Abstract
Physical exercise can induce various adaptation reactions in skeletal muscle tissue, such as sarcomere remodeling. The latter involves degradation of damaged sarcomere components, as well as de novo protein synthesis and sarcomere assembly. These processes are controlled by specific protease systems in parallel with molecular chaperones that assist in folding of newly synthesized polypeptide chains and their incorporation into sarcomeres. Since acute exercise induces oxidative stress and inflammation, leading to activation of the transcription factor NFκB (nuclear factor kappa B), we speculated that this transcription factor might also play a role in the regulation of long-term adaptation to regular exercise. Thus, we studied skeletal muscle adaptation to running exercise in a murine model system, with and without parallel treatment with the NFκB-inhibitory, anti-oxidant and anti-inflammatory drug pyrrolidine dithiocarbamate (PDTC). In control mice, 10 weeks of uphill (15° incline) treadmill running for 60 min thrice a week at a final speed of 14 m/min had differential, but only minor effects on many genes encoding molecular chaperones for sarcomere proteins, and/or factors involved in the degradation of the latter. Furthermore, there were marked differences between individual muscles. PDTC treatment modulated gene expression patterns as well, both in sedentary and exercising mice; however, most of these effects were also modest and there was little effect of PDTC treatment on exercise-induced changes in gene expression. Taken together, our data suggest that moderate-intensity treadmill running, with or without parallel PDTC treatment, had little effect on the expression of genes encoding sarcomere components and sarcomere-associated factors in murine skeletal muscle tissue.
Keywords: Mice, Treadmill running, Pyrrolidine dithiocarbamate (PDTC), Skeletal muscle, Gene expression
Introduction
Physical exercise induces a broad variety of molecular and cellular adaptations in skeletal muscle tissue. Besides metabolic alterations, structural adaptations, specifically within the sarcomeres, can occur (for review, see Seene et al. 2009).
Understanding the molecular basis of this adaptation process is crucial in order to obtain a better understanding of the cause-and-effect relationship between a specific exercise regimen and the resulting changes within skeletal muscle tissue.
The sarcomere is the contractile unit of a muscle. Whereas its basic structure mainly consists of actin and fiber type-specific myosin filaments, numerous proteins with specific functions are attached to and interact with the sarcomere. Specifically, a broad variety of proteins, the so-called “molecular chaperones,” coordinate folding of actin and myosin proteins and their proper incorporation into the sarcomere. One example is the heat shock protein Hsp90a, which, together with other factors such as the molecular chaperone Unc45b, assists in folding of the myosin protein (for review, see Crawford and Horowits 2011).
Other proteins, namely specific proteases, are involved in the degradation of misfolded or destroyed proteins, e.g., after eccentric exercise or in response to muscle injury. Important protein-degrading systems in the context of the sarcomere are the components of the ubiquitin proteasome system (UPS) and associated factors, such as the E3 ubiquitin ligases MuRF1 and atrogin-1 or the proteases of the calpain family (for review, see Bartoli and Richard 2005; Bell et al. 2016).
So far, little is known on the regulation of factors affecting sarcomere reorganization in response to exercise. However, single bouts of physical exercise can evoke oxidative stress and an inflammatory response, followed by temporary activation of the inflammation-activated ubiquitous transcription factor NFκB, despite the fact that in trained individuals, skeletal muscle NFκB activity appears to be unchanged or even lower when compared to sedentary controls (reviewed by Kramer and Goodyear 2007; Ji 2015, and references therein). Moreover, a link between NFκB activation and the degradation of sarcomere components has been well established in the context of the cachexia syndrome, which leads to severe muscle loss in a broad variety of chronic diseases, such as CHF, COPD, and many tumor entities. Here, a chronic, low-grade inflammatory situation induces the activation of skeletal muscle NFκB, which triggers upregulation of the UPS and other factors involved in the degradation of sarcomeric proteins (for review, see Bowen et al. 2015). Against this background, it is not surprising that at least acute physical exercise can lead to upregulation of specific components of the protein-degrading machinery, mainly the UPS, the E3 ubiquitin ligases MuRF1 and atrogin-1, and the calpains (for review, see Murton et al. 2008; Pasiakos and Carbone 2014). However, so far, it is not completely clear whether this is NFκB dependent or not.
Altogether, against this background, it is tempting to speculate whether exercise-induced inflammation and NFκB activation might also be involved in long-term skeletal muscle remodeling, specifically de novo folding of newly synthetized sarcomere proteins and their incorporation into the sarcomeric unit.
Thus, here, we analyzed skeletal muscle gene expression patterns, specifically genes encoding sarcomere components and sarcomere-associated factors, in response to a 10-week uphill running protocol, with and without parallel treatment with the NFκB inhibitor pyrrolidine dithiocarbamate (PDTC), in mice. Our results show that in hindlimb muscles, this protocol had differential, but only moderate effects. Furthermore, the effects appear to be very specific for individual muscles.
Experimental procedures
Animals
Male C57BL/6 (C57BL/6NCrl H-2b) mice (n = 32) were purchased from Charles River (Sulzfeld, Germany). They were housed and fed according to federal guidelines and were inspected by a veterinarian several times per week. Mice were randomly assigned to one of the four groups (sedentary, sedentary/PDTC, exercised, exercised/PDTC) (n = 8 each). All animal experiments were carried out in accordance with the German animal protection law (Regierungspräsidium Tübingen, M9/14).
PDTC treatment
Dithiocarbamates are anti-oxidants and potent NFκB inhibitors (Piette et al. 1997). PDTC specifically inhibits NFκB binding to its DNA consensus sequence by blocking ubiquitination and degradation of the NFκB inhibitor IκB, thus preventing NFκB release and nuclear translocation (Liu et al. 1999; Cuzzocrea et al. 2002; Schreck et al. 1992). PDTC is a well-established NFκB inhibitor and has been used in a broad variety of animal experiments for almost 20 years (Németh et al. 1998; Cuzzocrea et al. 2002, 2003; Parodi et al. 2005; Nai et al. 2006; Strait et al. 2008; Ma et al. 2008; Gu et al. 2009; Zhang et al. 2009; Mariappan et al. 2010; Sharma et al. 2011). In our study, PDTC was added to the drinking water at a concentration of 0.04 mg/mL. Based on an average drinking volume of approximately 6–8 mL/day/mouse (Bachmanov et al. 2002), this corresponds to a dosage of approximately 8–12 mg PDTC per day and kg body weight. Water bottles were light-protected and the water was changed every other day. PDTC treatment was started 4 days before the start of the training experiment.
Treadmill running
Six to seven week-old male C57BL/6 mice were first familiarized with the treadmill (Harvard Apparatus, Holliston, MA, USA) within a period of 2 weeks. For this purpose, they were initially allowed to explore the new surroundings for 3 × 10 min on three different days, with the treadmill switched off. Subsequently, they were run for 1 h on Mondays, Wednesdays, and Fridays every week, at an initial speed of 2 m/min and 0° incline, with gradually increasing speed and incline (3 m/min and 5° per week), until animals ran at a final speed of 14 m/min and 15° incline from the sixth week onwards. The training protocol was continued under these conditions for another 5 weeks. Each training session included a 10-min “warm up” period, during which speed and incline were gradually increased.
Skeletal muscle tissue preparation
Forty-eight hours after the last training session, mice were sacrificed, skeletal muscle tissue was dissected, snap-frozen in liquid nitrogen, and stored at − 80 °C or in RNAlater® (Thermo Fisher Scientific, Waltham, MA, USA) for RNA or protein isolation.
RNA isolation and qPCR
Muscle tissue was isolated and immediately stored in RNALater (Ambion) according to the manufacturer’s instructions. RNA was isolated using the Fibrous Tissue Kit (Qiagen, Hilden, Germany). RNA quantity and purity (260/280 ratio) was assessed using a BioPhotometer (Eppendorf AG, Hamburg, Germany). Of the total RNA, 500 ng was used for reverse transcription using M-MLV reverse transcriptase and random primers (Promega, Madison, WI, USA) in a total volume of 20 μL. Briefly, RNA samples were denatured for 5 min at 70 °C, and after adding reverse transcriptase and random primers, the mix was incubated at room temperature for 10 min, followed by 60 min at 50 °C. Subsequently, the enzyme was inactivated at 70 °C for 15 min, followed by cooling down the sample on ice. Semi-quantitative real-time PCR analysis was carried out using the C1000 Touch™ Thermal Cycler system together with the Eva Green Mastermix (Bio-Rad, Hercules, CA, USA). Three replicates of each RNA sample were analyzed. For detection of different transcripts, pre-designed (Qiagen QuantiTect Primer Assays) and self-designed oligonucleotide primers (exon-intron-spanning; Table 1) were used. In each experiment, melting curve analysis was routinely performed to verify that a single transcript was produced. Baseline correction and threshold setting were performed using the automatic calculation of the CFX Manager Software (Bio-Rad). Cq determination was performed with the same software using the “single-threshold” mode. Gene expression was normalized to the geometric means (GEOM) of the reference genes TBP, GAPDH, and HPRT using the qbase+ program (Biogazelle, Gent, Belgium), employing multiple reference gene normalization (geNorm algorithm). Relative gene expression was calculated using the comparative CT (2−ΔΔCT) method (Pfaffl 2001; Bustin et al. 2009). Non-RT- and non-template controls were run for all reactions.
Table 1.
Primers used for qPCR analysis
| Primer | Forward | Reverse |
|---|---|---|
| GAPDH | TGTGTCCGTCGTGGATCTGA | TTGCTGTTGAAGTCGCAGGAG |
| HPRT | AGTACAGCCCCAAAATGGTTAAG | CACAAACGTGATTCAAATCCCTG |
| TBP | AAGAGAGCCACGGACAACTGC | CTTCACATCACAGCTCCCCAC |
| PGC-1α | GCTCATTGTTGTACTGGTTGGATATG | CGTAGGCCCAGGTACGACAG |
| Myoglobin | CCCTGGAGGGTTGAGCACGGT | AGGCCACCTGGTCCTGAAGGG |
| SDHD | TCATGGGGACACCCTGCCGA | TCCACAGCATGGCAACCGCTC |
| CS | GCCCAGCAGCTGTAGGATGACC | GGGGCCTGTCCCTGGCGTA |
| MyH7 (Myosin Ib) | CCTTGGCACCAATGTCCCGGCTC | GAAGCGCAATGCAGAGTCGGTG |
| MyH2 (Myosin IIa) | ATGAGCTCCGACGCCGAG | TCTGTTAGCATGAACTGGTAGGCG |
| MyH1 (Myosin IIx) | GCTTCAAGTTTGGACCCACG | ACTTCCGGAGGTAAGGAGCA |
| MyH4 (Myosin IIb) | GTGATTTCTCCTGTCACCTCTC | GGAGGACCGCAAGAACGTGCTGA |
| Actg1 | TTCCTTCCTGGGCATGGAGT | GGCATACAGGTCTTTGCGGA |
| Murf1 | GCAGCTCATCAAGAGCATTGT | CCAAAGTCAATGGCCCTCAA |
| Atrogin-1 | GTGAGGACCGGCTACTGTGG | CAATCCAGCTGCCCTTTGTC |
| Traf6 | GCGAGAGATTCTTTCCCTGACG | TTGGCACTGGGGACAATTCAC |
| Calpain 1 | CTGGAGGCTGCAGGAACTAC | CTCCCGGTTGTCATAGTCGT |
| Calpain 3 | CGAGAAACGAGGGGAAACCC | ACAGATCTCCATGTCGTCGC |
| Calpastatin | TGGTGGTACAAGACGCATGTTA | ACATGTGGTCTGGAAGGCTG |
| MARCKS | CCAGTTCTCCAAGACCGCAG | CATTCTCCTGCCCATTTGCTTTG |
| Hsp70 | AGTCCGACATGAAGCACTGG | ATCTCCTCCGGGAAGAA |
| Hsp90aa | TGTTCATTCAGCCACGATGC | AACTGGGCAATTTCTGCCTG |
| Hsp90ab | CAGACCATGGTGAGCCCATT | CACCACTTCCTTGACCCTCC |
| Stub1 | CCCTGATAAGAGCCCGAGTG | ACAAGTGGGTTCCGAGTGATG |
| UFD2 | GTCACCAGACCGCAATCTCA | GGCTCCCAAACTAGACAACGA |
| CryaB | ACGTGAAGCACTTCTCTCCG | ATGTTCGTCCTGGCGTTCTT |
| TCP1 | TTAAACGGTCTCAGAGCGGA | GCAGCCATAACATTCTGGGA |
| FKBP8 | GCTGGGAGACTGCGATGTTA | TGTATGGGCTCCTGCCCT |
| skNAC | ACAGCCTCAGGCTGAGACAG | TTTGAAGTTCCTTTTGCCCAG |
| Smyd1 | GCATCTTCCCCAACCTGGGCCT | GGGCCCGGAGCTCAATCCTCAT |
List of all primers employed for semi-quantitative RT-PCR analysis in this study. Commercially designed primers (Qiagen QuantiTect Primer Assays): Titin, Nedd4, Mical, MyoD1, and Cmya4
Generation of protein extracts and western blotting
Whole-cell protein lysates from muscle tissue were generated as follows: snap-frozen muscle tissue was homogenized in 200 μL RIPA lysis buffer (Merck, Darmstadt, Germany) containing protease inhibitors (Pefabloc®, Roth, Karlsruhe, Germany) using a Beadbug homogenizer (Biozym, Hessisch Oldendorf, Germany). Alternatively, to detect only cytosolic Smyd1, a Triton X-100-containing lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 5 mM EDTA, 0.5% Triton X-100) was employed. Subsequently, samples were incubated on ice for 10 min. Samples were then sonicated on ice for five 5 s intervals, and after centrifugation, supernatants were collected. Protein concentration was determined using the BCA Kit (Thermo Fisher Scientific, Waltham, MA, USA). Samples were separated by SDS-PAGE according to Laemmli (Laemmli 1970) and subsequently transferred onto PVDF membranes (Bio-Rad). Membranes were blocked in 10% skim milk in 1× PBS (Roth, Karlsruhe, Germany) and incubated with antibodies as described (Adams et al. 2007; for a list of antibodies, see Table 2). Incubation with a mouse anti-GAPDH antibody (R&D Systems, Minneapolis, USA) and/or a rabbit anti-α-tubulin antibody (Cell Signaling, Danvers, MA, USA) served as a loading control. For detection, horseradish peroxidase-coupled anti-rabbit or anti-mouse secondary antibodies (Dianova, Hamburg, Germany) and the Amersham ECL detection system (GE Healthcare, Freiburg, Germany) were employed.
Table 2.
Antibodies used in this study
| Primary antibodies | |
|---|---|
| GAPDH | R&D System MAB5718 |
| α-Tubulin | Cell signaling #2125 |
| β-Actin | Cell signaling #4967 |
| IκBα | Santa Cruz sc-847 |
| Murf1 | ECM Bioscience MP3401 |
| Atrogin-1 | ECM Bioscience AP2041 |
| Traf6 | Santa Cruz sc-7221 |
| Unc45b | Abcam ab 178813 |
| CryaB | Abcam ab13497 |
| Smyd1 | Abcam ab181372 |
| Secondary antibodies | |
| Goat anti-mouse | Dianova 115–035-071 |
| Goat anti-rabbit | Dianova 111–035-045 |
Statistical analysis
Statistical analyses were performed using qbase+ statistical software (Biogazelle, Gent, Belgium). Data were analyzed by Mann-Whitney U test and REST 2009 software (Pfaffl 2001). Data were considered significant with p values of less than 0.05. All data are presented as means ± SD. “n” represents the number of animals in each group.
Results
Treadmill running and PDTC treatment
Treadmill running was well tolerated by all animals. Most mice complied easily; however, the majority did not run continuously at all times, but mostly did short interval sprints and then made themselves backslide for a few seconds. Furthermore, animals that received PDTC appeared healthy and active and were phenotypically indistinguishable from controls, suggesting that long-term PDTC treatment at a dosage of approximately 10 mg/kg/day was not harmful.
PDTC-mediated inhibition of NFκB activity in skeletal muscle tissue
To test whether PDTC was efficient in blocking NFκB activity, IκBα concentrations were assessed in M. tibialis anterior (TA) muscle tissue derived from all four animal groups. Determination of IκBα levels is an indirect method to evaluate NFκB activity, since there is an inverse correlation between these two parameters (for review, see Karin 1999). As shown in Fig. 1a, b, treadmill running had no effect on steady state IκBα levels in skeletal muscle tissue. By contrast, we found higher IκBα levels in PDTC-treated mice, both exercised and sedentary, in comparison to the respective non-PDTC-treated controls. These data suggest that PDTC was efficient in blocking NFκB activity throughout a 10-week training period, without major adverse effects. However, it should be pointed out that the effects on IκBα levels were not as pronounced in some of the other muscle types: specifically, there was no effect in M. soleus (S). Interestingly, we also found a trend towards elevated expression of the gene encoding the myogenic transcription factor MyoD1, which is known to be inhibited by NF-κB (Guttridge et al. 2000), in both M. quadriceps (Q) and M. gastrocnemius (G) after PDTC treatment, indicating that PDTC had indeed been efficient in reducing NF-κB-dependent effects on gene expression (Fig. 1c). However, due to the high inter-individual variability, these findings did not reach statistical significance.
Fig. 1.
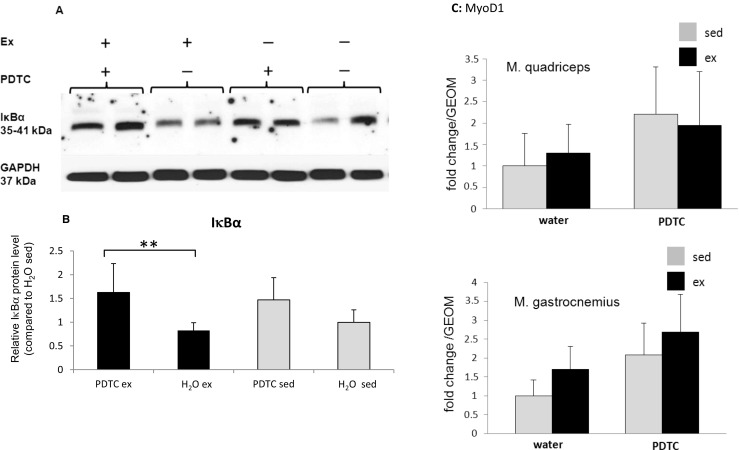
IκBα concentrations in individual skeletal muscles. a Representative Western blot analysis of IκBα concentrations in the M. tibialis anterior of sedentary and trained, PDTC-treated and control mice as indicated. GAPDH levels were analyzed as a control for equal loading and served as a standard for normalization of quantification. b Densitometric quantification of IκBα levels. Data are displayed as means ± SD (*p < 0.05, **p < 0.01; n = 8). c qPCR analysis of the gene encoding MyoD1 in Q and G. Data are displayed as geometric means (GEOM) ± SD (n = 8)
Metabolism-associated genes
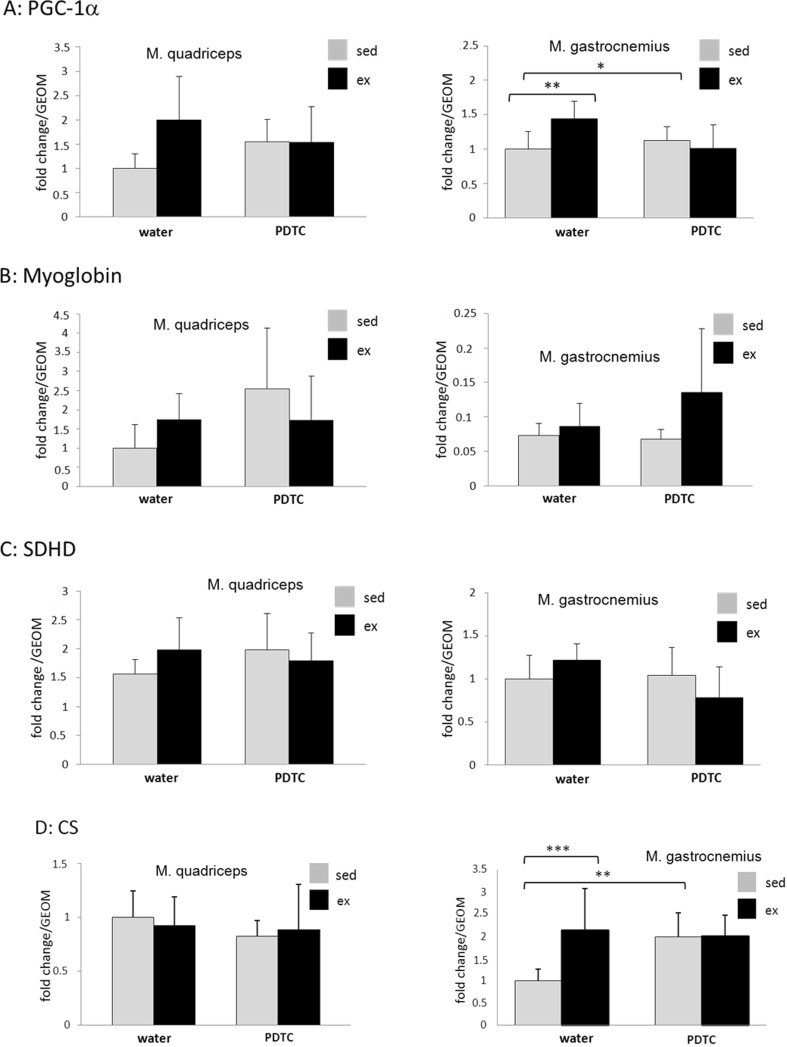
Next, we analyzed expression of metabolism-associated genes in samples derived from two different types of muscles (Q and G) of all four animal groups. As expected, induction of the gene encoding the “oxidative” transcriptional modulator PGC-1α was observed in both Q and G of exercised animals (Fig. 2a); however, only the data obtained for G reached statistical significance. In both muscles, PDTC treatment blocked Pgc-1α induction by exercise. A similar trend was also observed for the gene encoding the oxygen storage protein myoglobin; however, PDTC effects were not consistent in this case (Fig. 2b). The Sdhd gene, encoding subunit D of the succinate dehydrogenase complex, and the Cs gene, encoding citrate synthase, were also moderately induced by exercise in G (significance only for CS), Sdhd also in Q, effects which could not be observed in PDTC-treated animals (Fig. 2c, d). Finally, there was no major exercise-induced regulation of the genes encoding the “glycolytic” and "oxidative" lactate dehydrogenase isoforms (LDHA and LDHB), the Glut1 and Glut4 genes; the Ucp3 gene, encoding uncoupling protein 3, a factor known to be upregulated in response to exercise; or the Foxo1, sirtuin, Mef2c, and Nr4a3 genes, all encoding proteins involved in metabolic regulation in muscle cells (data not shown).
Fig. 2.
Expression analysis of genes encoding metabolic enzymes. qPCR analysis of the genes encoding PGC-1α (a), myoglobin (b), SDHD (c), and CS (d) as indicated. Data are displayed as geometric means (GEOM) ± SD,(*p < 0.05, **p < 0.01, ***p < 0.001; n = 8)
Expression of genes encoding MyHC isoforms
Next, we studied potential effects of training and/or NFκB inhibition on the expression of genes encoding fiber type-specific MyHC isoforms. As shown in Fig. 3a, in TA, MyH7, encoding the “slow oxidative” MyHC1, was slightly, but not significantly, enhanced in exercised animals. In Q and G, by contrast, expression of this gene was consistently, but also not significantly, downregulated. Furthermore, in Q, G, and TA of exercised animals, we found a modest and non-significant induction of MyH2, encoding the “fast oxidative” MyHC2A (Fig. 3b). Similarly, MyH1, encoding the fast MyHC2X, was slightly upregulated by exercise in G and TA (Fig. 3c). In all muscle types, exercise had little effect on expression of the gene encoding the rodent-specific “very fast glycolytic” MyHC2B (MyH4) (Fig. 3d). Parallel PDTC treatment modestly modulated expression of all MyH genes in all muscle types; however, there was no consistent pattern (Fig. 3a–d). Taken together, these data indicate that there were moderate, but no major effects on the expression of fiber type-specific MyH genes in the analyzed muscle tissues.
Fig. 3.
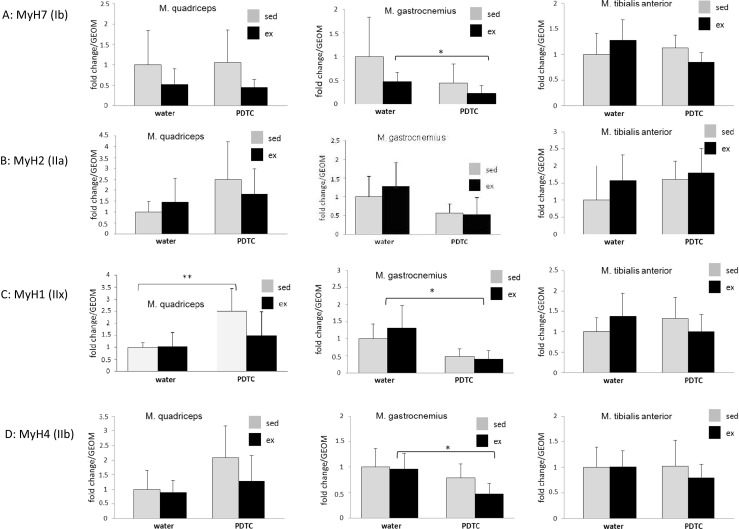
Expression analysis of genes encoding different myosin heavy chain isoforms. qPCR analysis of the genes encoding myosin Ib (a), IIa (b), IIx (c), and IIb (d) as indicated. Data are displayed as geometric means (GEOM) ± SD (*p < 0.05; n = 8)
Other structural sarcomere proteins
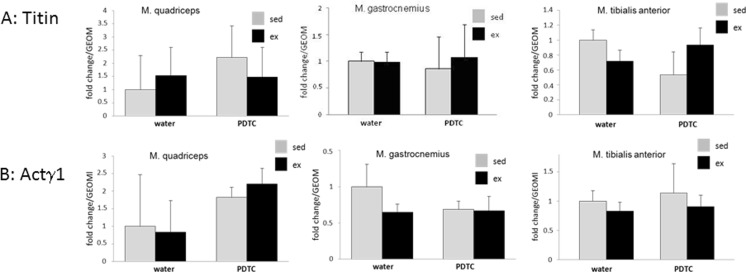
Sarcomeric rearrangements are crucial for training adaptation in response to physical exercise. Thus, we analyzed expression of a broad variety of genes encoding either sarcomere proteins or molecular chaperones assisting in their proper folding, trafficking, and incorporation into sarcomeric structures. As shown in Fig. 4, we found little effect of training on the expression of genes encoding the sarcomere molecular spring titin or actinin γ1, a protein of Z discs and costameres. In addition, we found little regulation of the genes encoding all three myomesin proteins, stabilizers of the sarcomere that interact with titin and are supposed to link M band structures and the intermediate filament cytoskeleton, in response to training (data not shown).
Fig. 4.
Other structural sarcomere proteins. qPCR analysis of the genes encoding titin (a) and actinin γ1 (b). Data are displayed as geometric means (GEOM) ± SD (n = 8)
E3 ubiquitin ligases involved in the degradation of sarcomeric proteins
Skeletal muscle adaptation to exercise is associated with turnover of sarcomere proteins. Specifically, damaged sarcomere proteins become degraded, and new proteins become synthesized and incorporated into sarcomeric structures. Thus, we analyzed expression of genes encoding different players involved in this process. First, we determined expression of the genes encoding the key E3 ubiquitin ligases MuRF1 and atrogin-1, both of which have been implicated in the degradation of sarcomere proteins during skeletal muscle atrophy, but also in exercise adaptation. As shown in Fig. 5a, there were only minor exercise-induced changes in MuRF1 expression in all analyzed muscle types, both at the messenger RNA (mRNA) and at the protein level. In addition, as shown in Fig. 5b, Atrogin-1 expression was slightly, but not significantly, downregulated in most muscle types of exercised mice, both at the mRNA and at the protein level. Interestingly, expression of the Traf6 gene, which encodes an E3 ubiquitin ligase involved in cachectic decay of skeletal muscle tissue (Paul et al. 2010), was slightly, but not significantly, repressed by training in all of the analyzed hindlimb muscles, but only at the protein, hardly at the mRNA level (Fig. 5c). In addition, the Nedd4 gene, which encodes the E3 ubiquitin ligase Nedd4, appeared to be slightly, but not significantly, downregulated by exercise at least in Q (Fig. 5d). Parallel treatment with PDTC had variable and non-consistent effects on the expression of all four genes (Fig. 5a–d).
Fig. 5.
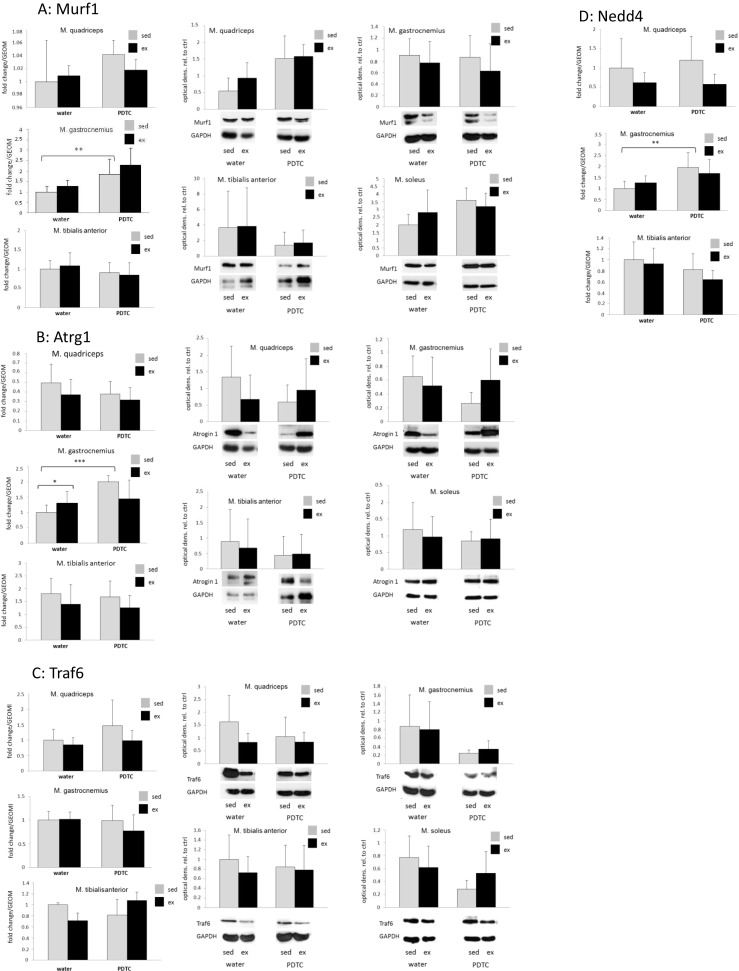
E3 ubiquitin ligases. qPCR analysis of the genes encoding MuRF1 (a), atrogin-1 (b), Traf6 (c), and Nedd4 (d). In a–c, Western blot analysis was also carried out to detect the respective factors at the protein level. Data are displayed as geometric means (GEOM) ± SD (*p < 0.05, **p < 0.01, ***p < 0.001; n = 8 (n = 4 for TA and MuRF1))
Calpain system
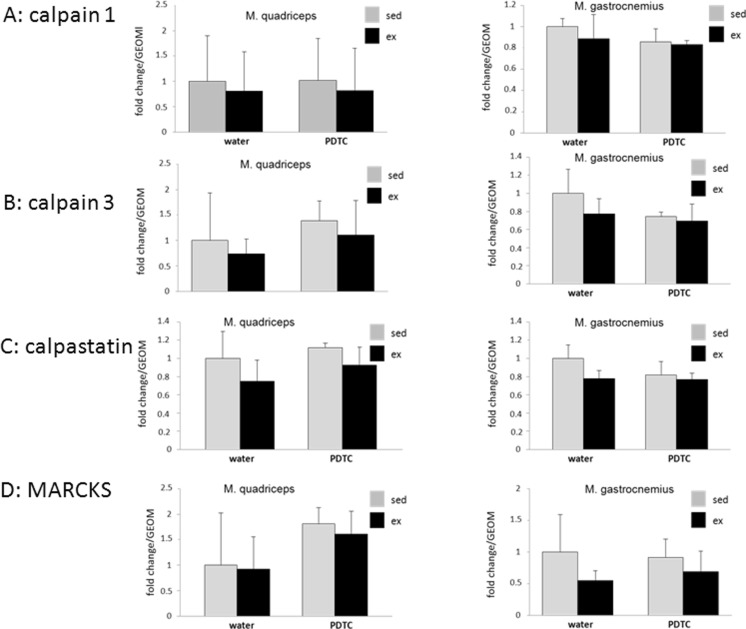
Next, we analyzed expression of genes encoding components of the calpain protease system, which is also involved in the degradation of sarcomere proteins. As shown in Fig. 6, even though there was a consistent trend towards downregulation with exercise in most cases, independently of whether animals received parallel PDTC treatment or not, we found no major effects on the expression of the calpain 1 and calpain 3 genes, the gene encoding the calpain inhibitor calpastatin, or the calpain substrate MARCKS, which, when accumulated, blocks myoblast migration and fusion, in any of the analyzed skeletal muscles.
Fig. 6.
Calpain system. qPCR analysis of the genes encoding calpain 1 (a), calpain 3 (b), calpastatin (c), and MARCKS (d) as indicated. Data are displayed as geometric means (GEOM) ± SD (n = 8 (n = 4 for Q in c, n = 4/3 for G in a–c))
Sarcomere-associated molecular chaperones
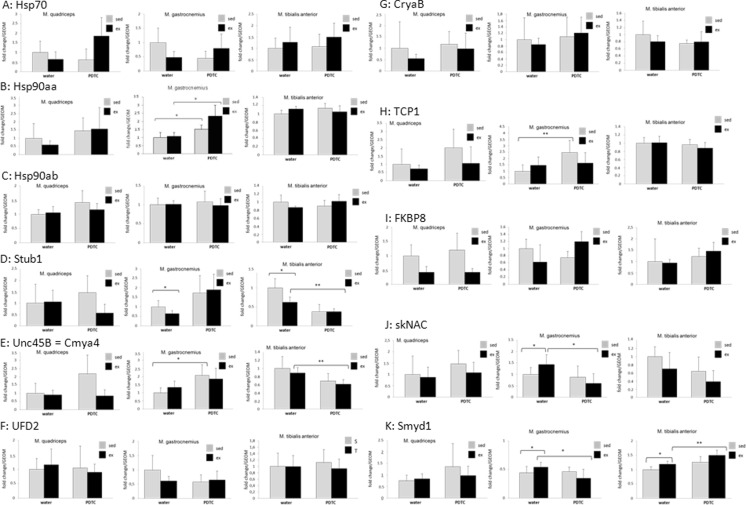
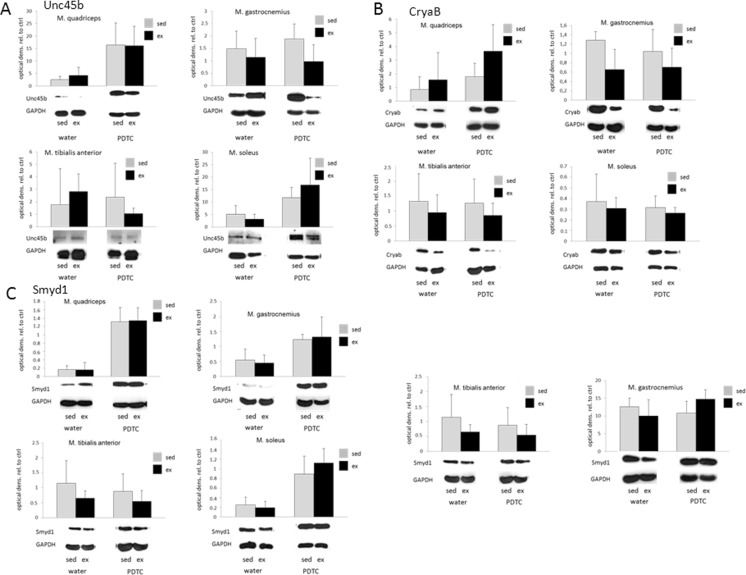
Next, we analyzed the regulation of genes encoding sarcomere-associated molecular chaperones. As shown in Fig. 7a, in Q and G, we found moderately, but not significantly, decreased expression levels of Hsp70 with exercise; in Q, this was also visible for Hsp90aa (Fig. 7b). The described effects were no longer detectable or even reverted under parallel PDTC treatment. By contrast, as expected, no regulation could be observed for the constitutively expressed Hsp90ab gene (Fig. 7c). Interestingly, the gene encoding the Hsp70/90 co-chaperone and E3 ligase Stub1/CHIP was significantly downregulated in response to exercise in both G and TA, with variable effects of PDTC treatment (Fig. 7d). The Unc45b gene, encoding a myosin chaperone and Hsp90 co-chaperone, was hardly regulated by exercise, both at the mRNA and at the protein level (Figs. 7e and 8a). Similar results were obtained for the gene encoding the Unc45b E4 multiubiquitinylation factor UFD2, which was only slightly, but not significantly, downregulated by exercise in G (Fig. 7f). The CryaB gene, encoding a chaperone important for myosin integrity, was also moderately, but not significantly, downregulated in response to exercise in all of the analyzed muscles; for Q, however, this effect could not be observed at the protein level (Figs. 7g and 8b). Again, PDTC treatment exerted variable and non-consistent effects on the expression of all genes. The TCP1 gene, encoding a further myosin-associated chaperone, was also slightly downregulated by exercise at least in Q (Fig. 7h). Downregulation by exercise, particularly in Q and G, was also observed for the FKBP8 gene (Fig. 7i), encoding a peptidyl-prolyl cis-trans isomerase (PPi) and molecular chaperone. All of these results, however, did not reach statistical significance. Interestingly, whereas we did not observe major regulation of the skNAC (skeletal muscle-specific variant of the alpha subunit of nascent polypeptide-associated complex) or Smyd1 (SET and MYND domain-containing protein 1) genes, encoding the two subunits of a putative molecular chaperone for sarcomere proteins, at the mRNA level (Fig. 7j, k), neither by exercise nor by PDTC treatment, we observed a strong increase in Smyd1 protein levels in the sarcoplasm of PDTC-treated animals, both sedentary and exercised (Fig. 8c, left panels), in all analyzed muscles except TA. This effect was no longer visible when we also lysed the cells’ nuclei (Fig. 8c, right panels), suggesting that PDTC treatment might alter subcellular distribution of the Smyd1 protein.
Fig. 7.
Chaperones - qPCR analysis. qPCR analysis of the genes encoding Hsp70 (a), Hsp90aa (b), Hsp90ab (c), Stub1 (d), Unc45b (e), UFD2 (f), CryaB (g), TCP1 (h), FKBP8 (i), skNAC (j), and Smyd1 (i). For the factors shown in e, g, k, Western blot analysis was also carried out as indicated. Data are displayed as geometric means (GEOM) ± SD (*p < 0.05, **p < 0.01; n = 8)
Fig. 8.
Chaperones - Western blot analysis. Western blot analysis of the genes encoding Unc45b (a), CryaB (b), and Smyd1 (c). Smyd1 was done with two different types of lysis buffers: one that predominantly extracts cytoplasmic proteins (left panels) and a “complete” lysis buffer (right panels)
Discussion
Our goal was to study the adaptive response of different murine hindlimb skeletal muscle types to a 10-week running exercise program, with a specific focus on genes encoding sarcomere or sarcomere-associated proteins. To test for a potential effect of the NFκB pathway on expression of the respective genes, a subset of animals was also treated with the NFκB inhibitor PDTC.
Despite contrary literature data (Németh et al. 1998; Cuzzocrea et al. 2002; Nai et al. 2006; Ma et al. 2008; Strait et al. 2008; Sharma et al. 2011), in our hands, single i.p. application of PDTC doses of 80–100 mg/kg in a parallel mouse cohort had led to severe and immediate neurological symptoms, warranting euthanasia. Against this background, when planning our 10-week training experiment, we applied a lower final PDTC dose. Throughout the experiment, all animals appeared healthy and were phenotypically indistinguishable from controls, suggesting that continuous PDTC application up to approximately 10 mg/kg/day is well tolerated by young male C57BL/6 mice.
When analyzing IκBα concentrations in different skeletal muscle tissues by Western blot, we found elevated levels in some individual muscles after PDTC treatment, indicating efficient inhibition of NFκB signaling. Exercise, by contrast, had no effect. However, strong IκBα induction was not visible in all muscle types. This might be due to low sensitivity of the IκBα Western blot. However, a NFκB p65-specific ELISA assay did not yield more prominent effects (data not shown). Alternatively, compensatory effects are a possibility. Furthermore, it is probable that absolute NFκB activity is already quite low in muscle tissue of resting, healthy mice, so that PDTC treatment induced little change. However, it is well possible that throughout the 10-week exercise regimen, individual training sessions might have temporarily induced NFκB activity, and that inhibiting this induction might have affected skeletal muscle adaptation to training. Nevertheless, of course, it is also feasible that higher PDTC doses, which, as explained in the preceding paragraph, could not be applied, might have induced more prominent effects on IκBα levels.
Furthermore, in general, when analyzing the results of PDTC treatment, it is important to keep in mind that these effects are not necessarily based on NFκB inhibition (alone). While PDTC might function as an inhibitor of IκB ubiquitin ligase activity (Hayakawa et al. 2003), and might also block NFκB activity indirectly by acting as an anti-oxidant (Piette et al. 1997), effects of this thiol-containing compound might also be the result of its acting on multiple other signaling pathways dependent on reactive oxygen species (ROS) action, not only the NFκB cascade.
Expression of metabolism-associated genes was moderately and mostly non-significantly affected by both exercise and PDTC treatment. Specifically, we found modest induction of genes associated with oxidative metabolic pathways, such as Pgc-1α, Myoglobin, Sdhd, and CS, suggesting adaptive reactions towards a more oxidative muscle phenotype at least to a certain degree. PDTC application, however, had inconsistent effects on the expression of these genes, and no general pattern could be identified. Finally, there was no significant exercise-induced regulation of the genes encoding the glycolytic and oxidative lactate dehydrogenase isoforms (LDHA and LDHB), the Glut1 and Glut4 genes, the Ucp3 gene, encoding uncoupling protein 3, a factor involved in fatty acid shuttling, suggesting that our exercise protocol did not enhance expression of all genes associated with a rather oxidative metabolism (or repress all glycolytic genes). Interestingly, consistent with our data, Peterson et al. (2008) could demonstrate that despite differences with regard to basal expression levels in individual muscles, a 9-week running exercise regimen did not induce Ucp3 expression in rats.
When studying expression of the genes encoding different fiber type-specific MyHC isoforms, we found moderate and mostly non-significant changes, but no consistent pattern. Remarkably, there was rather a tendency towards downregulation and not upregulation of MyH7, encoding the “slow” MyHC type 1 protein, with exercise, indicating that our regimen of 10 weeks of treadmill running did not induce a shift towards “slower” myosin isoforms in the muscles analyzed. This might be due to the fact that either the “endurance” component was not pronounced enough or to the fact that the potential to induce fiber type switches, as indicated by shifted patterns of isoform-specific MyH gene expression, in the adult murine muscle is quite limited.
Similarly, there were no consistent changes in expression of different genes encoding structural sarcomere or sarcomere-associated proteins, such as the elastic spring protein titin or actinin γ1, a protein of Z discs and costameres, indicating that expression of genes encoding structural sarcomere components might not be altered in the trained skeletal muscle.
There was a tendency towards downregulation of genes encoding E3 ubiquitin ligases involved in the degradation of sarcomeric proteins with exercise, namely, Atrogin-1. This is consistent with previous results. Whereas a single bout of exercise, both endurance- or resistance-oriented, in most, but not all, cases increases expression of the so-called “atrogenes” in skeletal muscles (Yang et al. 2006; Louis et al. 2007; Raue et al. 2007; Harber et al. 2009; Pasiakos et al. 2010; Dalbo et al. 2011; Fernandez-Gonzalo et al. 2013), long-term exercise training appears to lower their expression (Zanchi et al. 2009), suggesting that trained individuals might show a lower net rate of sarcomere protein degradation. However, these results did also not reach statistical significance.
Despite the known role of the calpain system in exercise-induced skeletal muscle remodeling (for review, see Murphy 2010; Pasiakos and Carbone 2014), we did not find significant changes with regard to the expression of genes encoding different calpains or their regulators. Again, however, there was a consistent trend towards downregulation of these genes with exercise, indicating that trained skeletal muscle tissue might be characterized by a less active calpain system. The fact that parallel PDTC treatment did not alter these effects indicates that they might not be NFκB dependent.
The effects of exercise on the expression of genes encoding sarcomere-associated molecular chaperones were also minor, mostly non-significant, variable, and very much dependent on the type of muscle analyzed, indicating that there is no major difference between “steady state” levels of skeletal muscle remodeling between trained and untrained individuals. Furthermore, in response to PDTC treatment, there was no consistent pattern of gene regulation, suggesting that basal levels of skeletal muscle remodeling, both in trained and in untrained individuals, are not highly dependent on NFκB activity. Interestingly, our data suggest that in all muscles except TA, PDTC might induce translocation of the Smyd1 protein to the sarcoplasm, where it might associate with the sarcomere (for review, see Du et al. 2014).
Against the background that in summary, we observed only modest changes in gene expression in response to our 10-week treadmill running protocol, it appears likely that the type, frequency, duration, or intensity of exercise (14.4 m/min, three times a week for 60 min over a period of 10 weeks) were too low to induce major effects in our cohort of young, healthy mice. Despite the fact that several studies on mouse treadmill running employed similar conditions (Rosa et al. 2005; Brooks et al. 2008; Glaser et al. 2010; Baker et al. 2011), Allen et al. (2001) and De Bono et al. (2006) could demonstrate in “voluntary wheel running” experiments that mice can reach velocities of 25–50 m/min without any problems and voluntarily run distances of 5–15 km/day. In addition, it is likely that even without a wheel supplied, the “sedentary” mice might already show a high degree of activity, thereby leveling out potential differences between “ex” and “sed” mice. Indeed, in our study, while treadmill running was well tolerated by all mice, only a few of them ran continuously at the adjusted pace at most times, whereas most animals switched between sprinting and backsliding/resting to some extent. This heterogeneity in running behavior might also explain part of the high variation found in some of our data.
Alternatively, it is possible that skeletal muscle plasticity with regard to endurance exercise in the murine system is limited—in other words, it is well possible that, for instance, the range within which expression patterns of genes encoding myosin isoforms and therefore metabolic characteristics and fiber types can be altered is quite narrow in mice.
Thus, due to behavioral as well as physiological aspects, mice might not be an ideal model system to perform endurance exercise training studies.
Taken together, our results indicate that 10 weeks of regular treadmill running (three times a week, 60 min each, with a maximum speed of 14.4 m/min reached after 5 weeks) induce only minor changes with regard to the expression of genes known to be involved in skeletal muscle plasticity. Furthermore, these changes were very much dependent on the type of muscle analyzed. Finally, whereas PDTC alone exerted persistent and in some cases profound effects on skeletal muscle gene expression patterns, there was little effect of this factor on exercise-induced changes in gene expression. Future research should determine the physiological significance of these subtle changes, as well as if and how they are dependent on the type, duration, and intensity of the chosen exercise regimen and on PDTC dosage.
Acknowledgements
We thank Thomas Beiter for the help with qPCR primer design and helpful discussions. This work was supported by a grant from the Monika-Kutzner-Stiftung, Berlin, Germany (to B.M.).
References
- Adams S, Pankow S, Werner S, Munz B (2007) Regulation of NF-kappaB activity and keratinocyte differentiation by the RIP4 protein: implivations for cutaneous wound repair. J Invest Dermatol 127:538–44 [DOI] [PubMed]
- Allen DL, Harrison BC, Maass A, Bell ML, Byrnes WC, Leinwand LA. Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J Appl Physiol. 2001;90:1900–1908. doi: 10.1152/jappl.2001.90.5.1900. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Reed DR, Beauchamp GK, Tordoff MG. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav Genet. 2002;32:435–443. doi: 10.1023/A:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JM, DeLisoi M, Parise G. Endurance exercise training promotes medullary hematopoiesis. FASEB J. 2011;25:4348–4357. doi: 10.1096/fj.11-189043. [DOI] [PubMed] [Google Scholar]
- Bartoli M, Richard I. Calpains in muscle wasting. Int J Biochem Cell Biol. 2005;37:2115–2133. doi: 10.1016/j.biocel.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Bell RA, Al-Khalaf M, Megeney LA. The beneficial role of proteolysis in skeletal muscle growth and stress adaptation. Skelet Muscle. 2016;6:16. doi: 10.1186/s13395-016-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen TS, Schuler G, Adams V. Skeletal muscle wasting in cachexia and sarcopenia: molecular pathophysiology and impact of exercise training. J Cachexia Sarcopenia Muscle. 2015;6:197–207. doi: 10.1002/jcsm.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SV, Vasilaki A, Larkin LM, McArdle A, Jackson MJ. Repeated bouts of aerobic exercise lead to reductions in skeletal muscle free radical generation and nuclear factor kappaB activation. J Physiol. 2008;596:3979–3990. doi: 10.1113/jphysiol.2008.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Crawford GL, Horowits R. Scaffolds and chaperones in myofibril assembly: putting the striations in striated muscle. Biophys Rev. 2011;3:25–32. doi: 10.1007/s12551-011-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzzocrea S, Chatterjee PK, Mazzon E, Dugo L, Serraino I, Britti D, Mazzullo G, Caputi AP, Thiememann C. Pyrrolidine dithiocarbamate attenuates the development of acute and chronic inflammation. Br J Pharmacol. 2002;135:496–510. doi: 10.1038/sj.bjp.0704463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzzocrea S, Rossi A, Pisano B, Di Paola R, Genovese T, Patel NS, Cuzzocrea E, Ianaro A, Sautebin L, Fulia F, Chatterjee PK, Caputi AP, Thiememann C. Pyrrolidine dithiocarbamate attenuates the development of organ failure induced by zymosan in mice. Intensive Care Med. 2003;29:2016–2025. doi: 10.1007/s00134-003-1887-8. [DOI] [PubMed] [Google Scholar]
- Dalbo VJ, Roberts MD, Hassell SE, Brown RD, Kerksick CM. Effects of age on serum hormone concentrations and intramuscular proteolytic signaling before and after a single bout of resistance training. J Strength Cond Res. 2011;25:1–9. doi: 10.1519/JSC.0b013e3181fc5a68. [DOI] [PubMed] [Google Scholar]
- De Bono JP, Adlam D, Paterson DJ, Channon KM. Novel quantitative phenotypes of exercise training in mouse models. Am J Phys Regul Integr Comp Phys. 2006;290:R926–R934. doi: 10.1152/ajpregu.00694.2005. [DOI] [PubMed] [Google Scholar]
- Du SJ, Tan X, Zhang J. SMYD proteins: key regulators in skeletal and cardiac muscle development and function. Anat Rec. 2014;297:1650–1662. doi: 10.1002/ar.22972. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalo R, Lundberg TR, Tesch PA. Acute molecular responses in untrained and trained muscle subjected to aerobic and resistance exercise training versus resistance training alone. Acta Physiol. 2013;209:283–294. doi: 10.1111/apha.12174. [DOI] [PubMed] [Google Scholar]
- Glaser BW, You G, Zhang M, Medler S. Relative proportions of hybrid fibres are unaffected by 6 weeks of running exercise in mouse skeletal muscles. Exp Physiol. 2010;95:211–221. doi: 10.1113/expphysiol.2009.049023. [DOI] [PubMed] [Google Scholar]
- Gu JW, Young E, Busby B, Covington J, Johnson JW. Oral administration of pyrrolidine dithiocarbamate (PDTC) inhibits VEGF expression, tumor angiogenesis and growth of breast cancer in female mice. Cancer Biol Ther. 2009;15:514–521. doi: 10.4161/cbt.8.6.7689. [DOI] [PubMed] [Google Scholar]
- Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- Harber MP, Crane JD, Dickinson JM, Jemiolo B, Raue U, Trappe TA, Trappe SW. Protein synthesis and the expression of growth-related genes are altered by running in human vastus lateralis and soleus muscles. Am J Phys Regul Integr Comp Phys. 2009;296:R708–R714. doi: 10.1152/ajpregu.90906.2008. [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Miyashita H, Sakamoto I, Kitagawa M, Tanaka H, Yasuda H, Karin M, Kikugawa K. Evidence that reactive oxygen species do not mediate NF-kappaB activation. EMBO J. 2003;22:3356–3366. doi: 10.1093/emboj/cdg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji LL. Redox signaling in skeletal muscle: role of aging and exercise. Adv Physiol Educ. 2015;39:352–359. doi: 10.1152/advan.00106.2014. [DOI] [PubMed] [Google Scholar]
- Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- Kramer HF, Goodyear LJ. Exercise, MAPK, and NF-kappaB signaling in skeletal muscle. J Appl Physiol. 2007;103:388–395. doi: 10.1152/japplphysiol.00085.2007. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu SF, Ye X, Malik AB. Inhibition of NF-kB activation by Pyrrolidine Dithiocarbamate prevents in vivo expression of proinflammatory genes. Circulation. 1999;100:1330–1337. doi: 10.1161/01.CIR.100.12.1330. [DOI] [PubMed] [Google Scholar]
- Louis E, Raue U, Yang Y, Jemiolo B, Trappe S. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J Appl Physiol. 2007;103:1744–1751. doi: 10.1152/japplphysiol.00679.2007. [DOI] [PubMed] [Google Scholar]
- Ma XL, Li YH, Geo JX, Li J, Guo L, Wu CZ. Expression of inducible nitric oxide synthase in the liver is under the control of nuclear factor kappa B in concanavalin A-induced hepatitis. J Gastroenterol Hepatol. 2008;23(7Pt2):e231–e235. doi: 10.1111/j.1440-1746.2007.05083.x. [DOI] [PubMed] [Google Scholar]
- Mariappan N, Elks CM, Sriramula S, Guggilam A, Liu Z, Borkhsenious O, Francis J. NF-kappaB-induced oxidative stress contributes to mitochondrial and cardiac dysfunction in type II diabetes. Cardiovasc Res. 2010;85:473–483. doi: 10.1093/cvr/cvp305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RM. Calpains, skeletal muscle function and exercise. Clin Exp Pharmacol Physiol. 2010;37:385–391. doi: 10.1111/j.1440-1681.2009.05310.x. [DOI] [PubMed] [Google Scholar]
- Murton AJ, Constantin D, Greenhaff PL. The involvement of the ubiquitin proteasome system in human skeletal muscle remodelling and atrophy. Biochim Biophys Acta. 2008;1782:730–743. doi: 10.1016/j.bbadis.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Nai YJ, Jianh Z-W, Wang Z, Li N, Li J-S. Prevention of cancer cachexia by pyrrolidine dithiocarbamate (PDTC) in colon 26 tumor-bearing mice. J Parenter Enter Nutr. 2006;31:18–25. doi: 10.1177/014860710703100118. [DOI] [PubMed] [Google Scholar]
- Németh ZH, Haskó G, Vizi ES. Pyrrolidine dithiocarbamate augments IL-10, inhibits TNF-alpha, MIP-1alpha, IL-12, and nitric oxide production and protects from the lethal effects of endotoxin. Shock. 1998;10:49–53. doi: 10.1097/00024382-199807000-00009. [DOI] [PubMed] [Google Scholar]
- Parodi FE, Mao D, Ennis TL, Bartoli MA, Thompson RW. Suppression of experimental abdominal aortic aneurysms in mice by treatment with pyrrolidine dithiocarbamate, an antioxidant inhibitor of nuclear factor-kB. J Vasc Surg. 2005;41:479–489. doi: 10.1016/j.jvs.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Pasiakos SM, Carbone JW. Assessment of skeletal muscle proteolysis and the regulatory response to nutrition and exercise. IUBMB Life. 2014;66:478–484. doi: 10.1002/iub.1291. [DOI] [PubMed] [Google Scholar]
- Pasiakos SM, McClung HL, McClung JP, Urso ML, Pikosky MA, Cloutier GJ, Fielding RA, Young AJ. Molecular responses to moderate endurance exercise in skeletal muscle. Int J Sport Nutr Exerc Metab. 2010;20:282–290. doi: 10.1123/ijsnem.20.4.282. [DOI] [PubMed] [Google Scholar]
- Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, Kumar A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol. 2010;191:395–411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JM, Bryner RW, Frisbee JC, Alway SE. Effects of exercise and obesity on UCP3 content in rat hindlimb muscles. Med Sci Sports Exerc. 2008;40:1616–1622. doi: 10.1249/MSS.0b013e31817702a4. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:45e. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J, Piret B, Bonizzi G, Schoonbroodt S, Merville MP, Legrand-Poels S, Bours V. Multiple redox regulation in NF-kappaB transcription factor activation. Biol Chem. 1997;378:1237–1245. [PubMed] [Google Scholar]
- Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Proteolytic gene expression differs at rest and after resistance exercise between young and old women. J Gerontol A Biol Sci Med Sci. 2007;62:1407–1412. doi: 10.1093/gerona/62.12.1407. [DOI] [PubMed] [Google Scholar]
- Rosa EF, Silva AC, Ihara SSM, Mora OA, Aboulafia J, Nouailhetas VLA. Habitual exercise program protects murine intestinal, skeletal, and cardiac muscles against aging. J Appl Physiol. 2005;99:1569–1575. doi: 10.1152/japplphysiol.00417.2005. [DOI] [PubMed] [Google Scholar]
- Seene T, Kaasik P, Umnova M. Structural rearrangements in contractile apparatus and resulting skeletal muscle remodelling: effect of exercise training. J Sports Med Phys Fitness. 2009;49:410–423. [PubMed] [Google Scholar]
- Sharma V, Gihotra R, Dhingra D, Gihotra N. Possible underlying influence of p38MAPK and NF-kappaB in the diminished anti-anxiety effect of diazepam in stressed mice. J Pharmacol Sci. 2011;116:257–263. doi: 10.1254/jphs.11026FP. [DOI] [PubMed] [Google Scholar]
- Schreck P, Meier B, Männel DN, Dröge W, Baeuerle PA (1992) Dithiocarbamates as potent inhibitors of nuclear factor kappa B in intact cells. J Exp Med 175:1181–94 [DOI] [PMC free article] [PubMed]
- Strait K, Li Y, Dillehay DL, Weitzmann MN. Supression of NFkB activation blocks osteclastic bone resorbtion during estrogen deficiency. Int J Mol Med. 2008;21:521–525. [PubMed] [Google Scholar]
- Yang Y, Jemiolo B, Trappe S. Proteolytic mRNA expression in response to acute resistance exercise in human single skeletal muscle fibers. J Appl Physiol. 2006;101:1442–1450. doi: 10.1152/japplphysiol.00438.2006. [DOI] [PubMed] [Google Scholar]
- Zanchi NE, de Siqueira Filho MA, Lira FS, Rosa JC, Yamashita AS, de Oliveira Carvalho CR, Seelaender M, Lancha-Jr AH. Chronic resistance training decreases MuRF-1 and atrogin-1 gene expression but does not modify Akt, GSK-3beta and p70S6K levels in rats. Eur J Appl Physiol. 2009;106:415–423. doi: 10.1007/s00421-009-1033-6. [DOI] [PubMed] [Google Scholar]
- Zhang X, Luhrs KJ, Ryff KA, Malik WT, Driscoll MJ, Culver B. Suppression of nuclear factor kappa B ameliorates astrogliosis but not amyloid burden in APPswe/PS1dE9 mice. Neuroscience. 2009;161:53–58. doi: 10.1016/j.neuroscience.2009.03.010. [DOI] [PubMed] [Google Scholar]