Abstract
Objectives
This study maps the lipid distributions based on magnetic resonance imaging (MRI) in-and opposed-phase (IOP) sequence and correlates the findings generated from lipid map to histological grading of glioma.
Methods
Forty histologically proven glioma patients underwent a standard MRI tumour protocol with the addition of IOP sequence. The regions of tumour (solid enhancing, solid non-enhancing, and cystic regions) were delineated using snake model (ITK-SNAP) with reference to structural and diffusion MRI images. The lipid distribution map was constructed based on signal loss ratio (SLR) obtained from the IOP imaging. The mean SLR values of the regions were computed and compared across the different glioma grades.
Results
The solid enhancing region of glioma had the highest SLR for both Grade II and III. The mean SLR of solid non-enhancing region of tumour demonstrated statistically significant difference between the WHO grades (grades II, III & IV) (mean SLRII = 0.04, mean SLRIII = 0.06, mean SLRIV = 0.08, & p < .01). A strong positive correlation was seen between WHO grades with mean SLR on lipid map of solid non-enhancing (ρ=0.68, p < .01).
Conclusion
Lipid quantification via lipid map provides useful information on lipid landscape in tumour heterogeneity characterisation of glioma. This technique adds to the surgical diagnostic yield by identifying biopsy targets. It can also be used as an adjunct grading tool for glioma as well as to provide information about lipidomics landscape in glioma development.
Keywords: Lipid distribution, In- and opposed phase, Signal loss ratio, Lipid map, Glioma, Grading
Highlights
-
•
In- and opposed-phase imaging is useful in gliomas characterisation and grading.
-
•
Signal loss ratio in the solid non-enhancing region is a potential imaging marker for discriminating between the WHO grades.
-
•
Lipid quantification via lipid distribution mapping provides useful information on lipid landscape in tumour heterogeneity.
1. Introduction
Gliomas comprise 27% of all brain tumours and 80% of all malignant brain tumours (Hess et al., 2004). The World Health Organisation (WHO) glioma classification system dichotomises the tumours to low-grade glioma (LGG) comprising of histological grades I and II tumours and high-grade glioma (HGG) comprising of histological grades III and IV tumours (Kleihues et al., 2002; Louis et al., 2016). Recent revision of the WHO guidelines incorporates molecular parameters alongside the histology in the classification of tumour entities (Louis et al., 2016). The complexity of the disease is often aggravated by tumour proliferation, localized invasion, metastasis, resistance to therapy (Swanson et al., 2003), and different responses to treatments among patients due to variation in genetic profiles of the tumours (Gupta et al., 2015; Pope et al., 2009).
Magnetic resonance imaging (MRI) is the generally preferred modality for brain imaging as it offers valuable information on overall tumour structure, composition, physiology and function (Guzman-De-Villoria et al., 2014). Tumour characteristics such as intensity distribution, enhancement, size, shape, structure, location, volume, border, focality, subventricular zone involvement, cystic changes, percentage of necrosis, and tumour volume are the conventional imaging parameters investigated for glioma characterisation (Lambin et al., 2012; Upadhyay and Waldman, 2011; Gutman et al., 2013; Nicolasjilwan et al., 2015; Ellingson, 2014). However, clinical translation using only conventional imaging assessments is difficult due to the heterogeneous nature of the tumours.
The detection and staging of glioma at an early stage are essential for early intervention to improve prognosis and minimize neurocognitive risks. Heterogeneity in neuroimaging, pathologic and molecular features had been the main existing problem for glioma diagnosis (Gutman et al., 2013). The determination of the grades and subtypes are challenging due to diverse characteristics, even within a single tumour. Tumour heterogeneity further complicates histopathological observations and interferes with treatment decisions and clinical management. Histopathology tests serve as the gold standard for glioma diagnosis but its usefulness is limited by several drawbacks such as high invasiveness, risk of infection and bleeding, subjected to inter- and intra-pathologists variability, and possibility of sampling error. Thus, there is a need for fast and non-invasive imaging biomarkers that could elucidate the underlying inter- and intra-tumoural histopathological changes.
An increased rate of lipid synthesis has long been recognised as an important characteristic in tumorigenesis. However, the contribution of lipids to the transformation, development, and progression of tumour are still not yet fully understood (Baenke et al., 2013). Lipids such as mobile fatty acids, triglycerides, phospholipids, cholesterol esters, sphingolipid, prostaglandins and steroid hormones serve as important components in necrosis (Fan, 2006), cellular membrane breakdown, (Li et al., 2002) and signal transduction. Lipogenesis is one of the main metabolism pathways regulated to sustain rapid tumour growth. Elevated lipid levels had been found to correspond with higher tumour grades and aggressiveness in characterisation of brain tumours (Guo et al., 2013; Ramli et al., 2015; Lim et al., 2011). The alteration in lipid levels of the brain is postulated to be due to the cell membrane perturbation induced by apoptosis and necrosis (Ramli et al., 2015; Van Cauter et al., 2014; Bieza and Krumina, 2013).
In-and opposed-phase (IOP) imaging is another alternative for lipid quantification other than magnetic resonance spectroscopy (MRS). The mechanism behind IOP imaging is the difference in resonance frequency between proton nuclei from water and fat content of the tissues. Water and fat experienced a slight difference in magnetic field strengths due to different chemical environments and surrounding electron density. IOP imaging has been used clinically for lipid detection in disorders such as adrenal adenoma, renal angiomyolipoma, and bone marrow pathology (Israel et al., 2005; Outwater et al., 1998; Seiderer et al., 1999; Namimoto et al., 2001). Recent usage is seen for detection of mobile lipids in intracranial lesions, including glioma (Ramli et al., 2015; Lim et al., 2011). The advantage of IOP imaging is that it provides an overall evaluation of lipid quantification of the tumour as compared to the smaller sampling area provided by MRS.
In this work, we aim to understand the lipid landscape of glioma. Different tumour regions were delineated using semi-automatic segmentation method. By providing the lipid map, we relate with conventional imaging components to advance our understanding of glioma heterogeneity.
2. Materials and methods
2.1. MRI data retrieval and review
This was a retrospective study of a prospectively collected pre-operative imaging data from 40 histologically proven glioma cases treated at our centre between December 2010 and October 2017. This study was approved by our institutional ethics board and all patients provided written informed consent. The glioma patients were subjected to MRI scanning using a 3 T MRI scanner (Signa HDx, General Electric, USA). All of them underwent brain tumour protocol, including T1-weighted (T1), T2-weighted (T2), post-contrast T1-weighted, diffusion-weighted imaging (DWI) with derived apparent diffusion coefficient (ADC), and additional chemical shift IOP sequence. The IOP imaging parameters were: 150 ms repetition time, 2.4 ms echo time for the in-phase, 5.8 ms echo time for the opposed-phase, flip angle of 80°, the number of averages is 1, 250 mm × 250 mm field of view, the matrix size was 256 × 256 and 5 mm slice thickness. The inclusion criteria for the study were: 1) histologically proven glioma (grade II-IV), and 2) availability of MRI images in IOP.
2.2. Delineation of tumour regions
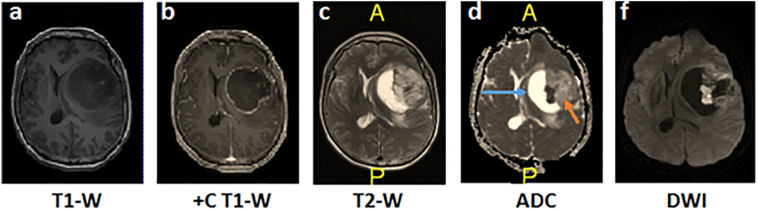
The regions of tumour (solid enhancing, solid non-enhancing, and cystic) were delineated on post-contrast T1 images using a semi-automated segmentation approach, i.e. snake model implemented in ITK-SNAP (Yushkevich and Gerig, 2011) with reference to structural MRI images (pre- and post-contrast T1, T2, and FLAIR) and diffusion images (ADC and DWI). The definition of the tumoural regions are described in Table 1 and the examples of the MRI images used for identification of tumoural regions are shown in Fig. 1. The tumour regions was segmented by a single user (PS) and the tumoural region masks was then verified by an experienced neuroradiologist (NR).
Table 1.
The definition of the tumoural regions in different MRI sequences in reference to grey matter.
| Tumour regions | MRI sequences | Signal comparison |
|---|---|---|
| Solid-enhancing | T1 | Iso to hypointense |
| post-contrast T1 | Hyperintense | |
| Solid non-enhancing | T1 | Iso to hypointense |
| T2 | Iso to hyperintense | |
| ADC | Iso to hypointense | |
| Cystic | T1 | Hypointense |
| T2 | Homogenous hyperintense areas | |
| DWI & ADC | Isointense to CSF |
CSF: cerebrospinal fluid.
Fig. 1.
The identification of tumour regions using different MRI sequences of a GBM patient. a) The pre-contrast T1 image, b) post-contrast T1 image where the solid enhancing region is the enhancing rim, c) T2 image, d) apparent diffusion coefficient (ADC) image with the blue arrow in ADC showed the cystic region while orange arrows depicted the solid non-enhancing region of the tumour and, f) diffusion-weighted image (DWI).
2.3. Image post-processing of the lipid maps and tumour region masks
The presence of lipid or percentages of fat is reflected by the quantification of relative signal loss in the in-phase and opposed-phase images, also termed the signal loss ratio (SLR) as defined in the following Eq. (26):
where signalin is the in-phase signal while signalopp is the opposed-phase signal. The lipid map is a two-dimensional visualization of the SLR.
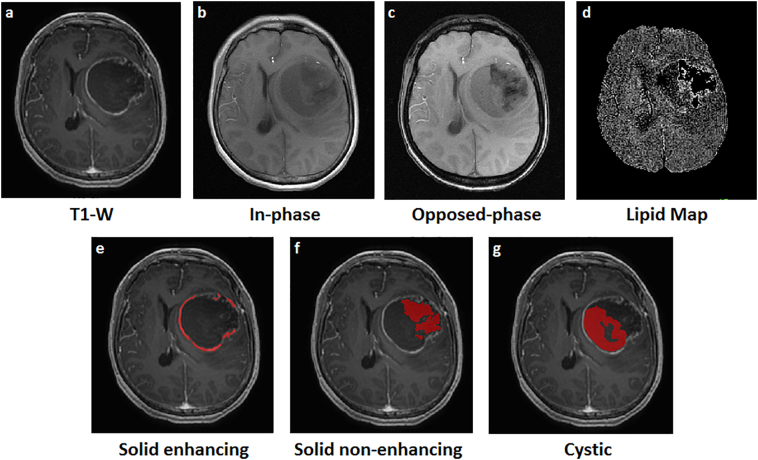
Image processing of the images and masks was performed using SPM12 (Statistical Parametric Software) (Wellcome Trust Centre for Neuroimaging, 2014) implemented in MATLAB (MathWorks, 2017). The lipid maps were first skull-stripped for brain extraction followed by the registration of the structural T1, lipid maps, and tumour region masks to standard MNI152 space (MNI152 is a template brain/space from Montreal Neurological Institute, stored within FSL (FMRIB Software Library) (FMRIB, 2012) (Fig. 2). The T1 images were then co-registered to in- and opposed-phase images. Blood vessels and capillaries were subtracted from the lipid map as they affect signal intensities. The mean SLR values of the regions were obtained by overlaying the region masks onto the lipid maps with reference to the co-registered T1 images.
Fig. 2.
The pre-processed images of a GBM patient. a) The raw T1 MRI image, b) in-phase sequence, c) opposed-phase sequence, d) skull stripped and co-registered lipid map, the co-registered masks (delineated in red) of e) solid enhancing region, f) solid non-enhancing region, and g) cystic region of the tumour.
2.4. Statistical analysis
Kruskal-Wallis test was carried out to establish significant differences in SLR of the tumour regions across the WHO grades. The statistical significant difference was declared at p < .05. The Spearman correlation test was performed to test the correlation between the mean SLR values and the tumour regions.
3. Results
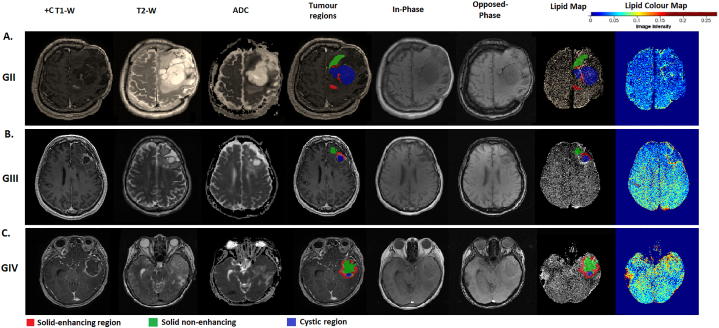
Table 2 shows the demographics of the glioma patients across the WHO tumour grades. The distribution of the SLR of solid non-enhancing region of the tumour demonstrated significant difference across the three grades (p < .01; Table 3) where increased SLR values were seen with increasing tumour grades (Fig. 3). Multiple comparisons between the different grades with correction showed significant difference between grade II and IV (p < .001) (Table 4). The lipid map in grade IV (GBM) showed higher extent of morphologic heterogeneity whereas lipid maps of low-grade gliomas were more homogeneous (Fig. 4). Increased heterogeneity was shown as positive skewness of all the grades in solid non-enhancing regions (skewness: GII = 0.98, GIII = 0.16 & GIV = 0.24). The solid enhancing and cystic regions showed negative skewness in grade III. A higher SLR value was seen from the lipid map in high-grade glioma, as shown as regions indicated in red. The lipid maps with tumour regions overlaid onto them were shown in Fig. 4 for different WHO grades. A strong positive correlation was seen between WHO grades with mean SLR on lipid map of solid non-enhancing region (ρ=0.68, p < .001).
Table 2.
Patients demographics across the tumour grades.
| Clinical parameters | WHO grade (n = 40) |
|||
|---|---|---|---|---|
| GII (n = 14) | GIII (n = 8) | GIV (n = 18) | ||
| Age (mean, (range)) | 37.36 (16–63) | 46.13 (25–71) | 51.78 (10–73) | |
| Gender | Male | 7 (50%) | 5 (62.5%) | 10 (55.6%) |
| Female | 7 (50%) | 3 (37.5%) | 8 (44.4%) | |
| Histology type | Diffuse astrocytoma | 5 | 0 | 0 |
| Pilomyxoid astrocytoma | 1 | 0 | 0 | |
| Pleomorphic xanthoastrocytoma | 1 | 0 | 0 | |
| Gemistocystic astrocytoma | 1 | 0 | 0 | |
| Fibrillary astrocytoma | 1 | 0 | 0 | |
| Oligoastrocytoma | 1 | 0 | 0 | |
| Oligodendroglioma | 4 | 1 | 0 | |
| Anaplastic oligodendroglioma | 0 | 5 | 0 | |
| Anaplastic astrocytoma | 0 | 2 | 0 | |
| Glioblastoma multiforme (GBM) | 0 | 0 | 17 | |
| Pontine GBM | 0 | 0 | 1 | |
| Tissue sampling | Total / partial resection | 12 | 8 | 14 |
| Stereotactic biopsy | 2 | 0 | 4 | |
| Tumour regions | With solid enhancing region | 12 (85.7%) | 4 (50%) | 18 (100%) |
| With solid non-enhancing region | 13 (92.9%) | 8 (100%) | 17 (94.4%) | |
| With cystic region | 9 (64.3%) | 5 (62.5%) | 14 (77.8%) | |
Table 3.
Descriptive data presented as the mean and standard deviation (SD) of the SLR values across WHO tumour grades.
| Markers | WHO grade |
Kruskal-Wallis test | ||
|---|---|---|---|---|
| Mean (SD) | ||||
| SLR | GII (n = 12) | GIII (n = 4) | GIV (n = 18) | |
| Solid enhancing | 0.074 (0.058) | 0.073 (0.021) | 0.075 (0.015) | 0.131 |
| SLR | GII (n = 13) | GIII (n = 8) | GIV (n = 17) | |
| Solid NE | 0.043 (0.017) | 0.061 (0.028) | 0.080 (0.019) | <0.01⁎ |
| SLR | GII (n = 9) | GIII (n = 5) | GIV (n = 14) | |
| Cystic | 0.080 (0.092) | 0.059 (0.022) | 0.061 (0.018) | 0.469 |
NE: non-enhancing.
p-values <.01.
Fig. 3.
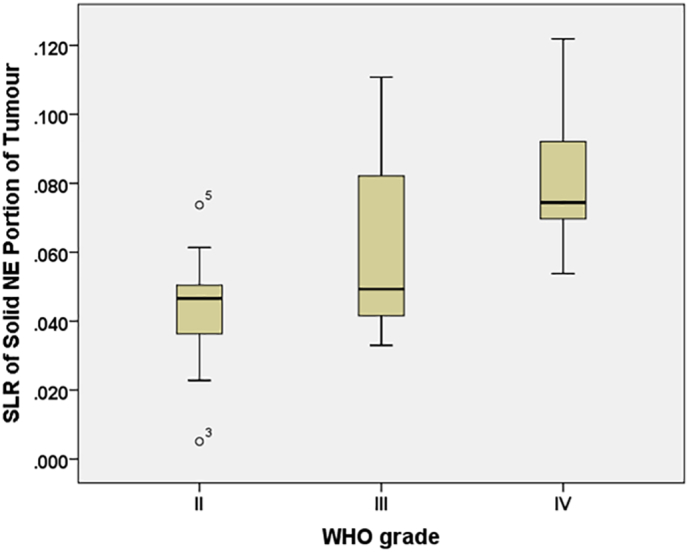
The SLR distributions across the WHO grades. The boxplots showing distributions of SLR in the solid non-enhancing region of the tumour across the WHO grades. NE: non-enhancing.
Table 4.
The multiple comparisons between the grades (II, III, & IV) for solid non-enhancing region of tumour.
| Group comparison | Test statistic | Standard error | Standard Test statistic | Significance | Adjusted significance |
|---|---|---|---|---|---|
| II-III | −6.43 | 4.99 | −1.29 | 0.198 | 0.593 |
| II-IV | −16.66 | 4.09 | −4.07 | >0.001⁎ | >0.001⁎ |
| III-IV | −10.23 | 4.77 | −2.15 | 0.032 | 0.095 |
Asymptotic significances (2-sided tests) are displayed. The significance level is 0.05. Significance values have been adjusted by the Bonferroni correction for multiple tests.
Fig. 4.
The structural and diffusion MRI images, tumour regions and lipid maps for three glioma patients of different grades (Grade II, III and IV). Examples of MRI images describes the T1, T2, ADC, semi-automatically segmented tumoural masks of the different regions (solid-enhancing, solid non-enhancing and cystic regions) overlaid onto the T1-W images, in-phase and opposed phase, the constructed lipid map with overlaid tumour region masks, and pseudo-colour lipid map displaying the lipid distribution of the brain for visualization purpose, of A) gemistocytic astrocytoma (grade II), B) anaplastic oligodendroglioma (grade III) and C) GBM (grade IV).
4. Discussion
In this study, we performed lipid mapping based on IOP MRI images, to landscape the lipid distribution of different grades of glioma. We postulated that the alteration of the structural lipid quantification obtained from lipid mapping could be attributed to the topographical distribution of the lipid molecules resulting from reprogramming of lipid metabolism. Our work is an initial attempt to understand the multifaceted landscape in terms of lipid distribution and microenvironment heterogeneity in gliomatous tumours. This work presents an improvement and extension of our earlier study on lipid quantification and glioma grading (Ramli et al., 2015). The improvement was carried out in three areas. Firstly, the current 3D ROI construction of the tumour regions implemented using a semi-automatic segmentation approach offers a more objective and consistent acquisition of the mean SLR values. Second, lipid quantification was evaluated in a more detailed manner by separating the solid region of the tumour into enhancing and non-enhancing regions. Finally, the SLR is presented in a 2D distribution (i.e. the lipid map) to better visualise lipid distribution in relation to the tumour morphology and glioma grades.
Our study showed that mean SLR of the solid non-enhancing region of the tumour is useful for discrimination of the tumour grades compared to solid-enhancing and cystic regions. The solid non-enhancing region of the tumour is more homogeneous than the solid enhancing and cystic regions. This region has more blood vessels (likely a combination of normal and newly formed vessels from neovascularization) where extravasation of blood plasma and contrast agents into the extravascular extracellular space occur due to blood-brain barrier (BBB) breakdown (Ahn et al., 2014; Jia et al., 2012); resulting in both intravascular enhancement and extravasation-related enhancement. The non-enhancing region of the tumour may harbour extracellular lipids from broken cellular membranes secondary to deficient blood perfusion, i.e. the opposite situation with the enhancing region. The lipid distribution at the non-enhancing region is better at differentiating glioma grades and may reflect the true aggressiveness of the tumoural tissues. Although lipid distributions (percentage of fat) are highest in the solid enhancing regions, they do not show any discriminating ability in this study as shown as the closeness of the mean SLR values across the grades. The lipid map outcome shows that the solid non-enhancing region of the tumour has the highest yield in the differentiation of glioma grading and therefore provides the best site for biopsy.
The SLR values in the lipid map reflect lipids fraction that arise from the lipid components in brain tissues. The lipid composition of the brain tissues offers insights about histological cell type, state of cellular growth, maturation, and differentiation (Eberlin et al., 2012), thus providing useful information for diagnosis and prognosis. The increase in lipid biosynthesis has been long recognised as an essential element of the metabolic reprogramming in cancer cells aside from glucose and glutamine metabolism (Baenke et al., 2013; Guo et al., 2013; Schulze and Harris, 2012). The metabolite reprogramming supports rapid proliferation of cancer cells to fulfill the increased demand for energy and nutrients. The alteration of visible lipids has been attributed to cellular processes such as proliferation, inflammation, malignancy, growth arrest, necrosis, apoptosis, migration, and invasion (Baenke et al., 2013; Griffin and Kauppinen, 2007). High expression of lipids is contributed by the prompt release from storage for cell growth and biological membranes generation, the breakdown of cellular membrane structure in response to growth stimuli resulting from malignant cell transformation, and increased numbers of cytoplasmic vesicles (Baenke et al., 2013; Guo et al., 2013; Griffin and Kauppinen, 2007).
The delineation of the brain tumour by manual segmentation provides essential distinction between lesions and normal tissues. Manual delineation of tumour is not only tedious and time-consuming but also subjected to intra- and inter-observer variability. This potentially leads to substantial inconsistency in the delineation of the region of interest (ROI). Furthermore, manual segmentation is limited by reproducibility and feasibility in labelling large cohorts of patients in clinical environment. These problems can be addressed by engaging automatic segmentation methods. The semi-automatic approach used in this study is a better way for delineation of the tumour regions as it is less time consuming and less dependent to observer's judgement. This method is applicable to the clinical practice where user interaction only requires the placement of the initial contour at the area of tumour regions.
Few limitations of this study include small sample size due to relatively quick progression of grade III to grade IV tumour and acquisition artefacts especially lesions near the air and bone interface. The IOP imaging clinically available in our study, is also known as Dixon method, and the measurement of SLR in this study is related to the two-point Dixon method used to separate fat and water signals (Dixon, 1984; Jingfei, 2008). Unfortunately, this approach is also sensitive to off-resonance effects that arise from magnetic field inhomogeneity (Jingfei, 2008). The magnetic susceptibility effects are pronounced especially in near air tissue-boundaries, implants, and iron depositions (Outwater et al., 1998). The three-point Dixon method acquires three imaging phases so that each acquisition occurs at a different phase between the water and fat signals (Pokharel et al., 2013). It has advantage of taking into account for magnetic field inhomogeneities but requires longer acquisition time compared to two-point Dixon method. Future work can be extended into the application of Iterative Decomposition with Echo Asymmetry and Least Squares Estimation (IDEAL), a robust reconstruction algorithm generalised from Dixon method that takes into account the transverse relaxation and intra-voxel dephasing effects (Bernard et al., 2008; Hu et al., 2010). IDEAL combines asymmetrically acquired echoes with an iterative least-squares decomposition algorithm to achieve maximum signal noise ratio in addressing issues related to magnetic field inhomogeneities (Takasu et al., 2015). Other factors that might affect the SLR values include blood flow, blood volume, haemorrhage, and susceptibility artefacts.
5. Conclusion
Lipid quantification using lipid distribution mapping with use of conventional imaging showed potential as a diagnostic tool, by providing sites for maximal differentiation in the grading of glioma. SLR value obtained from the lipid map is also a feasible imaging biomarker that can be used to reflect the lipid landscape of the glioma microenvironment. The knowledge accrued provides the basis for understanding the underlying tumoral histopathological changes that occur in the tumour regions and tumour development.
Acknowledgments
Acknowledgements
This study was made possible with financial assistance from the Fundamental Research Grant Scheme (FP009-2016) and Postgraduate Research Grant (PG219-2015B). The authors gratefully acknowledge the essential contributions of the research staff of University of Malaya Research Imaging Centre (UMRIC).
Declaration of interest
None.
References
- Ahn S.S., Na-Young S., Jong Hee C. Prediction of methylguanine methyltransferase promoter methylation in glioblastoma using dynamic contrast-enhanced magnetic resonance and diffusion tensor imaging. J. Neurosurg. 2014;121(2):367–373. doi: 10.3171/2014.5.JNS132279. [DOI] [PubMed] [Google Scholar]
- Baenke F., Peck B., Miess H. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 2013;6(6):1353–1363. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C.P., Liney G.P., Manton D.J. Comparison of fat quantification methods: a phantom study at 3.0T. J. Magn. Reson. Imaging. 2008;27(1):192–197. doi: 10.1002/jmri.21201. [DOI] [PubMed] [Google Scholar]
- Bieza A., Krumina G. The value of magnetic resonance spectroscopy and diffusion tensor imaging in characterization of gliomas growth patterns and treatment efficiency. J. Biomed. Sci. Eng. 2013;2013(6):518–526. [Google Scholar]
- Dixon W.T. Simple proton spectroscopic imaging. Radiology. 1984;153(1):189–194. doi: 10.1148/radiology.153.1.6089263. [DOI] [PubMed] [Google Scholar]
- Eberlin L.S., Norton I., Dill A.L. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72(3):645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson B.M. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr. Neurol. Neurosci. Rep. 2014;15(1):1–12. doi: 10.1007/s11910-014-0506-0. [DOI] [PubMed] [Google Scholar]
- Fan G. Magnetic resonance spectroscopy and gliomas. Cancer Imaging. 2006;6(1):113–115. doi: 10.1102/1470-7330.2006.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FMRIB FMRIB Software Library (FSL v.4.1.9) MNI152 template. 2012. https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation (Availabe via cited 2017)
- Griffin J., Kauppinen R. A metabolomics perspective of human brain tumours. FEBS J. 2007;274(5):1132–1139. doi: 10.1111/j.1742-4658.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- Guo D., Bell E., Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS oncology. 2013;2(3):289–299. doi: 10.2217/cns.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Young J.R., Shah D.A. Pretreatment dynamic susceptibility contrast MRI perfusion in glioblastoma: prediction of EGFR gene amplification. Clin. Neuroradiol. 2015;25(2):143–150. doi: 10.1007/s00062-014-0289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman D.A., Cooper L.A.D., Hwang S.N. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267(2):560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-De-Villoria J., Mateos-Perez J., Fernandez-Garcia P. Added value of advanced over conventional magnetic resonance imaging in grading gliomas and other primary brain tumors. Cancer Imaging. 2014;14(1):35. doi: 10.1186/s40644-014-0035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess K., Broglio K., Bondy M. Adult glioma incidence trends in the United States, 1977–2000. Cancer. 2004;101(10):2293–2299. doi: 10.1002/cncr.20621. [DOI] [PubMed] [Google Scholar]
- Hu H.H., Kim H.-W., Nayak K.S. Comparison of fat–water MRI and single-voxel MRS in the assessment of hepatic and pancreatic fat fractions in humans. Obesity. 2010;18(4):841–847. doi: 10.1038/oby.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel G., Hindman N., Hecht E. The use of opposed-phase chemical shift MRI in the diagnosis of renal angiomyolipomas. AJR Am. J. Roentgenol. 2005;184(6):1868–1872. doi: 10.2214/ajr.184.6.01841868. [DOI] [PubMed] [Google Scholar]
- Jia Z., Geng D., Xie T. Quantitative analysis of neovascular permeability in glioma by dynamic contrast-enhanced MR imaging. J. Clin. Neurosci. 2012;19(6):820–823. doi: 10.1016/j.jocn.2011.08.030. [DOI] [PubMed] [Google Scholar]
- Jingfei M. Dixon techniques for water and fat imaging. J. Magn. Reson. Imaging. 2008;28(3):543–558. doi: 10.1002/jmri.21492. [DOI] [PubMed] [Google Scholar]
- Kleihues P., Louis D., Scheithauer B. The WHO classification of tumors of the nervous system. J. Neuropathol. Exp. Neurol. 2002;61(3):215–225. doi: 10.1093/jnen/61.3.215. [DOI] [PubMed] [Google Scholar]
- Lambin P., Rios-Velazquez E., Leijenaar R. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lu Y., Pirzkall A. Analysis of the spatial characteristics of metabolic abnormalities in newly diagnosed glioma patients. J. Magn. Reson. Imaging. 2002;16(3):229–237. doi: 10.1002/jmri.10147. [DOI] [PubMed] [Google Scholar]
- Lim C., Ng K., Ramli N. Evaluation of the application of chemical shift for the detection of lipid in brain lesion. Radiography. 2011;17(1):43–48. [Google Scholar]
- Louis D.N., Perry A., Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- MathWorks . 2017. MATLAB version R2017a. (cited 2017) [Google Scholar]
- Namimoto T., Yamashita Y., Mitsuzaki K. Adrenal masses: quantification of fat content with double-Echo chemical shift in-phase and opposed-phase FLASH MR images for differentiation of adrenal adenomas. Radiology. 2001;218(3):642–646. doi: 10.1148/radiology.218.3.r01mr15642. [DOI] [PubMed] [Google Scholar]
- Nicolasjilwan M., Hu Y., Yan C. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J. Neuroradiol. 2015;42(4):212–221. doi: 10.1016/j.neurad.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outwater E., Blasbalg R., Siegelman E. Detection of lipid in abdominal tissues with opposed-phase gradient-echo images at 1.5 T: techniques and diagnostic importance. Radiographics. 1998;18(6):1465–1480. doi: 10.1148/radiographics.18.6.9821195. [DOI] [PubMed] [Google Scholar]
- Pokharel S.S., Macura K.J., Kamel I.R. Current MR imaging lipid detection techniques for diagnosis of lesions in the abdomen and pelvis. Radiographics. 2013;33(3):681–702. doi: 10.1148/rg.333125068. [DOI] [PubMed] [Google Scholar]
- Pope W.B., Kim H.J., Huo J. Recurrent glioblastoma multiforme: ADC histogram analysis predicts response to bevacizumab treatment. Radiology. 2009;252(1):282–289. doi: 10.1148/radiol.2521081534. [DOI] [PubMed] [Google Scholar]
- Ramli N., Khairy A.M., Seow P. Novel application of chemical shift gradient echo in- and opposed-phase sequences in 3 T MRI for the detection of H-MRS visible lipids and grading of glioma. Eur. Radiol. 2015;26:7. doi: 10.1007/s00330-015-4045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- Seiderer M., Staebler A., Wagner H. MRI of bone marrow: opposed-phase gradient-echo sequences with long repetition time. Eur. Radiol. 1999;9:652–661. doi: 10.1007/s003300050727. [DOI] [PubMed] [Google Scholar]
- Swanson K.R., Bridge C., Murray J.D. Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. J. Neurol. Sci. 2003;216(1):1–10. doi: 10.1016/j.jns.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Takasu M., Kaichi Y., Tani C. Iterative decomposition of water and fat with Echo asymmetry and least-squares estimation (IDEAL) magnetic resonance imaging as a biomarker for symptomatic multiple myeloma. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0116842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay N., Waldman A.D. Conventional MRI evaluation of gliomas. Br. J. Radiol. 2011;84(Spec Iss 2):S107–S111. doi: 10.1259/bjr/65711810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauter S., De Keyzer F., Sima D.M. Integrating diffusion kurtosis imaging, dynamic susceptibility-weighted contrast-enhanced MRI, and short echo time chemical shift imaging for grading gliomas. Neuro-Oncology. 2014;16(7):1010–1021. doi: 10.1093/neuonc/not304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Centre for Neuroimaging Statistical Parametric Mapping software package version 12 (SPM12) 2014. http://www.fil.ion.ucl.ac.uk/spm/software/spm12/ (Availabe via cited 2017)
- Yushkevich P.A., Gerig G. ITK-Snap version 3.6.0. 2011. http://www.itksnap.org (Available via cited 2017)