Abstract
Spermatogonial stem cells (SSCs) have significant applications in reproductive and regenerative medicine. However, nothing is known about genes in mediating human SSCs. Here we have explored for the first time the function and mechanism of P21-activated kinase 1 (PAK1) in regulating the proliferation and apoptosis of the human SSC line. PAK1 level was upregulated by epidermal growth factor (EGF), but not glial cell line-derived neurotrophic factor (GDNF) or fibroblast growth factor 2 (FGF2). PAK1 promoted proliferation and DNA synthesis of the human SSC line, whereas PAK1 suppressed its apoptosis in vitro and in vivo. RNA sequencing identified that PDK1, ZNF367, and KDR levels were downregulated by PAK1 knockdown. Immunoprecipitation and Western blots demonstrated that PAK1 interacted with PDK1. PDK1 and KDR levels were decreased by ZNF367-small interfering RNAs (siRNAs). The proliferation of the human SSC line was reduced by PDK1-, KDR-, and ZNF367-siRNAs, whereas its apoptosis was enhanced by these siRNAs. The levels of phos-ERK1/2, phos-AKT, and cyclin A were decreased by PAK1-siRNAs. Tissue arrays showed that PAK1 level was low in non-obstructive azoospermia patients. Collectively, PAK1 was identified as the first molecule that controls proliferation and apoptosis of the human SSC line through PDK1/KDR/ZNF367 and the ERK1/2 and AKT pathways. This study provides data on novel gene regulation and networks underlying the fate of human SSCs, and it offers new molecular targets for human SSCs in translational medicine.
Keywords: PAK1, human spermatogonial stem cells, proliferation, apoptosis, PDK1/KDR/ZNF367, ERK1/2 and AKT pathways
Introduction
Several lines of evidence highlight the most significant studies on spermatogonial stem cells (SSCs) due to their great plasticity. First of all, SSCs can be induced to generate mature and functional spermatozoa, and thus they can be utilized for reproductive medicine to treat male infertility. We have recently shown that human SSCs from cryptorchid patients can be coaxed to differentiate to haploid spermatids with fertilization and developmental ability.1 Second, SSCs are unique adult stem cells that transmit genetic information from one generation to subsequent offspring. Therefore, SSCs can be used as an important source for experimental modifications of the mammalian genome. Third, SSCs divide throughout life and they differentiate to spermatocytes, spermatids, and eventually mature spermatozoa, and thus they can be employed as an excellent model to uncover molecular mechanisms underlying the proliferation versus differentiation of stem cells. Fourth, it has recently been demonstrated that both rodent and human SSCs can acquire pluripotency to become embryonic stem cell (ESC)-like cells that differentiate to mature and functional cells of the three germ cell layers.2, 3, 4 Finally, we and peers have recently reported that mouse and human SSCs are able to directly transdifferentiate to cells with phenotypic and functional hepatocytes;5, 6, 7 neuron;8 and various kinds of tissues, including prostatic, uterine, and skin epithelium.9 As such, human SSCs have great applications in regenerative medicine to treat various kinds of human diseases.
The fate determinations of SSCs are regulated precisely by genetic and epigenetic factors. Much progress has recently been made in unveiling the proliferation, differentiation, and transdifferentiation of rodent SSCs. We and our peers have revealed that GDNF (glial cell line-derived neurotrophic factor) promotes DNA synthesis and proliferation of mouse SSCs via the Ras/ERK1/2 pathway10 and that FGF2 (fibroblast growth factor 2) is required for the survival and proliferation of mouse SSCs.11 BMP4 and SCF have been shown to induce the differentiation of mouse spermatogonia.12 With regard to epigenetic regulation, we have reported that microRNA (miRNA)-20 and miRNA-106a promote the self-renewal of mouse SSCs by targeting STAT3 and cyclin D1.13 MiRNA-21 has been shown to control the proliferation and maintenance of mouse SSCs by the regulation of transcription factor ETV5,14 while miRNA-221 and miRNA-222 maintain the undifferentiated status of mouse spermatogonia through the inhibition of KIT expression.15 In addition, overexpression of microRNA let-7 leads to the decrease of the germline stem cells in Drosophila.16
Recently, it has been demonstrated that miRNA-202 knockout results in the differentiation of mouse SSCs to c-KIT-expressing spermatogonia.17 We have uncovered a number of differentially expressed microRNAs among human spermatogonia, pachytene spermatocytes, and round spermatids.18 Notably, there are distinct cell types and biochemical phenotypes between rodent SSCs and human SSCs. First, cell identities for mouse SSCs and human SSCs are different. The As spermatogonia are generally regarded as the actual stem cells in mice, whereas the Apr and Aal spermatogonia are suggested to be the potential stem cells. In human, the Adark spermatogonia are believed to be the reserve stem cells, while the Apale spermatogonia are thought to be the renewing stem cells.19 Differences have been recently highlighted in spermatogonial subpopulations and kinetics between mice and human.20, 21 Second, human SSCs and mouse SSCs share some, but not all, phenotypic markers, as evidenced by the fact that human SSCs are negative for POU5F1 (also known as Oct4),22 which is a hallmark for rodent SSCs. Lastly, rodent spermatogenic lineage development is different from human. The mouse spermatogenic cycle has 12 stages, whereas there are six spermatogenic cycle stages in human, although a new classification of 12 spermatogenic stages in human spermatogenesis has recently been proposed.23 Consequently, the molecular mechanisms in regulating SSCs in human and other species including rodents could be different due to these significant differences mentioned above. For instance, the JAK/STAT-signaling pathway has been shown to promote the self-renewal of Drosophila SSCs.24 In contrast, STAT3 signaling pathway is essential for the differentiation of mouse SSCs.25 Nevertheless, almost nothing is known about genes and signaling pathways regulating the fate decisions of human SSCs.
To identify new genes required for the proliferation of human SSCs, we performed RNA sequencing, and notably, we found that the transcript of PAK1 (P21-activated kinase 1) was enhanced by 10% fetal bovine serum (FBS) in the human SSC line. Therefore, we hypothesized that PAK1 might play a role in regulating the proliferation and apoptosis of human SSCs. We have recently established a human SSC line with morphological, phenotypic, and functional features of human primary SSCs,26 and, therefore, this human SSC line was utilized to uncover the role and mechanism of PAK1. We observed that EGF (epidermal growth factor), but not GDNF or FGF2, elevated PAK1 level in the human SSC line. PAK1 promoted DNA synthesis and proliferation but inhibited apoptosis of the human SSC line. PAK1 regulated PDK1, ZNF367, and KDR, and, interestingly, PAK1 interacted with PDK1 while ZNF367 controlled PDK1 and KDR. Furthermore, PAK1 small interfering RNAs (siRNAs) inactivated the ERK1/2 and AKT pathway and decreased the levels of cyclin A rather than cyclin B1, cyclinD1, and CDK2. Additionally, we found that PAK1 levels were significantly lower in several types of non-obstructive azoospermia (NOA) patients than obstructive azoospermia (OA) patients with normal spermatogenesis. Therefore, this study offers new insights into molecular mechanisms underlying the proliferation and apoptosis of human SSCs, and it provides novel clues for the application of human SSCs in reproduction and regenerative medicine.
Results
The Human SSC Line Expresses a Number of Genes and Proteins for Human SSCs
We first verified the identity of the human SSC line. RT-PCR and Western blots showed that the cell line expressed SV40 mRNA (Figure S1A) and SV40 protein (Figure S1E). RT-PCR revealed that the human cell line expressed numerous genes for human germ cells and human spermatogonia, including VASA and MAGEA4 (MAGE family member A4) (Figure S1B), as well as markers for human SSCs, e.g., GPR125 (G protein-coupled receptor 125), GFRA1 (GDNF family receptor alpha 1), RET (Ret proto-oncogene), UCHL1 (ubiquitin C-terminal hydrolase L1), THY1, and PLZF (Figure S1C). In addition, SOX9 and GATA4 were detected in human Sertoli cells, whereas UCHL1, GPR125, GFRA1, THY1, RET, MAGEA4, and VASA were undetectable in these cells (Figure S1D), thus confirming the specific expression of the genes in the human SSC line. Western blots displayed that the proteins of GPR125 (Figure S1E), THY1 (Figure S1E), RET (Figure S1F), DAZ2 (Figure S1F), and UCHL1 (Figure S1F) were present in this cell line. Immunocytochemistry further revealed that the human cell line was positive for THY1 (Figure S1G), GPR125 (Figure S1H), and GFRA1 (Figure S1I). Replacement of primary antibodies with isotype rabbit or goat immunoglobulin Gs (IgGs) was used as negative controls (Figures S1J and S1K), and no immunostaining was observed, thus verifying specific staining of the antibodies mentioned above in the cell line. Together, these results indicate that the human cell line is human SSCs phenotypically.
PAK1 Is Elevated by EGF, but Not GDNF or FGF2, and It Is Expressed in Human SSCs
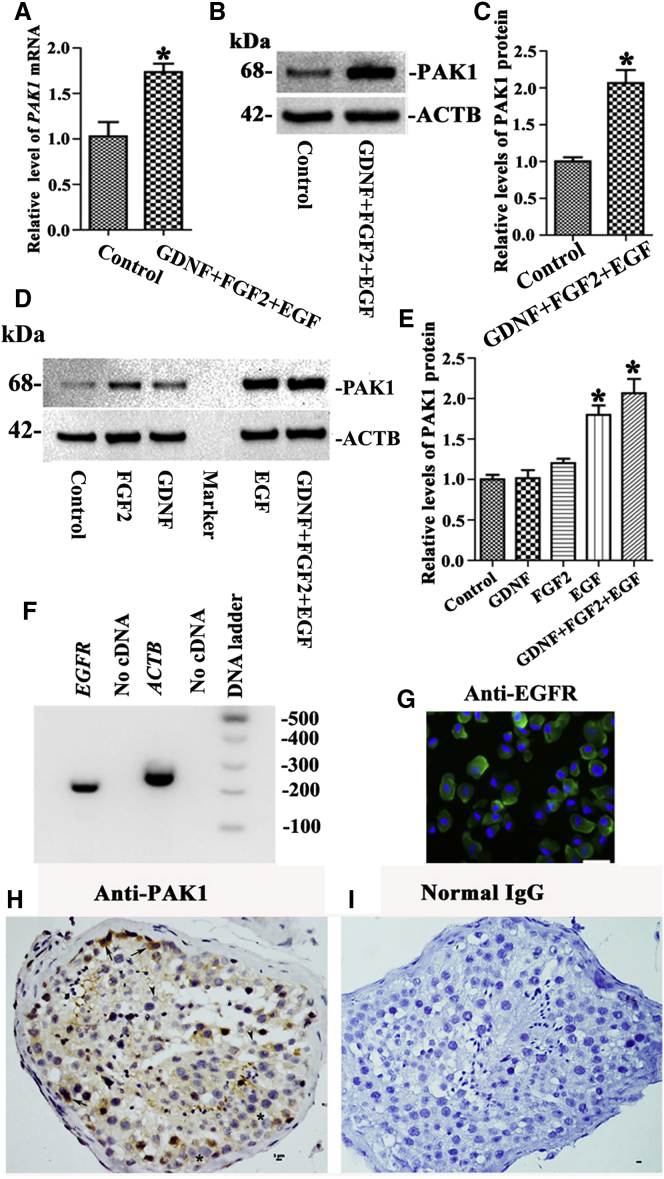
To identify novel genes that are essential for the proliferation of human SSCs, we conducted RNA sequencing showing that PAK1 transcript was elevated at 2.218-fold by 10% FBS compared to 0.5% FBS in the human SSC line. Real-time PCR and Western blots demonstrated that PAK1 mRNA and PAK1 protein were enhanced by 10% FBS compared with 0.5% FBS in the human SSC line, respectively (Figures S2A–S2C). Since FBS contains several growth factors, we determined whether the levels of PAK1 were changed by the defined growth factors. Real-time PCR revealed that PAK1 mRNA was upregulated by growth factors EGF, FGF2, and GDNF at 10 hr of the treatment in the human SSC line (Figure 1A), and Western blots indicated that PAK1 protein was enhanced by these growth factors at 24 hr of the treatment in the human SSC line (Figures 1B and 1C). To ascertain which growth factor regulates PAK1, we performed Western blots showing that the level of PAK was elevated by EGF, but not by GDNF or FGF2, in the human SSC line (Figures 1D and 1E). These data suggest that PAK1 is regulated by EGF rather than GDNF or FGF2 in the human SSC line. RT-PCR and immunocytochemistry demonstrated that the EGF receptor EGFR mRNA (Figure 1F) and EGFR protein (Figure 1G) were present in the human SSC line.
Figure 1.
The Effect of EGF, FGF2, and GDNF on PAK1, EGFR Presence in the Human SSC Line, and the Expression of PAK1 in Human Primary SSCs
(A) Real-time PCR displayed mRNA changes of PAK1 by growth factors EGF, FGF2, and GDNF in the human SSC line. (B and C) Western blots showed protein changes of PAK1 (B) and its relative level (C) by these growth factors mentioned above in the human SSC line. Culture medium without EGF, FGF2, or GDNF was used as the control. (D and E) Western blots demonstrated the protein changes of PAK (D) or its relative level (E) by EGF, GDNF, or FGF2 or EGF + GDNF + FGF2 in the human SSC line. *Statistically significant differences (p < 0.05) between the individual growth factor, EGF + FGF2 + GDNF-treated cells, and the control (A, C, and E). (F and G) RT-PCR and immunocytochemistry showed the expression of EGFR mRNA (F) and EGFR protein (G) in the human SSC line. Scale bar, 10 μm (G). (H) Immunohistochemistry revealed cellular localization of PAK1 in human spermatogonia (arrows), but not pachytene spermatocytes (asterisks) or round spermatids (arrow heads). (I) Replacement of anti-PAK1 with isotype IgG was used as a negative control. Scale bars, 5 μm (H and I).
We determined whether PAK1 was expressed in human primary SSCs. Immunohistochemistry revealed that PAK1 was expressed in human spermatogonia along the basement of seminiferous tubules in human testes (Figure 1H), whereas it was undetected in pachytene spermatocytes or round spermatids (Figure 1H). Primary antibody was replaced with isotype IgGs to serve as negative controls, and no immunostaining was seen (Figure 1I). Collectively, these results imply that PAK1 is present in human SSCs.
PAK1 Knockdown Leads to Decreases in the Proliferation and DNA Synthesis and an Increase in the Apoptosis in the Human SSC Line
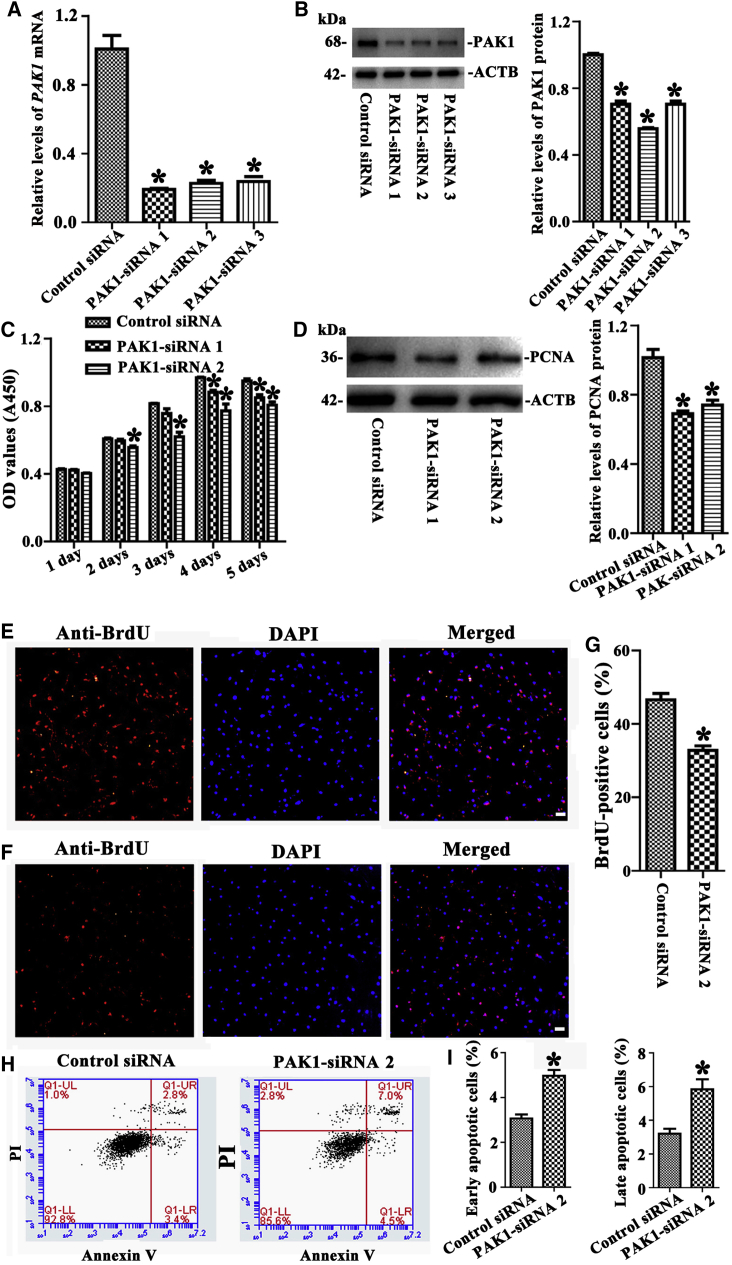
Next, we explored the roles of PAK1 in regulating the proliferation and apoptosis of human SSCs in vitro. To improve the efficiency of PAK1 knockdown, three pairs of siRNAs targeting different regions of PAK1 mRNA were designed, and control siRNA didn’t bind any sequence of PAK1. Real-time PCR revealed that PAK1 mRNA was significantly decreased by PAK1-siRNA 1, 2, and 3 in the human SSC line at 24 hr after transfection (Figure 2A). Western blots showed that the protein of PAK1 was reduced by PAK1-siRNA 1, 2, and 3 in the human SSC line, while the interfering effect of PAK1-siRNA 2 was the most prominent (Figure 2B). CCK-8 assay was executed from 24 to 120 hr after the transfection of PAK1-siRNA 2 and 1 in the human SSC line, and both siRNAs evidently reduced the proliferation of the human SSC line at 48–120 hr (Figure 2C). Western blots indicated that the level of PCNA, a hallmark for cell proliferation, was decreased by PAK1-siRNA 2 and 1 (Figure 2D). Similarly, the percentage of bromodeoxyuridine (BrdU)-positive cells was reduced in the human SSC line by PAK1-siRNA 2 (Figures 2E and 2G) compared to the control siRNA (Figures 2F and 2G). Furthermore, annexin V/propidium iodide (PI) staining and flow cytometry implicated that the early and late apoptosis of the human SSC line was increased by PAK1-siRNA 2 compared to the control siRNA (Figures 2H and 2I). Considered together, these data indicate that PAK1 promotes DNA synthesis and the proliferation and inhibits the apoptosis of the human SSC line.
Figure 2.
The Influence of PAK1 Knockdown on the Proliferation, DNA Synthesis, and Apoptosis of the Human SSC Line
(A) Real-time PCR showed mRNA changes of PAK1 by PAK1-siRNA 1, 2, and 3 in the human SSC line. (B) Western blots revealed the protein changes of PAK1 by PAK1-siRNA 1, 2, and 3 in the human SSC line. (C) CCK-8 assay demonstrated the proliferation of the human SSC line after transfection of PAK1-siRNA 1 and 2. (D) Western blots displayed the changes of PCNA protein by PAK1-siRNA 1 and 2 in human SSCs. (E–G) BrdU incorporation assay showed the percentages of BrdU-positive cells affected by control siRNA (E and G) and PAK1-siRNA 2 (F and G) in the human SSC line. Scale bars, 20 μm (E and F). (H and I) Annexin V/PI staining and flow cytometry displayed the percentages of early (I, left panel) and late (I, right panel) apoptosis in the human SSC line affected by PAK1-siRNA 2 (H, right panel) and control siRNA (H, left panel). *Statistically significant differences (p < 0.05) between PAK1-siRNA -treated cells and the control siRNA (A–D, G, and I).
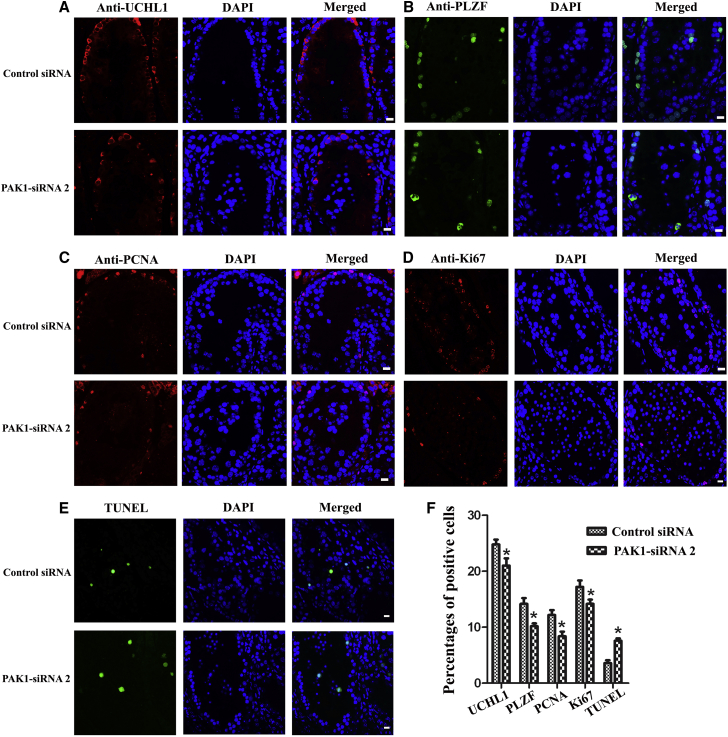
We also determined the role of PAK1 in regulating human SSCs in vivo. Xenotransplantation was performed with the human SSC line by transfecting PAK1-siRNA 2 or control siRNA. Immunohistochemistry revealed that PAK1-siRNA 2 resulted in decrease in the percentages of UCHL1-, PLZF-, PCNA-, and Ki67-positive cells and the enhancement of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells (Figures 3A–3F). These data implicate that PAK1 knockdown inhibits the proliferation and stimulates the apoptosis of the human SSC line in vivo.
Figure 3.
PAK1-siRNA 2 Knockdown Led to the Reduction of Proliferation and the Increase of Apoptosis in the Recipient Mice Grafted with the Human SSC Line
(A–F) Immunohistochemistry showed the expression of UCHL1- (A), PLZF- (B), PCNA- (C), Ki67- (D), and TUNEL- (E) positive cells and their percentages (F) in the recipient nude mice grafted with the human SSC line by PAK1-siRNA 2 or control siRNA transfection. Scale bars, 10 μm (A–E). *Statistically significant differences (p < 0.05) between PAK1-siRNA 2-treated cells and the control siRNA. Notes in (F): the percentage of TUNEL-positive cells was calculated by TUNEL-positive positive cells from all cells within the seminiferous tubule. The percentage of UCHL1-, PLZF-, PCNA-, and Ki67-positive cells were counted by the positive cells from the cells along the basement membrane. At least 3 integrated seminiferous tubules were randomly selected in each testicular section, and at least 5 sections were counted.
PDK1, KDR, and ZNF367 Are Targets of PAK1 in the Human SSC Line, and PAK1 Interacts with PDK1
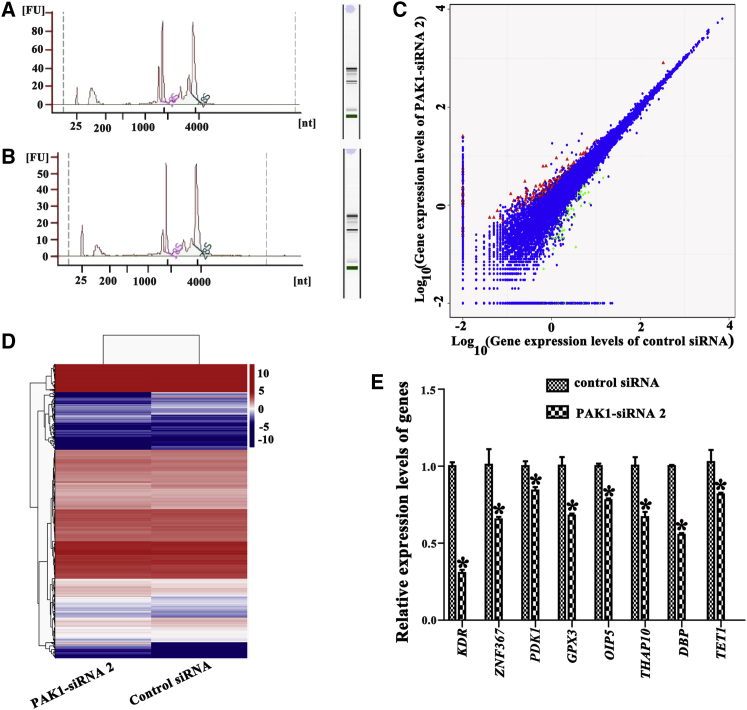
To identify the targets of PAK1 in regulating human SSCs, RNA sequencing was performed to screen the changes of transcription profiles in the human SSC line transfected with PAK1-siRNA 2 or control siRNA. The value of RIN (RNA Integrity Number) was 8.3, and electropherogram by an Agilent bioanalyzer displayed the concentrations and nucleotides (nt) of RNA isolated from the human SSC line with control siRNA (Figure 4A) and PAK1-siRNA 2 (Figure 4B). There were 16,961 genes expressed in the human SSC line with control siRNA and 16,912 genes present in the human SSC line with PAK1-siRNA 2. Scatterplots and hierarchical clustering showed that 97 genes were downregulated and 140 genes were upregulated by PAK1-siRNA 2 (Figures 4C and 4D). Gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses indicated that the changed genes, including PDK1, KDR, and ZNF367, by PAK1-siRNA 2 were involved in various kinds of signaling pathways and biological process (Figures S3–S5; Table 1), which are essential for cell proliferation and survival. Notably, RNA sequencing revealed that a number of genes, including PDK1, KDR, and ZNF367, were downregulated after PAK1 knockdown (Table 2). Real-time PCR showed that the transcripts of randomly chosen genes, including PDK1, KDR, ZNF367, GPX3, OIP5, THAP10, DBP, and TET1, were decreased by PAK1-siRNA 2 (Figure 4E), which was consistent with the data of RNA sequencing.
Figure 4.
Identification of PDK1, KDR, and ZNF367 as the Targets for PAK1 in Human SSCs
(A and B) Electropherogram showed the concentrations and nucleotides (nt) of RNA isolated from the human SSC line with the control siRNA (A) and PAK1-siRNA 2 (B). (C and D) Scatterplots (C) and hierarchical clustering (D) illustrated the differentially expressed genes (DEGs) between PAK1-siRNA 2 and control siRNA. Red dots and green dots in (C) represented upregulated and downregulated genes, respectively. (E) Real-time PCR revealed the changes of PDK1, KDR, ZNF367, GPX3, OIP5, THAP10, DBP, and TET1 mRNA by PAK1-siRNA 2 compared to the control siRNA. *Statistically significant differences (p < 0.05) between PAK1-siRNA 2-treated cells and the control siRNA.
Table 1.
Gene Ontology Analysis of the DEGs between PAK1-siRNA 2 and Control siRNA
| Biological Process | Gene Symbol |
|---|---|
| Signaling transduction | PAK1, KDR, PDGFRL, RAB4B-EGLN2, FGF7P6, ISOC1, CEP126, TIAM1, GTF2IP4 |
| CTNND1, PAIP2B, CSNK1E, GHR, WDR17, EDNRA, PARD6G, TIAM1, DGKI, LINC00910, CNTF, CDH1, PDK1 | |
| Gene transcription | ZNF367, GTF2IP4, POLR2J2, GTF2H2, GTF2IP1, LOC388436, LOC79999 |
| Cell growth, aging, and death | TUBA3FP, SFXN2, STAG3L2, STAG3L1, RNASEK, CCZ1B, WASH3P, GSTTP2 |
Table 2.
RNA Sequencing Showed the DEGs between PAK1-siRNA 2 and Control siRNA in the Human SSC Line
| Gene Symbol | Gene Description | Fold Changes (PAK1-siRNA 2/Control-siRNA) | p Value |
|---|---|---|---|
| PAK1 | p21-activated kinase 1 | 0.268 | 0.000000 |
| KDR | kinase insert domain receptor | 0.347 | 0.000075 |
| ZNF367 | zinc finger protein 367 | 0.366 | 0.000137 |
| PDK1 | 3-phosphoinositide dependent protein kinase 1 | 0.795 | 0.000481 |
| GPX3 | glutathione peroxidase 3 | 0.409 | 0.000459 |
| OIP5 | Opa interacting protein 5 | 0.324 | 0.000445 |
| THAP10 | THAP domain containing 10 | 0.335 | 0.001493 |
| DBP | albumin D-box binding protein | 0.424 | 0.001132 |
| TET1 | tet methylcytosine dioxygenase 1 | 0.5 | 0.000088 |
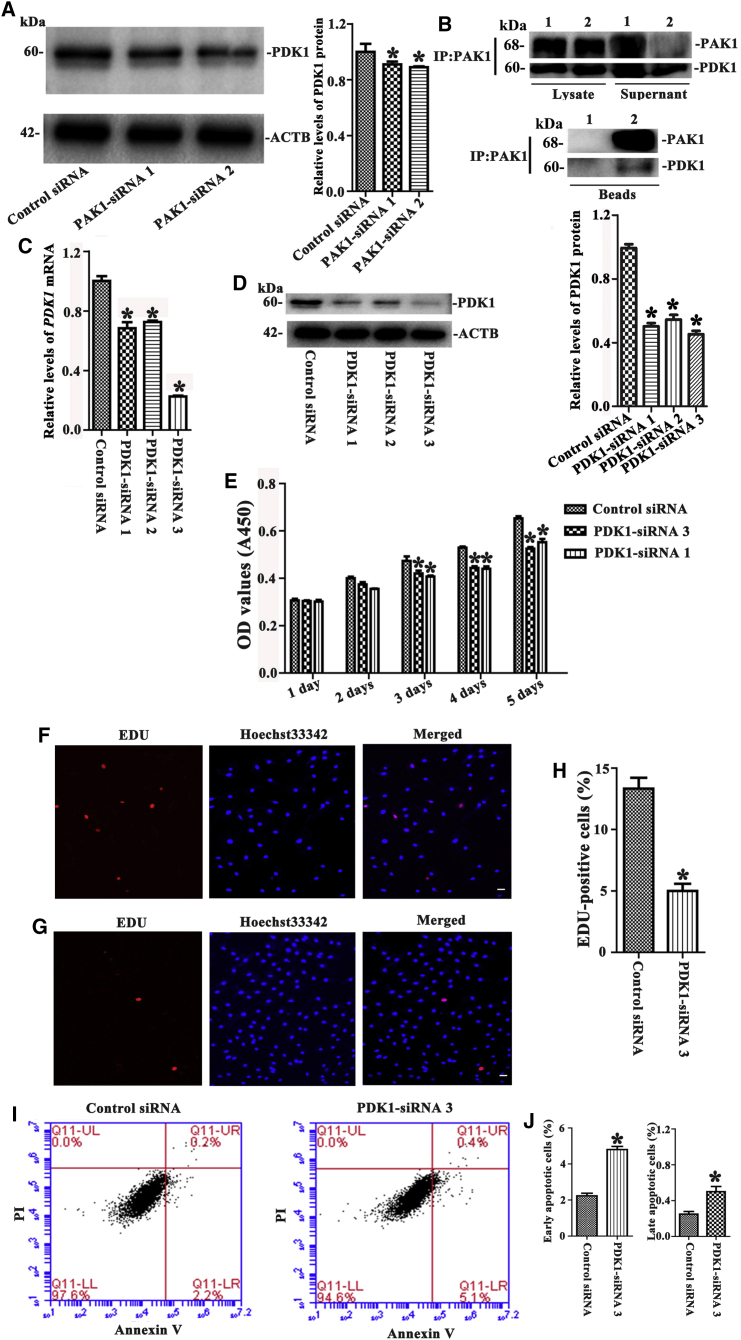
We examined whether there was an interaction between PAK1 and PDK1 in the human SSC line. Western blots showed that the level of PDK1 protein was decreased by PAK1-siRNA 2 and PAK1-siRNA 1 (Figure 5A). Significantly, immunoprecipitation assay and Western blots further revealed that PAK1 could bind to PDK1 in the human SSC line, whereas IgG couldn’t pull down PDK1 (Figure 5B).
Figure 5.
The Influence of PDK1 Knockdown on the Proliferation, DNA Synthesis, and Apoptosis of the Human SSC Line
(A) Western blots showed protein changes of PDK1 by PAK1-siRNA 1 and 2 in the human SSC line. *Statistically significant differences (p < 0.05) between PAK1-siRNA 1- and 2-treated cells and the control siRNA. (B) Immunoprecipitation assay and Western blots demonstrated the binding of anti-PAK1 to PDK1 in the human SSC line. 1, IgG; 2, anti-PAK1. (C) Real-time PCR revealed the mRNA changes of PDK1 by PDK1-siRNA 1, 2, and 3 in the human SSC line. (D) Western blots revealed the protein changes of PDK1 by PDK1-siRNA 1, 2, and 3 in the human SSC line. (E) CCK-8 assay showed the proliferation of the human SSC line after transfection of PDK1-siRNA 1 and 3. (F–H) EDU incorporation assay demonstrated the percentages of EDU-positive cells affected by control siRNA (F and H) and PDK1-siRNA 3 (G and H) in the human SSC line. Scale bars, 20 μm (F and G). (I and J) Annexin V/PI staining and flow cytometry demonstrated the percentages of early (J, left panel) and late apoptosis (J, right panel) in the human SSC line treated with PDK1-siRNA 3 (I, right panel) and control siRNA (I, left panel). *Statistically significant differences (p < 0.05) between PDK1-siRNA-treated cells and the control siRNA (C–E, H, and J).
PDK1 Knockdown Results in Decreases in the Proliferation and DNA Synthesis and an Increase in the Apoptosis in the Human SSC Line
Real-time PCR and Western blots revealed that PDK1 mRNA and PDK1 protein were decreased by PDK1-siRNA 1, 2, and 3 in the human SSC line (Figures 5C and 5D), while the interfering effect of PDK1-siRNA 3 was the most (Figures 5C and 5D). CCK-8 and 5-Ethynyl-2′-deoxyuridine (EDU) assays indicated that the proliferation of the human SSC line and EDU-positive cells were decreased by PDK1-siRNA 3 (Figures 5E–5H). Additionally, the early and late apoptosis of the human SSC line was increased by PDK1-siRNA 3 compared to the control siRNA (Figures 5I and 5J).
KDR Silencing Causes the Decrease in Proliferation and an Enhancement in the Apoptosis in the Human SSC Line
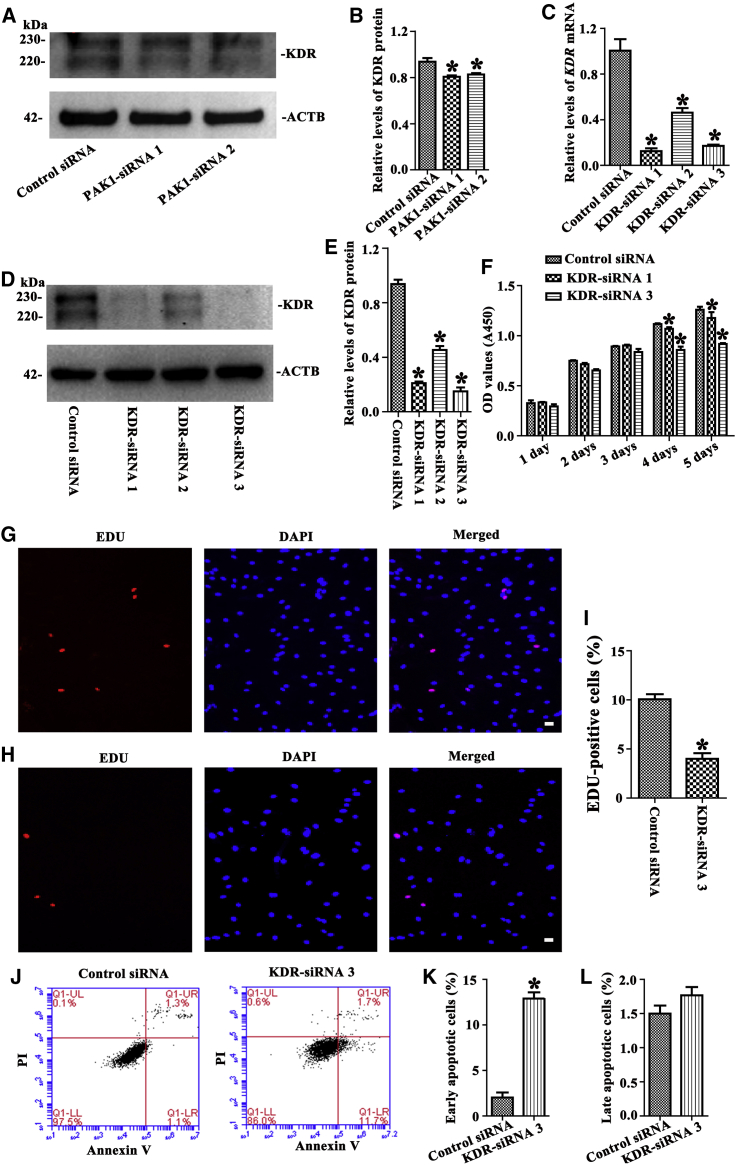
As shown in Figures 6A and 6B, KDR protein was decreased by PAK1-siRNA 1 and PAK1-siRNA 2, suggesting that KDR is a target of PAK1 in the human SSC line. In consideration of these changes after PAK1 knockdown, we investigated the effect of KDR on the proliferation and apoptosis of the human SSC line. Real-time PCR and Western blots demonstrated that the transcript of KDR and translation of KDR were reduced significantly by KDR-siRNA 1, 2, and 3 (Figures 6C–6E). CCK-8 and EDU assays showed that the proliferation (Figure 6F) and EDU-positive cells (Figures 6G–6I) were decreased in the human SSC line by KDR-siRNA 1 and KDR-siRNA 3. Annexin V/PI staining and flow cytometry indicated that KDR-siRNA 3 increased the early apoptotic percentage of the human SSC line (Figures 6J and 6K), although the change of later apoptosis was not significant in the human SSC line without or with KDR-siRNA 3 (Figures 6J and 6L).
Figure 6.
The Role of KDR Silencing on the Proliferation, DNA Synthesis, and Apoptosis of the Human SSC Line
(A and B) Western blots showed protein changes of KDR (A) and its relative level (B) by PAK1-siRNA 1 and 2 in the human SSC line. *Statistically significant differences (p < 0.05) between PAK1-siRNA 1- and 2-treated cells and the control siRNA. (C) Real-time PCR showed mRNA changes of KDR by KDR-siRNA 1, 2, and 3 in the human SSC line. (D and E) Western blots demonstrated protein changes of KDR (D) and its relative level (E) by KDR-siRNA 1, 2, and 3 in the human SSC line. (F) CCK-8 assay showed the proliferation of the human SSC line after transfection of KDR-siRNA 3 and KDR-siRNA 1. (G–I) EDU incorporation assay illustrated the percentages of EDU-positive cells affected by the control siRNA (G and I) and KDR-siRNA 3 (H and I) in the human SSC line. Scale bars, 10 μm (G and H). (J–L) Annexin V/PI staining and flow cytometry demonstrated the percentages of early (K) and late (L) apoptosis in the human SSC line treated with KDR-siRNA 3 (J, right panel) and control siRNA (J, left panel). *Statistically significant differences (p < 0.05) between KDR-siRNA-treated cells and the control siRNA (C, E, F, I, K, and L).
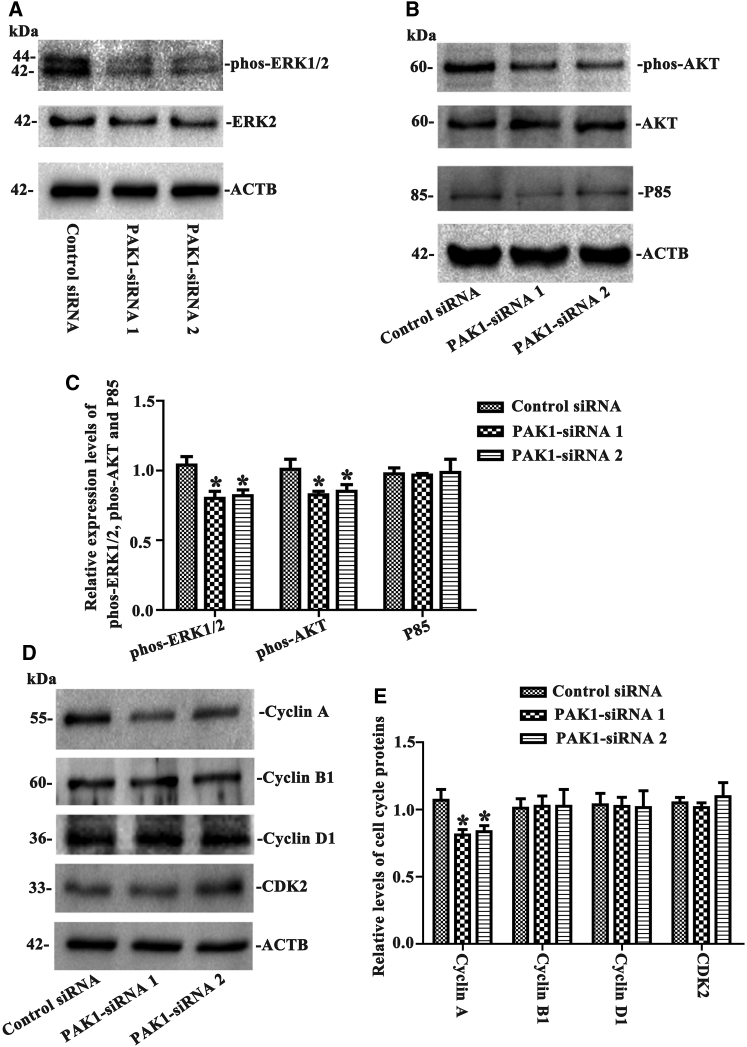
PAK1 Knockdown Leads to the Inactivation of phos-ERK1/2 and phos-AKT and the Decrease of Cyclin A in the Human SSC Line
To seek which signaling pathways were activated by PAK1 in the human SSC line, we assessed the changes of phos-ERK1/2, phos-AKT, and P85. Western blots showed that the levels of phos-ERK1/2 (Figures 7A and 7C) and phos-AKT (Figures 7B and 7C) were obviously reduced by PAK1-siRNA 1 and PAK1-siRNA 2, while there was no obvious change of P85 protein in the human SSC line without or with PAK1-siRNA 1 and PAK1-siRNA 2 (Figures 7B and 7C).
Figure 7.
The Influence of PAK1-siRNAs on the Levels of phos-ERK1/2, phos-AKT, P85, and Cell Cycle Proteins in the Human SSC Line
(A–E) Western blots showed the level changes of phos-ERK1/2 (A and C), phos-AKT (B and C), P85 (B and C), cyclin A (D and E), cyclin B1 (D and E), cyclin D1 (D and E), and CDK2 (D and E) in the human SSC line with PAK1-siRNA 1 and 2 and control siRNA. ERK2, AKT, and ACTB were used as controls of loading proteins. *Statistically significant differences (p < 0.05) between PAK1-siRNA 1- and 2-treated cells and the control siRNA.
Furthermore, we checked out how PAK1 affected the cell cycle-related proteins, including cyclin A, cyclin B1, cyclin D1, and CDK2. As shown in Figures 7D and 7E, PAK1 knockdown resulted in the decrease in cyclin A, but not cyclin B1, cyclin D1, or CDK2, in the human SSC line.
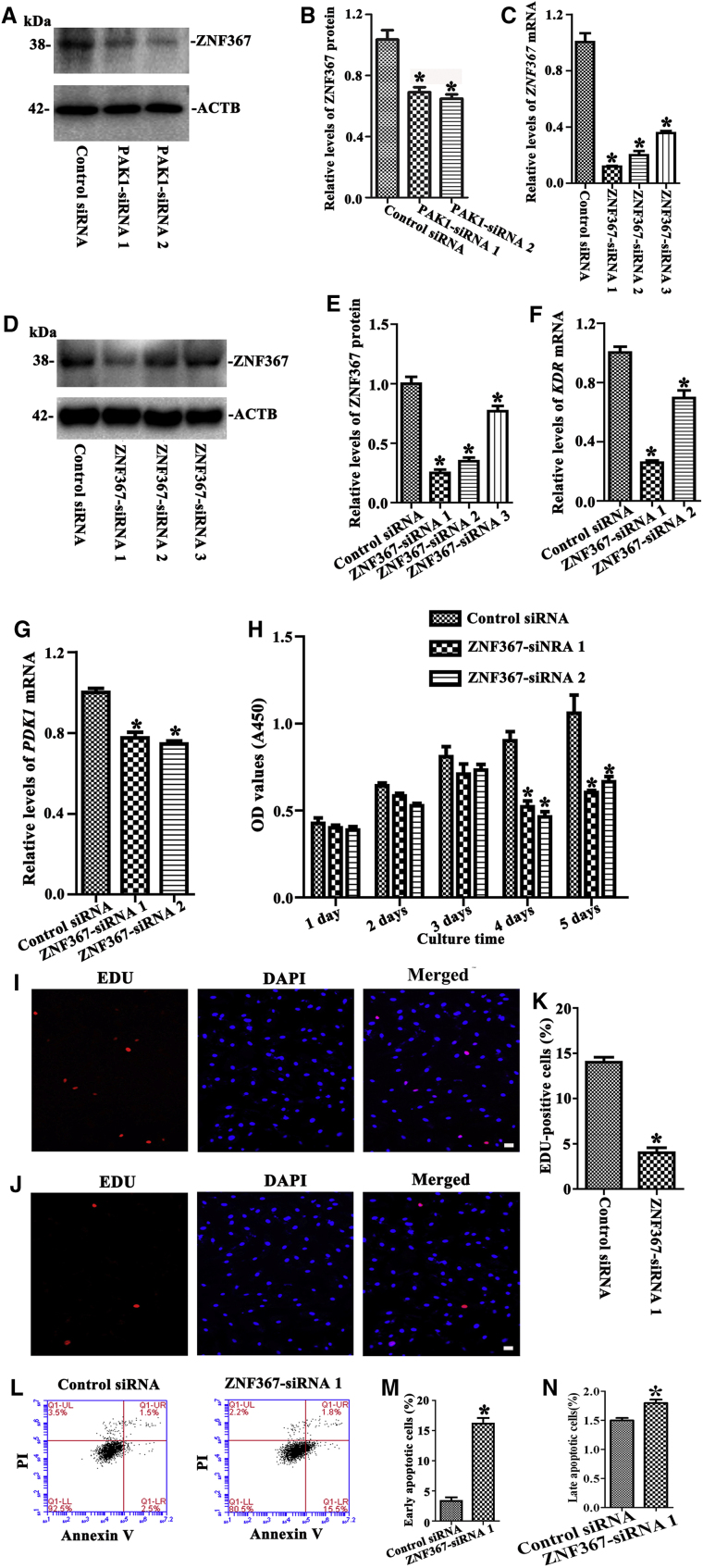
Transcription Factor ZNF367 Silencing Results in the Reduction in the Proliferation and the Increase in the Apoptosis in the Human SSC Line
The level of ZNF367 protein was decreased by PAK1-siRNA 1 and PAK1-siRNA 2 (Figures 8A and 8B). We next explored the effect of transcription factor ZNF367 on the proliferation and the apoptosis in the human SSC line. Real-time PCR and Western blots showed that ZNF367 mRNA (Figure 8C) and ZNF367 protein (Figures 8D and 8E) were diminished by ZNF367-siRNA 1, 2, and 3 in the human SSC line. Interestingly, the transcripts of KDR (Figure 8F) and PDK1 (Figure 8G) were reduced by ZNF367-siRNA 1 and ZNF367-siRNA 2 in the human SSC line, reflecting that ZNF367 regulates KDR and PDK1 in the human SSC line.
Figure 8.
The Effectiveness of ZNF367 Silencing on the Proliferation, DNA Synthesis, and Apoptosis of the Human SSC Line
(A and B) Western blots showed protein changes of ZNF367 (A) and its relative level (B) by PAK1-siRNA 1 and 2 in the human SSC line. *Statistically significant differences (p < 0.05) between PAK1-siRNA 1- and 2-treated cells and the control siRNA. (C) Real-time PCR revealed mRNA changes of ZNF367 by ZNF367-siRNA 1, 2, and 3 in the human SSC line. (D and E) Western blots showed protein changes of ZNF367 (D) and its relative level (E) by ZNF367-siRNA 1, 2, and 3 in the human SSC line. (F and G) Real-time PCR revealed mRNA changes of KDR (F) and PDK1 (G) by ZNF367-siRNA 1 and 2 in the human SSC line. (H) CCK-8 assay showed the proliferation of the human SSC line after transfection of ZNF367-siRNA 1 and 2. (I–K) EDU incorporation assay illustrated the percentages of EDU-positive cells affected by control siRNA (I and K) and ZNF367-siRNA 1 (J and K) in the human SSC line. Scale bars, 20 μm (I and J). (L–N) Annexin V/PI staining and flow cytometry demonstrated the percentages of early (M) and late (N) apoptosis in the human SSC line treated with ZNF367-siRNA 1 (L, right panel) and control siRNA (L, left panel). *Statistically significant differences (p < 0.05) between ZNF367-siRNA-treated cells and the control siRNA (C, E–H, K, M, and N).
CCK-8 and EDU assays demonstrated that the proliferation (Figure 8H) and EDU-positive cells (Figures 8I–8K) were decreased in the human SSC line by ZNF367-siRNA 1 and znf367-siRNA 2. In contrast, the early and late apoptotic percentages of the human SSC line were enhanced by ZNF367-siRNA 1 (Figures 8L–8N).
PAK1 Abnormal Expression Is Associated with NOA Patients
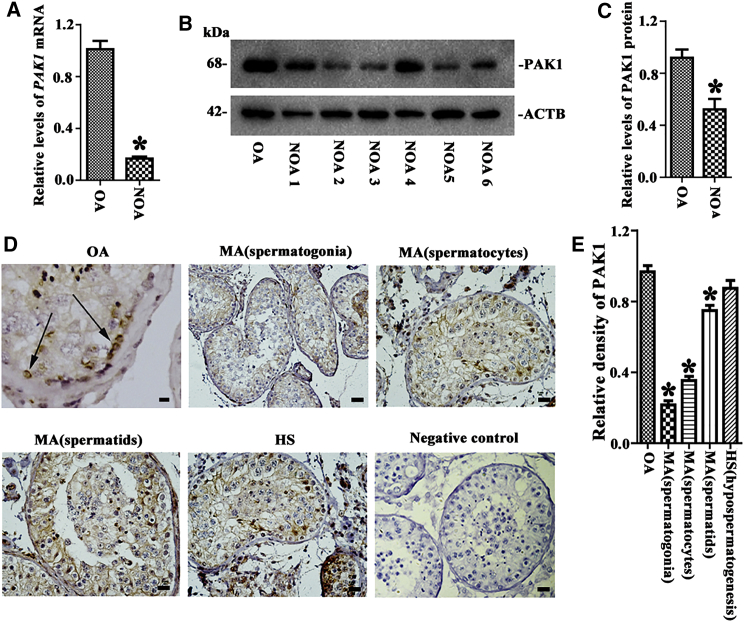
NOA is a serious disease affecting human reproduction, and it can be classified into several subtypes, including maturation arrest (MA) at different stages of spermatids, spermatocytes, or spermatogonia and hypo-spermatogenesis (HS). We finally compared the levels of PAK1 among subtypes of NOA patients and OA patients with normal spermatogenesis. NOA 1, 4, and 5 were patients with hypo-spermatogenesis, while NOA 2, 3, and 6 were patients with MA. Male germ cells, including spermatogonia, spermatocytes, and spermatids, were separated from the testicular tissues of OA and NOA patients using the two-step enzymatic digestion and differential plating. The isolated cells were positive for VASA (Figure S6A) and DAZL (Figure S6B) and negative for WT1 (Figure S6C) or GATA4 (Figure S6D), reflecting that these cells are male germ cells without contamination of human Sertoli cells. Real-time PCR and Western blots demonstrated that the transcript of PAK1 (Figure 9A) and PAK1 protein (Figures 9B and 9C) were lower in human male germ cells of NOA patients than OA patients.
Figure 9.
The Expression of PAK1 in Male Germ Cells and the Testis of OA Patients and Certain Subtypes of NOA Patients
(A–C) Real-time PCR and Western blots revealed the transcripts of PAK1 (A) and PAK1 protein (B and C) in male germ cells of NOA patients and OA patients. (D and E) Tissue arrays showed the levels of PAK1 protein (D) and its relative level (E) in MA at spermatogonia, spermatocytes, or spermatids, HS patients, and OA patients. Specific immunostaining of PAK1 in human spermatogonia (arrows) was shown in an OA patient. *Statistically significant differences (p < 0.05) between NOA patients and OA patients (C and E). The quantification of PAK1 immunostaining was determined by calculating the number scores of PAK1-positive cells and the intensity scores of staining.
Tissue arrays further revealed that the levels of PAK1 in spermatogonia were lower in MA at spermatogonium, spermatocyte, and spermatid stages than OA patients (Figures 9D and 9E), while there was no significant difference in PAK1 expression level between HS patients and OA patients (Figures 9D and 9E), suggesting that PAK1 abnormal expression is associated with MA at spermatogonium, spermatocyte, and spermatid stages.
Discussion
Basic studies and clinical applications of human SSCs have been seriously hampered due to a limited number of human primary SSCs and the difficulty of obtaining human testicular tissues.26 To solve these urgent problems, we have established for the first time a stable human SSC line with an unlimited proliferation potential and high safety.26 In this study, we verified the identity of the human SSC line as human SSCs at both transcriptional and translational levels for numerous makers of primary human SSCs. This human SSC line could survive, proliferate, and colonize in vivo after xenotransplantation to the seminiferous tubules.26 Therefore, the human SSC line could be utilized to obtain a sufficient number of human SSCs for uncovering the function and signaling pathway of novel genes in regulating the fate decisions of human SSCs. Here we have identified PAK1 as the first molecule that controls the proliferation, DNA synthesis, and apoptosis of human SSCs.
Growth factors have been shown to be indispensible for mediating the proliferation and survival of rodent SSCs. It has been demonstrated that GDNF is essential for the maintenance of mouse undifferentiated spermatogonia,27 and we have revealed that GDNF upregulates c-Fos transcription and induces CREB-1, ATF-1, and CREM-1 phosphorylation to stimulate mouse SSC proliferation.10 The balance between GDNF and FGF2 levels has a vital effect on SSC self-renewal in vivo.28 EGF is composed of 53 amino acid residues and it is produced by various kinds of tissues, including testis.29 Sialoadenectomy in male mice leads to the reduction of circulating EGF and impaired spermatogenesis, and, conversely, the administration of EGF can reverse this phenomenon. The average concentrations of EGF in blood plasma are significantly lower in infertile males.30 EGF can promote the proliferation and DNA synthesis of rat spermatogonia,31 and it facilitates the growth of rat SSCs.32 To identify novel genes that regulate the proliferation of human SSCs, we chose GDNF, FGF2, and EGF for culturing and expanding the human SSC line. Interestingly, we found that PAK1 transcript and PAK1 protein were elevated by EGF, but not by GDNF or FGF2, implicating that PAK1 is regulated by EGF in the human SSC line.
We found that PAK1 was expressed in the human SSC line and human primary spermatogonia in human testis. PAK1 has been reported to play an important role in regulating cell proliferation and apoptosis in many other kinds of human normal cells and cancer cells.33, 34 However, the roles and molecular mechanisms of PAK1 in controlling human SSCs remain unknown. To elucidate the function of PAK1, we performed CCK-8 assay, BrdU incorporation assay, and annexin V/PI staining and flow cytometry in the human SSC cell line with PAK1-siRNAs. Specifically, we have demonstrated that PAK1 silencing leads to the reduction in proliferation and DNA synthesis of human SSCs as well as the increase in apoptosis in the human SSC line.
To identify the targets of PAK1, we conducted RNA sequencing to compare the changes of global gene expression levels between PAK1-siRNA and control siRNA in human SSCs. Significantly, we observed that both transcripts and proteins of PDK1, KDR, and ZNF367 were decreased by PAK1-siRNAs. PDK1 conditional knockout in germline cells results in the decrease in fertility.35 KDR is a type III receptor tyrosine kinase and it is one of receptors for vascular endothelial growth factor (VEGF). It has been demonstrated that VEGFA regulates the maintenance and survival of SSCs.36 ZNF367 belongs to the zinc-finger protein family, e.g., Plzf, a crucial transcription factor for the self-renewal of mouse SSCs.37, 38 Notably, we found that PAK1 could interact with PDK1 since PAK1 binds to PDK1, as shown by immunoprecipitation, and that ZNF367 could regulate PDK1 and KDR. Furthermore, we demonstrated that the silencing of PDK1, KDR, and ZNF367 resulted in the suppression of proliferation and DNA synthesis and an enhancement of apoptosis in the human SSC line. These results suggest that PDK1, KDR, and ZNF367 are targets for PAK1 and that PDK1, KDR, and ZNF367 play important roles in regulating the proliferation and apoptosis of the human SSC line.
PAK1 acts via different kinds of signaling pathways, including the PI3K/AKT pathway, MAPK pathway, and β-catenin pathway, in different human cell types.39, 40 In this study, we found that the levels of phos-ERK1/2 and phos-AKT rather than P85 were decreased by PAK1-siRNAs in the human SSC line, indicating that PAK1 works through the ERK1/2- and AKT-signaling pathways. We also found that PAK1 mediated cyclin A, but not cyclin B1, cyclin D1, or CDK2. Furthermore, we have unveiled an association between the levels of PAK1 and several subtypes of NOA patients. The pathogenesis of NOA patients remains largely unknown. It has been reported that the number of human SSCs in NOA patients is significantly reduced compared with men with normal spermatogenesis.41 Our data suggest that a low level of PAK1 might lead to the abnormal spermatogenesis of NOA patients.
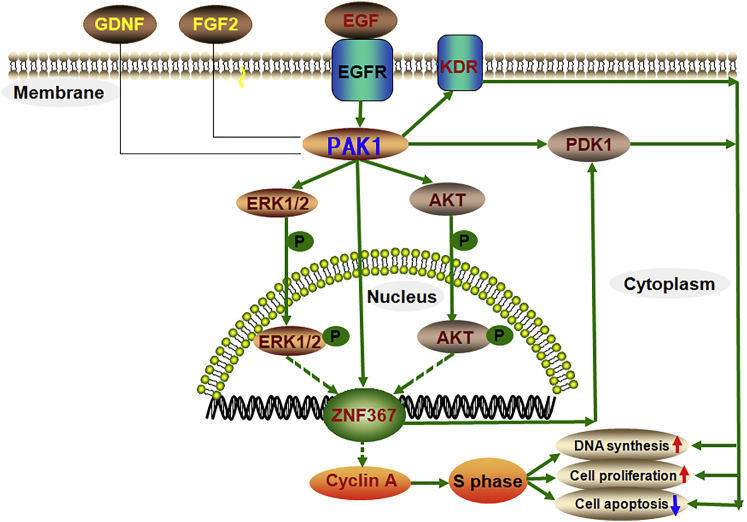
In summary, we have demonstrated for the first time that PAK1 is expressed in the human SSC line and primary human SSCs and that it regulates the proliferation, DNA synthesis, and apoptosis of the human SSC line. The function and mechanism of PAK1 as well as its numerous downstream effectors and networks in human SSCs has been illustrated and summarized in Figure 10. Specifically, we have revealed that PAK1 is mediated by EGF and it controls PDK1, ZNF367, and KDR in the human SSC line. PAK1 interacts with PDK1, while ZNF367 regulates PDK1 and KDR. PAK1 activates ERK1/2- and AKT-signaling pathways and regulates cyclin A to facilitate the human SSC line entering S phase for DNA synthesis and cellular proliferation. Collectively, PAK1 has been identified as the first gene that regulates the proliferation, DNA synthesis, and apoptosis of human SSCs through PDK1/KDR/ZNF367 and the ERK1/2 and AKT pathways. This study offers data on the novel genes and signaling pathways as well as regulatory networks controlling the fate decisions of human SSCs, and it provides new molecular targets for the applications of human SSCs in treating human disease.
Figure 10.
Schematic Diagram Illustrates and Summarizes the Role and Signaling Pathway of PAK1 in Regulating Human SSCs
PAK1 is mediated by EGF, but not by GDNF or FGF2; PDK1, ZNF367, and KDR are the targets of PAK1. PAK1 binds to PDK1, and ZNF367 regulates PDK1 and KDR. PAK1 enhances the phosphorylation (phos-) of ERK1/2 and AKT, which facilitates directly or indirectly the entrance of phos-ERK1/2 and phos-AKT from cytoplasm to nuclei. PAK1 increases cyclin A level and stimulates human SSCs to enter S phase for DNA synthesis and cellular proliferation. Solid arrows, promote directly; dotted arrows, stimulate directly or indirectly; P in a circle, phosphorylate.
Materials and Methods
Human SSC Line and Cell Culture
The human SSC line was established in our laboratory by transfecting human primary SSCs with a plasmid, namely, Lenti-EF1α-SV40LargeT-IRES-EGFP, which expresses the SV40 large T antigen under the control of the EF1α promoter.26 The identity of the human SSC line was verified by the expression of numerous genes and proteins for human primary SSCs.
The human SSC line was cultured with DMEM/Nutrient Mixture F12 (DMEM/F12, Gibco, Grand Island, NY) supplemented with 10% FBS (Gibco) and 100 unit/mL penicillin and streptomycin (Invitrogen). The cells were passaged every 3–4 days using 0.05% trypsin and 0.53 mM EDTA (Invitrogen), and they were maintained at 34°C in a humidified 5% CO2 incubator. To ascertain which growth factor mediates PAK1, cells were cultured with the medium with 10 ng/mL GDNF (R&D Systems), 10 ng/mL FGF2 (R&D Systems), 10 ng/mL EGF (Sigma), or the combination of the three growth factors.
RNA Extraction and RT-PCR
Total RNA was extracted from the human SSC line and human Sertoli cells using RNAiso Plus reagent (Takara, Kusatsu, Japan), and the concentrations and quality of isolated RNA were determined by Nanodrop (Thermo Scientific, MA, USA). RNA with the ratio of A260/A280 = 1.9–2.0 was utilized to ensure good quality. DNase I was used to remove potential contamination of genomic DNA. RT was conducted using the First Strand cDNA Synthesis Kit (Thermo Scientific, USA), and PCR of the cDNA was carried out according to the protocol described previously.42 The primers of genes, including SV40, VASA, MAGEA4, GPR125, GFRA1, RET, UCHL1, THY1, PLZF, EGFR, ACTB, and GAPDH, were designed and are listed in Table 3. The PCR reaction started at 95°C for 5 min and was performed as follows: denaturation at 95°C for 30 s, annealing at 52°C–60°C as indicated in Table 3 for 30 s, elongation at 72°C for 45 s, for 35 cycles. The PCR samples were incubated for an additional 7 min at 72°C. PCR products were separated on 2% agarose gels, which were stained with Safer Ethidium Bromide Alternatives-GelGreen (Biotium, USA), and the images were captured by Image Analyzer ChemiDoc XRS+ (Bio-Rad).
Table 3.
The Sequences of the Primers of Genes for RT-PCR
| Gene Name | Primer Sequences (5′–3′) | Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| SV40 | forward: GAACAGCCCAGCCACTATAA | 248 | 58 |
| reverse: ACTCCAGCCATCCATTCTTC | |||
| VASA | forward: GCAGAAGGAGGAGAAAGTAGTG | 289 | 56 |
| reverse: CTCGTCCTGCAAGTATGATAGG | |||
| MAGEA4 | forward: CTTACCCACTACCATCAGCTTC | 212 | 58 |
| reverse: CTCGTCCTGCAAGTATGATAGG | |||
| RET | forward: CTCGTTCATCGGGACTTG | 126 | 56 |
| reverse: ACCCTGGCTCCTCTTCAC | |||
| PLZF | forward: CGGTTCCTGGATAGTTTGC | 317 | 54 |
| reverse: GGGTGGTCGCCTGTATGT | |||
| TYH1 | forward: ATCGCTCTCCTGCTAACAGTC | 135 | 52 |
| reverse: CTCGTACTGGATGGGTGAACT | |||
| UCHL1 | forward: AGCTGAAGGGACAAGAAGTTAG | 265 | 60 |
| reverse: TTGTCATCTACCCGACATTGG | |||
| GFRA1 | forward: CGGGTGGTCCCATTCATATC | 411 | 60 |
| reverse: TGGCTGGCAGTTGGTAAA | |||
| GPR125 | forward: GCGTCATTACGGTCTTTGGAA | 199 | 60 |
| reverse: ACGGCAATTCAAGCGGAGG | |||
| SOX9 | forward: AGGTGCTCAAAGGCTACGACTG | 322 | 58 |
| reverse: TGCCCGTTCTTCACCGACT | |||
| GATA4 | forward: GCCTCCTCTGCCTGGTAAT | 120 | 54 |
| reverse: CAGTCCCATCAGCGTGTAAA | |||
| PAK1 | forward: GGTGGTGGCTGCACAGTAG | 215 | 58 |
| reverse: TCTGAGGCAGGAGGTGGTAA | |||
| ACTB | forward: CATGTACGTTGCTATCCAGGC | 250 | 60 |
| reverse: CTCCTTAATGTCACGCACGAT | |||
| GAPDH | forward: AATCCCATCACCATCTTCC | 382 | 58 |
| reverse: CATCACGCCACAGTTTCC |
Isolation of Male Germ Cells from Testis Biopsies of OA Patients and NOA Patients
Testicular biopsies were obtained from OA patients and NOA patients who underwent microdissection testicular sperm extraction at Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine. The OA patients were caused by inflammation or vasoligation, and they had normal spermatogenesis. NOA patients included MA, i.e., spermatogenesis arrest at spermatogonium, spermatocyte, or spermatid stage, and HS. The testicular tissues from OA patients and NOA patients were washed twice with DMEM (Gibco) containing 2% penicillin and streptomycin (Gibco). The testis tissues were minced by scissors to become a semi-liquid state. The seminiferous tubules were isolated by enzyme I containing 2 mg/mL collagenase IV (Life Technologies) and 1 μg/μL DNase I (Roche) in oscillating water bath at 34°C, 100 rpm for 15 min. Male germ cells and Sertoli cells were obtained from seminiferous tubules by enzyme II containing 4 mg/mL collagenase IV, 2.5 mg/mL hyaluronidase (Sigma), 2 mg/mL trypsin (Sigma), and 1 μg/μL DNase I for 10–12 min. Finally, male germ cells were acquired through the differential plating pursuant to the procedure previously described.43 This study was approved by the Institutional Ethical Review Committee of Ren Ji Hospital (license number of ethics statement: 2012-01), Shanghai Jiao Tong University School of Medicine, and the informed consent for testicular biopsies was obtained from the donors for research only.
Real-Time PCR
Total RNA was extracted from the human SSC line with 10% or 0.5% FBS, without or with treatment of PAK1-siRNAs, PDK1-siRNAs, KDR-siRNAs, or ZNF367-siRNAs, and male germ cells from OA patients and NOA patients using Trizol (Takara, Kusatsu, Japan). The quality and concentrations of total RNA were measured by Nanodrop (Thermo Scientific), and the ratios of A260/A280 of total RNA were set as 1.9–2.0 to ensure good quality. The First Strand cDNA Synthesis Kit (Thermo Scientific) was used to conduct RT of total RNA. The primer sequences of genes used for real-time PCR were designed and are listed in Table 4. Real-time PCR reactions were conducted using Power SYBR Green PCR Master Mix (Applied Biosystems, Woolston Warrington, UK) and a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). To quantify the PCR products, the comparative CT (threshold cycle) method was used as described previously.44 The CT values of genes were normalized to those of housekeeping gene ACTB (ΔCT = CT(GENE) − CT(ACTB)), and the relative expression of genes to the control was calculated by the formula 2−ΔΔCT(ΔΔCT = ΔCT(GENE) − ΔCT(control)).
Table 4.
The Sequences of Gene Primers for Real-Time PCR
| Gene Name | Primer Sequences(5′-3′) | PCR Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| PAK1 | forward: CGTGGCTACATCTCCCATTT | 91 | 60 |
| reverse: AGGCTTCTTCTTCTGCTTCTC | |||
| ZNF367 | forward: TGTGTGACTATCCAGACTGTGG | 131 | 60 |
| reverse: TGCATGGGTGAATCTGCTCAG | |||
| KDR | forward: GGCCCAATAATCAGAGTGGCA | 109 | 60 |
| reverse: CCAGTGTCATTTCCGATCACTTT | |||
| PDK1 | forward: GGAACAGCGCAGTACGTTTCT | 132 | 60 |
| reverse: CTCGTTTCCAGCTCGGAATGG | |||
| GPX3 | forward: GAGCTTGCACCATTCGGTCT | 94 | 60 |
| reverse: GGGTAGGAAGGATCTCTGAGTTC | |||
| OIP5 | forward: TGAGAGGGCGATTGACCAAG | 189 | 60 |
| reverse: AGCACTGCGTGACACTGTG | |||
| THAP10 | forward: ACCCCATGCTGATAATCCATCT | 123 | 60 |
| reverse: TGAATACCCACACTACGGTGA | |||
| DBP | forward: CTGATCTTGCCCTATCAAGCATT | 58 | 60 |
| reverse: CGATGTCTTCGAGGGTCAAA | |||
| GPR78 | forward: CCACCAGGAAGATTGGCATTG | 142 | 60 |
| reverse: CTTGCTGTAGGTCAGGCACT | |||
| TET1 | forward: CATCAGTCAAGACTTTAAGCCCT | 87 | 60 |
| reverse: CGGGTGGTTTAGGTTCTGTTT | |||
| ACTB | forward: CACTCTTCCAGCCTTCCTTC | 104 | 60 |
| reverse: GTACAGGTCTTTGCGGATGT |
Western Blots
The human SSC line with 10% or 0.5% FBS, without or with treatment of PAK1-siRNAs, PDK1-siRNAs, KDR-siRNAs, or ZNF367-siRNAs, was lysed with whole-cell lysis buffer containing protease inhibitor cocktail (MedChemExpress, USA). After 30-min lysis on ice, cell lysates were cleared by centrifugation at 12,000 × g, and the concentrations of protein were measured by BCA kit (Dingguo, China). From each sample, 34 μg of cell lysate were used for SDS-PAGE (Bio-Rad, Richmond, CA), and Western blots were performed according to the protocol we described previously.45 The detailed information of the chosen antibodies, including PAK1, PDK1, KDR, ZNF367, Phos-ERK1/2, ERK2, phos-AKT, AKT, cyclin A, cyclin B1, cyclin D1, CDK2, and ACTB, for Western blots was listed in Table 5. The blots were detected by chemiluminescence (Chemi-Doc XRS, Bio-Rad, Hercules, CA, USA), and integrated density values (IDVs) were calculated by comparing the signals of target proteins with housekeeper ACTB or their own non-phosphorylated forms.
Table 5.
The Detailed Information on Primary Antibodies Used for Western Blots, Immunocytochemistry, and Immunohistochemistry
| Antibody | Vendor | Source | Working Dilution |
|---|---|---|---|
| PAK1 | CST | rabbit | WB: 1:1,000, ICC:1:400 |
| KDR | Abcam | rabbit | WB: 1:1,000 |
| PDK1 | CST | rabbit | WB: 1:1,000 |
| ZNF367 | Abcam | rabbit | WB: 1:500 |
| Phos-ERK1/2 | CST | rabbit | WB: 1:1,000 |
| ERK2 | Santa Cruz Biotechnology | mouse | WB: 1:200 |
| Phos-AKT | Abcam | rabbit | WB: 1:1,000 |
| P85 | Santa Cruz Biotechnology | goat | WB: 1:200 |
| AKT | Abcam | rabbit | WB: 1:1,000 |
| PCNA | Abcam | rabbit | WB: 1:1,000, IHC:200 |
| Cyclin A | Santa Cruz Biotechnology | rabbit | WB: 1:200 |
| Cyclin B1 | Santa Cruz Biotechnology | rabbit | WB: 1:200 |
| Cyclin D1 | Santa Cruz Biotechnology | rabbit | WB: 1:200 |
| Cyclin E | Santa Cruz Biotechnology | mouse | WB: 1:200 |
| CDK2 | Santa Cruz Biotechnology | rabbit | WB: 1:200 |
| GPR125 | Abcam | rabbit | ICC:1:200 |
| GFRA1 | Abcam | goat | ICC:1:200 |
| CD90 | Abcam | rabbit | ICC:1:200 |
| UCHL1 | AbDSerotec | mouse | ICC and IHC:1:200 |
| RET | Santa Cruz Biotechnology | rabbit | WB: 1:200 |
| DAZ2 | Abcam | rabbit | WB: 1:1,000 |
| DAZL | Abcam | rabbit | IF:1:100 |
| VASA | Santa Cruz Biotechnology | goat | IF:1:100 |
| WT1 | Santa Cruz Biotechnology | rabbit | IF:1:100 |
| GATA4 | Santa Cruz Biotechnology | goat | IF:1:100 |
| Ki67 | BD Biosciences | mouse | IHC:1:200 |
| PLZF | Santa Cruz Biotechnology | rabbit | IHC:1:200 |
| SV40 | Santa Cruz Biotechnology | mouse | WB: 1:200 |
| ACTB | Proteintech | mouse | WB: 1:5,000 |
WB, Western blots; ICC, immunocytochemistry; IHC, immunohistochemistry.
Immunoprecipitation and Western Blots
A total of 106 human SSC line was lysed in 500 μL freshly prepared complete whole-cell lysis buffer containing protease inhibitor cocktail (MedChemExpress, NJ, USA) for 30 min on ice. Cell lysates were cleared by centrifugation at 12,000 × g for 15 min at 4°C, and the protein concentrations were measured by BCA kit Pierce BCA Protein Assay Kit (Thermo Scientific). Rabbit antibody against PAK1 (Cell Signaling Technology, 2602S) or rabbit IgG was added to cell lysate and incubated overnight at 4°C on a rotator. Protein G magnetic beads (25 μL, Active Motif, USA) were added to cell lysate and supernatant and incubated for 2 hr at 4°C. After washes three time with 500 μL washing buffer, bead pellets were resuspended with 50 μL 1× loading buffer and boiled at 95°C for 5 min. Western blots were performed as follows: cell lysate and supernatant from each sample were separated using 10% SDS-PAGE (Bio-Rad, Hercules, USA) and transferred to nitrocellulose membranes for 2 hr at room temperature. The membranes were blocked using 5% nonfat dry milk in TBS-T for 1 hr at room temperature. After washing with TBS-T, the membranes were incubated with antibodies against PAK1 and PDK1 overnight at 4°C. The detailed information on antibodies against PAK1 and PDK1 is given in Table 5. After extensive washes, the membranes were incubated with horseradish peroxidase-conjugated IgG (Santa Cruz Biotechnology) at a 1:2,000 dilution for 1 hr at room temperature. The membranes were detected by Chemi-Doc XRS system (Bio-Rad, Hercules, USA), and densitometric analyses were processed with ImageJ software.
Immunocytochemistry
The human SSC line was fixed using 4% paraformaldehyde (PFA) for 30 min, and these cells were washed twice with PBS (Medicago, Uppsala, Sweden). After blocking with 3% BSA at room temperature for 1 hr, cells were incubated with the primary antibodies, including PAK1 (CST, catalog: 2602S, 1:200), THY1 (CD90, Abcam, ab133350, 1:200), GPR125 (Abcam, ab51705, 1:200), GFRA1 (Abcam, ab8026, 1:200), and EGFR (Abcam, ab52894, 1:200), at 4°C overnight. The detailed information of antibodies was given in Table 5. Rhodamine-conjugated secondary antibody (Invitrogen, USA) was used as the secondary antibody, and the nuclei were stained with DAPI. Replacement of primary antibodies with isotype IgGs served as negative controls. The images were captured with a Nikon microscope (Tokyo, Japan).
Immunohistochemistry
Testis sections of OA patients and recipient mice grafted with human SSCs by PAK1-siRNA 2 or the control siRNA transfection were deparaffinized by xylene twice, hydrated with a series of graded alcohol, and treated with 3% H2O2 (Boster Biological Technology, Guangzhou, China) for 15 min at room temperature to block the endogenous peroxidase activity. After blocking with 5% BSA for 1 hr at room temperature, the sections were incubated with primary antibodies (Table 5) at 4°C overnight. After extensive washes with PBS, the sections were incubated with HRP-conjugated second antibody for 1 hr at room temperature. After extensive washing with PBS, DAB (3, 3-diaminobenzidine) (Vector Lab, Burlingame, USA) was used to label PAK1 protein. Finally, the sections were stained with hematoxylin and observed under a Nikon microscope.
RNAi of PAK1, PDK1, KDR, and ZNF367
The siRNA sequences targeting human PAK1, PDK1, KDR, and ZNF367 mRNA were synthesized from GenePharma (Suzhou, China) and were listed in Table 6. The siRNAs without targeting any sequence of these genes were utilized as negative controls. For PAK1, KDR, PDK1, and ZNF367 knockdown, 100 nM siRNA or control siRNA was transfected into human SSCs using Lipofectamine 3000 (Life Technologies, Carlsbad, USA), according to the manufacturer’s protocol. At 48 hr after transfection, cells were harvested to evaluate level changes of genes and proteins by real-time PCR and Western blots as mentioned above.
Table 6.
The Sequences for Various Kinds of siRNAs
| siRNA Name | siRNA Sequence |
|---|---|
| PAK1-siRNA 1 | sense 5′-GGCGAUCCUAAGAAGAAAUTT-3′ |
| antisense 5′-AUUUCUUCUUAGGAUCGCCTT-3′ | |
| PAK1-siRNA 2 | sense 5′-GCAUCAAUUCCUGAAGAUUTT-3′ |
| antisense 5′-AAUCUUCAGGAAUUGAUGCTT-3′ | |
| PAK1-siRNA 3 | sense 5′-GCAUCAAUUCCUGAAGAUUTT-3′ |
| antisense 5′-UAUUCCGGGUCAAAGCAUCTT-3′ | |
| PDK1-siRNA 1 | sense 5′-UUCCCUAAGGCAAGAGACC TT-3′ |
| antisense 5′-GGUCUCUUGCCUUAGGGAA TT-3′ | |
| PDK1-siRNA 2 | sense 5′-UGCCCCUCCCGCUGUGGUCTT-3′ |
| antisense 5′-GACCACAGCGGGAGGGGCATT-3′ | |
| PDK1-siRNA 3 | sense 5′-AAAUUCUUCCCUAAGGCAATT-3′ |
| antisense 5′-UUGCCUUAGGGAAGAAUUUTT-3′ | |
| ZNF367-siRNA 1 | sense 5′-GCCUGAGCAGAUUCACCCATT-3′ |
| antisense 5′-UGGGUGAAUCUGCUCAGGCTT-3′ | |
| ZNF367-siRNA 2 | sense 5′-GGACACACUCAGCAAACAUTT-3′ |
| antisense 5′-AUGUUUGCUGAGUGUGUCCTT-3′ | |
| ZNF367-siRNA 3 | sense 5′-GCAGGACCCUCUGGAAUACTT-3′ |
| antisense 5′-GUAUUCCAGAGGGUCCUGCTT-3′ | |
| KDR-siRNA 1 | sense 5′-GACGGACAGUGGUAUGGUUTT-3′ |
| antisense 5′-AACCAUACCACUGUCCGUCTG-3′ | |
| KDR-siRNA 2 | sense 5′-CCAUCGUCAUGGAUCCAGATT-3′ |
| antisense 5′-UCUGGAUCCAUGACGAUGGAC-3′ | |
| KDR-siRNA 3 | sense 5′-CAUGUUCUCUAAUAGCACATT-3′ |
| antisense 5′-UGUGCUAUUAGAGAACAUGGT-3′ |
BrdU Incorporation Assay
In total, 1 × 105 cells/well of the human SSC line were placed on 12-well plates with DMEM/F12 containing 10% FBS and cultured for 12 hr. The cells were starved in DMEM/F12 for 24 hr, and 30 μg/mL BrdU (Sigma, St. Louis, MO) was added to the culture medium. After 16 hr of culture, cells were fixed in 4% PFA for 30 min. Immunocytochemistry was conducted using anti-BrdU (Sigma) according to the method described previously.10 After three times washing with PBS, rhodamine-conjugated secondary antibody (Invitrogen, Carlsbad, USA) was added to cells and incubated for 1 hr at room temperature. Cells were observed under fluorescence microscopy (Nikon, Tokyo, Japan). At least 500 cells were counted to calculate the percentages of BrdU-positive cells from three independent experiments.
EDU Incorporation Assay
The human SSC line was seeded in 96-well plate containing DMEM/F12 medium with 50 μM EDU (RiboBio, Guangzhou, China). After 12 hr of culture, the cells were washed with DMEM and fixed with 4% PFA. Cells were neutralized with 2 mg/mL glycine and permeabilized with 0.5% Triton X-100 for 10 min at room temperature. EDU immunostaining was performed with Apollo staining reaction buffer. The nuclei of cells were stained with Hoechst 33342, and the EDU-positive cells were counted under fluorescence microscopy (Nikon).
CCK-8 Assay
The human SSC line was plated in 96-well plates at a density of 5,000 cells/well, and they were tranfected without or with 100 nM PAK1-siRNAs, PDK1-siRNAs, KDR-siRNAs, ZNF367-siRNAs, or control siRNAs. The proliferation ability of human SSCs was detected by CCK-8 assay (Dojin Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. In brief, 10 μL of CCK-8 reagents were added into each well of the cells; 3 hr later, a microplate reader (Thermo Scientific) was used to measure the absorbance at the wavelength of 450 nm.
Annexin V/PI Staining and Flow Cytometry
The percentages of apoptosis in the human SSC line transfected without or with PAK1-siRNAs, PDK1-siRNAs, KDR-siRNAs, ZNF367-siRNAs, or control siRNAs were determined using the Annexin V-APC/PI apoptosis detection kit and flow cytometry according to the protocol described previously.44 Human SSC cells were seeded at 80,000 cells/well in 6-well plates. Cells were collected and washed twice with PBS at the third day after siRNA transfection. The cells were simultaneously stained with Annexin V-fluorescein isothiocyanate (FITC) (green fluorescence) and the non-vital dye PI (red fluorescence), which allowed the discrimination of intact cells (FITC−PI−), early apoptotic cells (FITC+PI−), and late apoptotic cells (FITC+PI+).
Xenotransplantation of the Human SSC Line
To clarify the role of PAK1 in controlling the human SSC line in vivo, we conducted the xenotransplantation assay. Ten male nude mice at 6 weeks old were obtained from Shanghai Laboratory Animal Center, Chinese Academy of Sciences, Shanghai, China, and they were housed in a specific-pathogen-free (SPF) facility and treated according to the experimental animal guidelines and regulation of Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine. Busulfan (Sigma, St. Louis, MO, USA) was delivered to nude mice by intraperitoneal injection at the concentration of 35 mg/kg body weight to remove male germ cells. The human SSC line was treated with PAK1-siRNA 2 or the control siRNA for 36 hr. The cells were resuspended in DMEM/F12 at the concentration of 107 cells/mL. Approximately 20 μL cell suspension with PAK1-siRNA 2 was transplanted into the seminiferous tubules of one testis via the efferent duct, while the other testis was grafted with the cells after control siRNA transfection. At 4 weeks after cell transplantation, the mice were sacrificed and their testes were used to prepare the paraffin sections. Finally, immunocytochemistry was conducted to check the expression levels of proteins for markers of human SSCs and cell proliferation, and TUNEL was carried out to detect cell apoptosis.
TUNEL Assay
Testis paraffin sections of recipient mice were deparaffinized using xylene, hydrated with graded alcohol, and they were treated with 20 μg/mL proteinase K. After equilibration with 1× equilibration buffer for 20 min, the sections were incubated in FITC-12-dUTP Labeling Mix (Yeasen, Shanghai, China) at 37°C for 60 min. The cell nuclei were stained with DAPI, and the TUNEL-positive cells were counted under fluorescence microscopy (Nikon).
RNA Sequencing
Total RNA was extracted from the human SSC line with PAK1-siRNA 2 or control siRNA or culture with 10% FBS or 0.5% FBS. RNA was treated with DNase I to remove potential DNA contamination. The mRNA was enriched by using the oligo (dT) magnetic beads. The first strand of cDNA was synthesized by using random hexamer-primer, and dNTPs RNase H and DNA polymerase I were utilized to synthesize the second strand of cDNA. The double-strand cDNA was purified with magnetic beads, and end reparation and 3′ end single-nucleotide A (adenine) addition were performed. Next, sequencing adaptors were ligated to the fragments that were enriched by PCR amplification. The cDNA libraries were used for sequencing on an Illumina HiSeq 4000 instrument. RNA sequencing libraries were established for two RNA sequencing samples at BGI (Shenzhen, China), and sequencing data were mapped to the human genome (hg19) with default parameters. NOISeq46 was used to produce biologically meaningful rankings of differentially expressed genes (DEGs). Subsequently, those DEGs were assessed by RNA-seq experiments (RSEMs),47 and the significantly DEGs were selected according to the following criteria: p < 0.05. GO enrichment analysis and KEGG pathway enrichment analysis were further performed on the DEGs in human SSCs between PAK1-siRNA 2 and the control siRNA.
Testicular Tissue Arrays
Human testicular tissue arrays were performed to compare the levels of PAK1 protein in paraffin-embedded testicular samples of OA patients and several subtypes of NOA patients, including MA at a different stage of spermatid, spermatocyte, or spermatogonium and HS. After H&E staining, the 1.6-mm core from each sample was inserted into a grid of paraffin block with tissue arrays (Beecher Instruments). The tissue array blocks were cut to 4-μm sections and stained with antibody against PAK1 at a dilution of 1:200 according to method.22 For data analysis, every section was evaluated by two pathologists in terms of the uniform pre-established criteria as described previously.48 The expressions of PAK1 in subtypes of NOA patients were quantified relatively by comparing with the immunostaining intensities of OA patients.
Statistical Analysis
Experiments were repeated at least three times. All values were presented as mean (bars) ± SD (error bars). GraphPad Prism 5.0 was used for statistical analysis, and the test of normality and homogeneity of variances was conducted before being analyzed with t test and one-way ANOVA. p < 0.05 was considered statistically significant.
Author Contributions
H.F. performed the experiments, wrote the manuscript, and helped with data analysis. W.Z. assisted with the experiments. Q.Y. and M.N. assisted with the experiments and ordered reagents. F.Z., Q.Q., G.M., H.W., L.W. and M.S. assisted with the experiments. Z.L. assisted with the testicular tissue collection and experiments. Z.H. was responsible for the conception and design, supervision of all aspects of the laboratory experiments, data analysis, writing the manuscript, and final approval of the manuscript. All authors approved the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by grants from the National Nature Science Foundation of China (31872845, 31230048 and 31671550), the Chinese Ministry of Science and Technology (2016YFC1000606 and 2014CB943101), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (2012.53), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152511), and the Shanghai Hospital Development Center (SHDC12015122).
Footnotes
Supplemental Information includes six figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.06.006.
Supplemental Information
References
- 1.Yang S., Ping P., Ma M., Li P., Tian R., Yang H., Liu Y., Gong Y., Zhang Z., Li Z., He Z. Generation of haploid spermatids with fertilization and development capacity from human spermatogonial stem cells of cryptorchid patients. Stem Cell Reports. 2014;3:663–675. doi: 10.1016/j.stemcr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanatsu-Shinohara M., Inoue K., Lee J., Yoshimoto M., Ogonuki N., Miki H., Baba S., Kato T., Kazuki Y., Toyokuni S. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Seandel M., James D., Shmelkov S.V., Falciatori I., Kim J., Chavala S., Scherr D.S., Zhang F., Torres R., Gale N.W. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kossack N., Meneses J., Shefi S., Nguyen H.N., Chavez S., Nicholas C., Gromoll J., Turek P.J., Reijo-Pera R.A. Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells. 2009;27:138–149. doi: 10.1634/stemcells.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z., Gong Y., Guo Y., Hai Y., Yang H., Yang S., Liu Y., Ma M., Liu L., Li Z. Direct transdifferentiation of spermatogonial stem cells to morphological, phenotypic and functional hepatocyte-like cells via the ERK1/2 and Smad2/3 signaling pathways and the inactivation of cyclin A, cyclin B and cyclin E. Cell Commun. Signal. 2013;11:67. doi: 10.1186/1478-811X-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z., Sun M., Yuan Q., Niu M., Yao C., Hou J., Wang H., Wen L., Liu Y., Li Z., He Z. Generation of functional hepatocytes from human spermatogonial stem cells. Oncotarget. 2016;7:8879–8895. doi: 10.18632/oncotarget.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Niu M., Sun M., Yuan Q., Yao C., Hou J., Wang H., Wen L., Fu H., Zhou F. Transdifferentiation of human male germline stem cells to hepatocytes in vivo via the transplantation under renal capsules. Oncotarget. 2017;8:14576–14592. doi: 10.18632/oncotarget.14713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H., Liu Y., Hai Y., Guo Y., Yang S., Li Z., Gao W.Q., He Z. Efficient Conversion of Spermatogonial Stem Cells to Phenotypic and Functional Dopaminergic Neurons via the PI3K/Akt and P21/Smurf2/Nolz1 Pathway. Mol. Neurobiol. 2015;52:1654–1669. doi: 10.1007/s12035-014-8966-4. [DOI] [PubMed] [Google Scholar]
- 9.Simon L., Ekman G.C., Kostereva N., Zhang Z., Hess R.A., Hofmann M.C., Cooke P.S. Direct transdifferentiation of stem/progenitor spermatogonia into reproductive and nonreproductive tissues of all germ layers. Stem Cells. 2009;27:1666–1675. doi: 10.1002/stem.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Z., Jiang J., Kokkinaki M., Golestaneh N., Hofmann M.C., Dym M. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells. 2008;26:266–278. doi: 10.1634/stemcells.2007-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Wang S., Wang X., Liao S., Wu Y., Han C. Endogenously produced FGF2 is essential for the survival and proliferation of cultured mouse spermatogonial stem cells. Cell Res. 2012;22:773–776. doi: 10.1038/cr.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng L.X., Chen Y., Dettin L., Pera R.A., Herr J.C., Goldberg E., Dym M. Generation and in vitro differentiation of a spermatogonial cell line. Science. 2002;297:392–395. doi: 10.1126/science.1073162. [DOI] [PubMed] [Google Scholar]
- 13.He Z., Jiang J., Kokkinaki M., Tang L., Zeng W., Gallicano I., Dobrinski I., Dym M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205–2217. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu Z., Goodyear S.M., Rao S., Wu X., Tobias J.W., Avarbock M.R., Brinster R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q.E., Racicot K.E., Kaucher A.V., Oatley M.J., Oatley J.M. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–290. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toledano H., D’Alterio C., Czech B., Levine E., Jones D.L. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., Cai T., Zheng C., Lin X., Wang G., Liao S., Wang X., Gan H., Zhang D., Hu X. MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Res. 2017;45:4142–4157. doi: 10.1093/nar/gkw1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., Niu M., Yao C., Hai Y., Yuan Q., Liu Y., Guo Y., Li Z., He Z. Fractionation of human spermatogenic cells using STA-PUT gravity sedimentation and their miRNA profiling. Sci. Rep. 2015;5:8084. doi: 10.1038/srep08084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clermont Y. The cycle of the seminiferous epithelium in man. Am. J. Anat. 1963;112:35–51. doi: 10.1002/aja.1001120103. [DOI] [PubMed] [Google Scholar]
- 20.Boitani C., Di Persio S., Esposito V., Vicini E. Spermatogonial cells: mouse, monkey and man comparison. Semin. Cell Dev. Biol. 2016;59:79–88. doi: 10.1016/j.semcdb.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Di Persio S., Saracino R., Fera S., Muciaccia B., Esposito V., Boitani C., Berloco B.P., Nudo F., Spadetta G., Stefanini M. Spermatogonial kinetics in humans. Development. 2017;144:3430–3439. doi: 10.1242/dev.150284. [DOI] [PubMed] [Google Scholar]
- 22.He Z., Kokkinaki M., Jiang J., Dobrinski I., Dym M. Isolation, characterization, and culture of human spermatogonia. Biol. Reprod. 2010;82:363–372. doi: 10.1095/biolreprod.109.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muciaccia B., Boitani C., Berloco B.P., Nudo F., Spadetta G., Stefanini M., de Rooij D.G., Vicini E. Novel stage classification of human spermatogenesis based on acrosome development. Biol. Reprod. 2013;89:60. doi: 10.1095/biolreprod.113.111682. [DOI] [PubMed] [Google Scholar]
- 24.Kiger A.A., Jones D.L., Schulz C., Rogers M.B., Fuller M.T. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 25.Oatley J.M., Kaucher A.V., Avarbock M.R., Brinster R.L. Regulation of mouse spermatogonial stem cell differentiation by STAT3 signaling. Biol. Reprod. 2010;83:427–433. doi: 10.1095/biolreprod.109.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou J., Niu M., Liu L., Zhu Z., Wang X., Sun M., Yuan Q., Yang S., Zeng W., Liu Y. Establishment and Characterization of Human Germline Stem Cell Line with Unlimited Proliferation Potentials and no Tumor Formation. Sci. Rep. 2015;5:16922. doi: 10.1038/srep16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng X., Lindahl M., Hyvönen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 28.Takashima S., Kanatsu-Shinohara M., Tanaka T., Morimoto H., Inoue K., Ogonuki N., Jijiwa M., Takahashi M., Ogura A., Shinohara T. Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Reports. 2015;4:489–502. doi: 10.1016/j.stemcr.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radhakrishnan B., Oke B.O., Papadopoulos V., DiAugustine R.P., Suarez-Quian C.A. Characterization of epidermal growth factor in mouse testis. Endocrinology. 1992;131:3091–3099. doi: 10.1210/endo.131.6.1446643. [DOI] [PubMed] [Google Scholar]
- 30.Adekunle A.O., Falase E.A., Ausmanus M., Kopf G.S., Van-Arsdalen K.N., Teuscher C. Comparative analysis of blood plasma epidermal growth factor concentrations, hormonal profiles and semen parameters of fertile and infertile males. Afr. J. Med. Med. Sci. 2000;29:123–126. [PubMed] [Google Scholar]
- 31.Wahab-Wahlgren A., Martinelle N., Holst M., Jahnukainen K., Parvinen M., Söder O. EGF stimulates rat spermatogonial DNA synthesis in seminiferous tubule segments in vitro. Mol. Cell. Endocrinol. 2003;201:39–46. doi: 10.1016/s0303-7207(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 32.Chen J.X., Xu L.L., Wang X.C., Qin H.Y., Wang J.L. Involvement of c-Src/STAT3 signal in EGF-induced proliferation of rat spermatogonial stem cells. Mol. Cell. Biochem. 2011;358:67–73. doi: 10.1007/s11010-011-0922-2. [DOI] [PubMed] [Google Scholar]
- 33.Cagnet S., Faraldo M.M., Kreft M., Sonnenberg A., Raymond K., Glukhova M.A. Signaling events mediated by α3β1 integrin are essential for mammary tumorigenesis. Oncogene. 2014;33:4286–4295. doi: 10.1038/onc.2013.391. [DOI] [PubMed] [Google Scholar]
- 34.Martin E., Ouellette M.H., Jenna S. Rac1/RhoA antagonism defines cell-to-cell heterogeneity during epidermal morphogenesis in nematodes. J. Cell Biol. 2016;215:483–498. doi: 10.1083/jcb.201604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goertz M.J., Wu Z., Gallardo T.D., Hamra F.K., Castrillon D.H. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J. Clin. Invest. 2011;121:3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sargent K.M., Clopton D.T., Lu N., Pohlmeier W.E., Cupp A.S. VEGFA splicing: divergent isoforms regulate spermatogonial stem cell maintenance. Cell Tissue Res. 2016;363:31–45. doi: 10.1007/s00441-015-2297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain M., Zhang L., Boufraqech M., Liu-Chittenden Y., Bussey K., Demeure M.J., Wu X., Su L., Pacak K., Stratakis C.A., Kebebew E. ZNF367 inhibits cancer progression and is targeted by miR-195. PLoS ONE. 2014;9:e101423. doi: 10.1371/journal.pone.0101423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovelace D.L., Gao Z., Mutoji K., Song Y.C., Ruan J., Hermann B.P. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016;143:1893–1906. doi: 10.1242/dev.132761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shrestha Y., Schafer E.J., Boehm J.S., Thomas S.R., He F., Du J., Wang S., Barretina J., Weir B.A., Zhao J.J. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene. 2012;31:3397–3408. doi: 10.1038/onc.2011.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M.J., Wu D.W., Wang Y.C., Chen C.Y., Lee H. PAK1 confers chemoresistance and poor outcome in non-small cell lung cancer via β-catenin-mediated stemness. Sci. Rep. 2016;6:34933. doi: 10.1038/srep34933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hentrich A., Wolter M., Szardening-Kirchner C., Lüers G.H., Bergmann M., Kliesch S., Konrad L. Reduced numbers of Sertoli, germ, and spermatogonial stem cells in impaired spermatogenesis. Mod. Pathol. 2011;24:1380–1389. doi: 10.1038/modpathol.2011.97. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y., Hai Y., Yao C., Chen Z., Hou J., Li Z., He Z. Long-term culture and significant expansion of human Sertoli cells whilst maintaining stable global phenotype and AKT and SMAD1/5 activation. Cell Commun. Signal. 2015;13:20. doi: 10.1186/s12964-015-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Y., Liu L., Sun M., Hai Y., Li Z., He Z. Expansion and long-term culture of human spermatogonial stem cells via the activation of SMAD3 and AKT pathways. Exp. Biol. Med. (Maywood) 2015;240:1112–1122. doi: 10.1177/1535370215590822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H., Yuan Q., Sun M., Niu M., Wen L., Fu H., Zhou F., Chen Z., Yao C., Hou J. BMP6 Regulates Proliferation and Apoptosis of Human Sertoli Cells Via Smad2/3 and Cyclin D1 Pathway and DACH1 and TFAP2A Activation. Sci. Rep. 2017;7:45298. doi: 10.1038/srep45298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Z., Jiang J., Hofmann M.C., Dym M. Gfra1 silencing in mouse spermatogonial stem cells results in their differentiation via the inactivation of RET tyrosine kinase. Biol. Reprod. 2007;77:723–733. doi: 10.1095/biolreprod.107.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pallares J., Bussaglia E., Martínez-Guitarte J.L., Dolcet X., Llobet D., Rue M., Sanchez-Verde L., Palacios J., Prat J., Matias-Guiu X. Immunohistochemical analysis of PTEN in endometrial carcinoma: a tissue microarray study with a comparison of four commercial antibodies in correlation with molecular abnormalities. Mod. Pathol. 2005;18:719–727. doi: 10.1038/modpathol.3800347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.