid
- HPLC
high performance liquid chromatography
- RI
refractive index
- Opt
codon-optimized
- GPD1 and GPD2
NAD-dependent glycerol 3-phosphate dehydrogenase
- CYC1
Cytochrome c, isoform 1
- TDH3
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), isozyme 3
Contributor Information
Yosuke Kobayashi, Email: kobayashi.y@biomt.onmicrosoft.com.
Takehiko Sahara, Email: t-sahara@aist.go.jp.
Satoru Ohgiya, Email: s.ohgiya@aist.go.jp.
Yoichi Kamagata, Email: y.kamagata@aist.go.jp.
Kazuhiro E. Fujimori, Phone: +81-29-849-1228, Email: k-fujimori@aist.go.jp
References
- Bamba T, Hasunuma T, Kondo A. Disruption of PHO13 improves ethanol production via the xylose isomerase pathway. AMB Express. 2016;6(1):4. doi: 10.1186/s13568-015-0175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya AS, Bhattacharya A, Pletschke BI. Synergism of fungal and bacterial cellulases and hemicellulases: a novel perspective for enhanced bio-ethanol production. Biotechnol Lett. 2015;37(6):1117–1129. doi: 10.1007/s10529-015-1779-3. [DOI] [PubMed] [Google Scholar]
- Björkqvist S, Ansell R, Adler L, Lidén G. Physiological response to anaerobicity of glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae. Appl Environ Microbiol. 1997;63(1):128–132. doi: 10.1128/aem.63.1.128-132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat D, Boles E, Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75(8):2304–2311. doi: 10.1128/AEM.02522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard F, Baldin F, Gerin I, Bolsée J, Noël G, Graff J, Veiga-da-Cunha M, Stroobant V, Vertommen D, Houddane A, Rider MH, Linster CL, Van Schaftingen E, Bommer GT. A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nat Chem Biol. 2016;12(8):601–607. doi: 10.1038/nchembio.2104. [DOI] [PubMed] [Google Scholar]
- Eriksson P, André L, Ansell R, Blomberg A, Adler L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol Microbiol. 1995;17(1):95–107. doi: 10.1111/j.1365-2958.1995.mmi_17010095.x. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Gonçalves DL, Matsushika A, de Sales BB, Goshima T, Bon EP, Stambuk BU. Xylose and xylose/glucose co-fermentation by recombinant Saccharomyces cerevisiae strains expressing individual hexose transporters. Enzyme Microb Technol. 2014;63:13–20. doi: 10.1016/j.enzmictec.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Henningsen BM, Hon S, Covalla SF, Sonu C, Argyros DA, Barrett TF, Wiswall E, Froehlich AC, Zelle RM. Increasing anaerobic acetate consumption and ethanol yields in Saccharomyces cerevisiae with NADPH-specific alcohol dehydrogenase. Appl Environ Microbiol. 2015;81(23):8108–8117. doi: 10.1128/AEM.01689-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail KS, Sakamoto T, Hasunuma T, Kondo A. Time-based comparative transcriptomics in engineered xylose-utilizing Saccharomyces cerevisiae identifies temperature-responsive genes during ethanol production. J Ind Microbiol Biotechnol. 2013;40(9):1039–1050. doi: 10.1007/s10295-013-1293-3. [DOI] [PubMed] [Google Scholar]
- Johansson B, Hahn-Hägerdal B. The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res. 2002;2(3):277–282. doi: 10.1111/j.1567-1364.2002.tb00095.x. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Garcia Sanchez R, Hahn-Hägerdal B, Gorwa-Grauslund MF. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2007;6:5. doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi H, Hasunuma T, Ogino C, Kondo A. Bioprocessing of bio-based chemicals produced from lignocellulosic feedstocks. Curr Opin Biotechnol. 2016;42:30–39. doi: 10.1016/j.copbio.2016.02.031. [DOI] [PubMed] [Google Scholar]
- Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin YS. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS ONE. 2013;8(2):e57048. doi: 10.1371/journal.pone.0057048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Xu H, Lesmana A, Kuzmanovic U, Au M, Florencia C, Oh EJ, Zhang G, Kim KH, Jin YS. Deletion of PHO13, encoding haloacid dehalogenase type IIA phosphatase, results in upregulation of the pentose phosphate pathway in Saccharomyces cerevisiae. Appl Environ Microbiol. 2015;81(5):1601–1609. doi: 10.1128/AEM.03474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Sahara T, Suzuki T, Kamachi S, Matsushika A, Hoshino T, Ohgiya S, Kamagata Y, Fujimori KE. Genetic improvement of xylose metabolism by enhancing the expression of pentose phosphate pathway genes in Saccharomyces cerevisiae IR-2 for high-temperature ethanol production. J Ind Microbiol Biotechnol. 2017 doi: 10.1007/s10295-017-1912-5. [DOI] [PubMed] [Google Scholar]
- Kötter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1993;38(6):776–783. doi: 10.1007/BF00167144. [DOI] [Google Scholar]
- Kötter P, Amore R, Hollenberg CP, Ciriacy M. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet. 1990;18(6):493–500. doi: 10.1007/BF00327019. [DOI] [PubMed] [Google Scholar]
- Kuriyama H, Seiko Y, Murakami T, Kobayashi H, Sonoda Y. Continuous ethanol fermentation with cell recycling using flocculating yeast. J Ferment Technol. 1985;63(2):159–165. [Google Scholar]
- Kuyper M, Hartog MM, Toirkens MJ, Almering MJ, Winkler AA, van Dijken JP, Pronk JT. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005;5(4–5):399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Lee SM, Jellison T, Alper HS. Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels. 2014;7(1):122. doi: 10.1186/s13068-014-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Inoue H, Kodaki T, Sawayama S. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol. 2009;84(1):37–53. doi: 10.1007/s00253-009-2101-x. [DOI] [PubMed] [Google Scholar]
- Naik SN, Goud VV, Rout PK, Dalai AK. Production of first and second generation biofuels: a comprehensive review. Renew Sustain Energy Rev. 2010;14:578–597. doi: 10.1016/j.rser.2009.10.003. [DOI] [Google Scholar]
- Paulova L, Patakova P, Branska B, Rychtera M, Melzoch K. Lignocellulosic ethanol: technology design and its impact on process efficiency. Biotechnology. 2014;33(6Pt2):1091–1107. doi: 10.1016/j.biotechadv.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Qi X, Zha J, Liu GG, Zhang W, Li BZ, Yuan YJ. Heterologous xylose isomerase pathway and evolutionary engineering improve xylose utilization in Saccharomyces cerevisiae. Front Microbiol. 2015;6:1165. doi: 10.3389/fmicb.2015.01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider Apel A, Ouellet M, Szmidt-Middleton H, Keasling JD, Mukhopadhyay A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Sci Rep. 2016;6:19512. doi: 10.1038/srep19512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara T, Fujimori KE, Nezuo M, Tsukahara M, Tochigi Y, Ohgiya S, Kamagata Y. Draft genome sequence of Saccharomyces cerevisiae IR-2, a useful industrial strain for highly efficient production of bioethanol. Genome Announc. 2014;2(1):e01160–e02013. doi: 10.1128/genomeA.01160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak M, Ho NW. Characterization of the effectiveness of hexose transporters for transporting xylose during glucose/xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast. 2004;21(8):671–684. doi: 10.1002/yea.1060. [DOI] [PubMed] [Google Scholar]
- Shen Y, Chen X, Peng B, Chen L, Hou J, Bao X. An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl Microbiol Biotechnol. 2012;96(4):1079–1091. doi: 10.1007/s00253-012-4418-0. [DOI] [PubMed] [Google Scholar]
- Träff KL, Otero Cordero RR, van Zyl WH, Hahn-Hägerdal B. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol. 2001;67(12):5668–5674. doi: 10.1128/AEM.67.12.5668-5674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vleet JH, Jeffries TW. Yeast metabolic engineering for hemicellulosic ethanol production. Curr Opin Biotechnol. 2009;20(3):300–306. doi: 10.1016/j.copbio.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Van Vleet JH, Jeffries TW, Olsson L. Deleting the para-nitrophenyl phosphatase (pNPPase), PHO13, in recombinant Saccharomyces cerevisiae improves growth and ethanol production on d-xylose. Metab Eng. 2008;10(6):360–369. doi: 10.1016/j.ymben.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Vilela Lde F, de Araujo VP, Paredes Rde S, Bon EP, Torres FA, Neves BC, Eleutherio EC. Enhanced xylose fermentation and ethanol production by engineered Saccharomyces cerevisiae strain. AMB Express. 2015;5:16. doi: 10.1186/s13568-015-0102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson M, Hallborn J, Penttilä M, Keränen S, Hahn-Hägerdal B. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol. 1995;61(12):4184–4190. doi: 10.1128/aem.61.12.4184-4190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson M, Bao X, Anderlund M, Lilius G, Bülow L, Hahn-Hägerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol. 1996;62(12):4648–4651. doi: 10.1128/aem.62.12.4648-4651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Kim S, Sorek H, Lee Y, Jeong D, Kim J, Oh EJ, Yun EJ, Wemmer DE, Kim KH, Kim SR, Jin YS. PHO13 deletion-induced transcriptional activation prevents sedoheptulose accumulation during xylose metabolism in engineered Saccharomyces cerevisiae. Metab Eng. 2016;34:88–96. doi: 10.1016/j.ymben.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Yamanaka K. Inhibition of d-xylose isomerase by pentitols and d-Xylose. Arch Biochem Biophys. 1969;131(2):502–506. doi: 10.1016/0003-9861(69)90422-6. [DOI] [PubMed] [Google Scholar]
- Young E, Lee SM, Alper H. Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol Biofuels. 2010;3:24. doi: 10.1186/1754-6834-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Oligonucleotide sequences used for synthesis of DNA fragments for homologous recombination.
Additional file 2: Table S2. Metabolic characteristics of yeast strains grown in YPDX medium.
Additional file 3: Figure S1. Fermentation profiles of XR-XDH-inserted and XI-inserted strains with pho13Δ on glucose/xylose co-fermentation.
Additional file 4: Figure S2. Fermentation profiles of XR-XDH-inserted and XI-inserted strains with pho13Δ on xylose containing medium.
Data Availability Statement
Not applicable (raw fermentation profiles were attached as supporting materials).