Abstract
Adaptive radiation research typically relies on the study of evolution in retrospective, leaving the predictive value of the concept hard to evaluate. Several radiations, including the cichlid fishes in the East African Great Lakes, have been studied extensively, yet no study has investigated the onset of the intraspecific processes of niche expansion and differentiation shortly after colonization of an adaptive zone by cichlids. Haplochromine cichlids of one of the two lineages that seeded the Lake Victoria radiation recently arrived in Lake Chala, a lake perfectly suited for within-lake cichlid speciation. Here, we infer the colonization and demographic history, quantify phenotypic, ecological and genomic diversity and diversification, and investigate the selection regime to ask if the population shows signs of diversification resembling the onset of adaptive radiation. We find that since their arrival in the lake, haplochromines have colonized a wide range of depth habitats associated with ecological and morphological expansion and the beginning of phenotypic differentiation and potentially nascent speciation, consistent with the very early onset of an adaptive radiation process. Moreover, we demonstrate evidence of rugged phenotypic fitness surfaces, indicating that current ecological selection may contribute to the phenotypic diversification.
Keywords: adaptive radiation, cichlid fish, incipient speciation, fitness surfaces, disruptive selection, niche expansion
1. Background
Adaptive radiations, when multiple species with different adaptations arise rapidly from a single ancestral population, provide iconic showcases of evolution. Adaptive radiation is thought to occur when a population from an ecologically versatile lineage with propensity to speciate encounters ecological opportunity [1–3], for instance upon colonization of habitats, unoccupied by incumbent species. Response to ecological opportunity is thought to begin with intraspecific ecological niche expansion, followed by a burst of phenotypic differentiation associated with speciation [2,4,5]. Celebrated examples of adaptive radiation are the Antillean Anolis lizards, Galapagos finches, Hawaiian silverswords, lobeliads and honeycreepers, and the cichlid fishes of the African Great Lakes [6–11].
Even though many of these radiations are extensively studied, our knowledge about the very beginnings of the adaptive radiation process remains limited and mainly confined either to theoretical exploration [2,5,12–14] or to small radiations in taxa that are only very distantly related to those that made the classical large radiations [15]. Some important questions regarding the mechanisms such as whether ecological diversification precedes or follows speciation can hardly be inferred that way. Ecological speciation work has revealed that divergence into phenotypically distinct and partially reproductively isolated incipient species can sometimes occur surprisingly fast, i.e. in less than 100 generations [16,17]. To understand the processes during the initiation of adaptive radiation, it is thus essential to study the very first generations after the colonization of a new adaptive zone. For cichlid fishes, radiation-prone adaptive zones are deep equatorial clear-water lakes, and cichlid lineages with a special propensity for speciation are those that are sexually dimorphic and ecologically versatile [18,19], especially haplochromines [19].
Lake Chala, a small (4.2 km2) but deep (approx. 90 m) equatorial clear water (Secchi depth approx. 6.6 m) crater lake in Tanzania, harbours four cichlid species: the endemic Oreochromis hunteri, an undescribed Oreochromis species (Oreochromis sp. ‘blue head’, sometimes erroneously referred to as Oreochromis korogwe), Coptodon rendalli and a haplochromine cichlid of the Astatotilapia bloyeti complex [20]. The latter three most likely were introduced after 1951 [21,22].
By occurring in a relatively deep and well-oxygenated lake [23] that receives high solar energy, and has very clear waters, the sexually colour-dimorphic Astatotilapia of Lake Chala meets all requirements for undergoing intralacustrine diversification [18]. Closely related members of this genus were founders of many of the largest and fastest African cichlid fish radiations [24–26], and have been shown to undergo incipient speciation in another crater lake within a few thousand years [27]. In contrast to all of these cases [24–27], the Astatotilapia in Lake Chala most likely arrived only decades ago [21,22], making them a perfectly suited system to investigate the very onset of the processes associated with adaptive radiation.
Here, we study the phenotypic, ecological and genomic variation within Astatotilapia of Lake Chala, a few decades after their ancestors arrived in the lake. We test the hypotheses that the population has expanded its ecological niche, and that it has begun to diversify ecologically, phenotypically and genetically. We reconstruct the colonization history, assess ecological, phenotypic and genomic variation and differentiation between the lake cichlids and their closest riverine relatives, and within the lake. Finally, we estimate fitness surfaces and ask whether they are consistent with the hypothesis that the population currently experiences disruptive natural selection which might initiate an adaptive radiation process.
2. Methods
(a). Fish sampling
We sampled Astatotilapia from three locations in Lake Chala in November 2014. At each location, we sampled the depth gradient down to the greatest depth that still yielded fish (55 m) using gill nets (mesh sizes: 12–22 mm knot-to-knot). For each fish, the capture depth was recorded and a standardized cuvette picture, a fin clip (for DNA extraction) and muscle tissue (for stable isotope analyses) were taken. To infer the colonization history of all Lake Chala cichlids and to investigate potential interspecific competitors, we additionally sampled all other cichlid species in Lake Chala (O. hunteri, O. sp. ‘blue head’ and C. rendalli), as well as Astatotilapia, Oreochromis (all species) and Coptodon from close-by water bodies (electronic supplementary material, appendix S1; [20]).
(b). DNA extraction and sequencing
DNA was extracted from fin clips of Astatotilapia from Lake Chala (n = 90, electronic supplementary material, tables S1 and S2), Nyumba ya Mungu (NYM) (n = 6), Lake Babati (n = 22), and C. rendalli from Lake Chala (n = 4) and Nyumba ya Mungu (n = 2). We used the phenol chloroform DNA extraction protocol outlined in Sambrook & Russell [28] and sequenced 830 bp of the mitochondrial D-loop region.
For Astatotilapia, we further performed restriction-site-associated DNA (RAD) sequencing to reconstruct genome-wide relationships with haplochromines in other East African water bodies and to reconstruct the demographic history of and investigate genomic differentiation within Lake Chala. RAD-libraries were prepared following the protocol by Baird et al. [29], with slight modifications [17]. Sequencing was performed on an Illumina HiSeq2500 platform at the Centre of Integrative Genomics, University of Lausanne. For processing of the Illumina sequence data, see the electronic supplementary material, appendix S2.
(c). Phylogenetic reconstructions
For reconstructing the phylogeographic relationships of Astatotilapia in northeast Tanzania, we aligned our mitochondrial sequences in BioEdit v. 7.2.5 together with sequences from GenBank of the northeast Tanzanian A. sparsidens/bloyeti clade [26]. Templeton, Crandall and Sing (TCS) networks were built and visualized in popART [30]. We did the same for Coptodon, together with GenBank sequences of most other Coptodon species occurring in northeastern Tanzania [31,32]. The same has been done for Oreochromis in [20]. We used RAxML 8.0.0 [33] to build a maximum-likelihood tree for Astatotilapia of northeastern Tanzania using concatenated sequences derived from RAD sequencing, including samples of A. sparsidens (Lake Manyara), A. cf. bloyeti (Pangani catchment), A. sp. from Lake Babati, Pundamilia spp. and Enterochromis spp. of Lake Victoria and Metriaclima zebra (Lake Malawi) as outgroup [26].
(d). Demographic analyses
We performed demographic modelling with fastsimcoal 2.6 [34] fitting the models to the observed multidimensional site frequency spectra computed in Arlequin [35], for testing four different scenarios. Lake Chala was modelled with (i) constant population size, (ii) a bottleneck directly after colonization, (iii) a continuous expansion starting directly after colonization, or (iv) a bottleneck directly after colonization, followed by a continuous expansion (parameters provided in the electronic supplementary material, table S6). Each model was combined once with a simple split between Lake Chala and the Pangani Astatotilapia and once with an additional second colonization event. Colonization time was inferred from each model combination using three different and complementary calibration approaches (further details in the electronic supplementary material, appendix S3).
To test whether haplochromines from the Lake Victoria region or Astatotilapia from nearby Lake Manyara or the Ruaha River may have contributed to the genomic variation in Astatotilapia of Lake Chala, we performed tests of introgression (ABBA-BABA tests) with Admixtools 1.1 ([36], electronic supplementary material, appendix S4).
(e). Morphology and trophic ecology
For 284 male Astatotilapia, 23 O. hunteri, 72 O. ‘blue head’ and nine C. rendalli, we measured 15 linear morphometric distances that have shown to be powerful to quantify subtle ecologically and taxonomically relevant morphometric variation among closely related species of haplochromine cichlids [37,38] using digital callipers: standard length (SL), head length (HL), head width (HW), body depth (BD), lower jaw length (LJL) and width (LJW), snout length (SnL), snout width (SnW), cheek depth (ChD), preorbital depth (POD) and width (POW), interorbital width (IOW), eye length (EyL) and depth (EyD), and premaxillary pedicel length (PPL; electronic supplementary material, figure S1). These distances were log-transformed and size-corrected by using the standardized residuals of the linear regression of each log-transformed trait against log-SL within each genus. Subsequent analyses of Astatotilapia sp. were restricted to sexually mature males to not confound diversification with ontogenetic or sex-related variation.
To test whether Astatotilapia of Lake Chala has diverged morphologically from its closest relatives outside the lake, A. cf. bloyeti from the Pangani River (see results on the phylogeography), we ran ANOVAs for each morphological trait. Furthermore, we tested whether colonization of the deep and clear Lake Chala (from the shallow and murky river and its reservoir) was associated with the predicted expansion of the realized morphospace correcting for different sample sizes using permutation tests (electronic supplementary material, appendix S5). All analyses, if not stated differently, were performed in R 3.2.1 [39].
To quantify phenotypic and ecological variation within Astatotilapia sp. ‘Chala’, we quantified nuptial coloration for all mature males that still showed nuptial coloration after capture (n = 84, electronic supplementary material, appendix S6, figure S2 and table S4), and analysed stable isotopic ratios (δ13C and δ15N; n = 133) and stomach contents (n = 67; electronic supplementary material, appendix S7). We tested for ecology-related individual variation in morphology by performing ANOVAs for each single trait against water depth and against two isotopic ratios reflecting feeding ecology (δ13C, δ15N).
To test whether the Astatotilapia were in the process of differentiating into ecologically and phenotypically divergent groups within the lake, we visually sorted our fish into phenotypic groups based on an integrated visual assessment of body shape and coloration (electronic supplementary material, table S3). We ended up with nine potential phenotypic groups, subsequently referred to as ‘morphs’ (BBE, blue belly; GAL, gaurochromis-like; LEA, lean; LEO, lean orange; LMO, large mouth; OSH, orange shoulder; PLB, planktivore blue; PLR, planktivore red; YBE, yellow belly). If individuals that we grouped to morphs on the basis of their visual appearance alone are significantly differentiated from each other ecologically and/or genomically, this would indicate that Lake Chala cichlids may have begun to diversify. To see whether the phenotypic groupings indeed represented morphologically distinguishable groups, we performed linear discriminant analyses (LDA), calculated Bhattacharyya distances between the morph clusters in the morphospace and tested for significance with Hotelling's T2-test. We would like to point out that our use of the term ‘morph’ does in no way imply discrete non-overlapping groups, such as have been described in trophic polymorphisms of north-temperate fishes [40].
(f). Population genomics
To study genetic differentiation between the Astatotilapia from Lake Chala and Astatotilapia from Nyumba ya Mungu (upper Pangani), we used 2770 single nucleotide polymorphisms retained after filtering our RAD-sequencing data to calculate F-statistics in Arlequin 3.5.1.4 [35]. To test for genetic structure within Lake Chala, we additionally used ParallelStructure [41] and ran principle component analyses (PCA) [42] in R (see the electronic supplementary material, appendix S2). As we did not find genetic structure between sampling locations within Lake Chala (best-supported K = 1, non-significant FST between all locations (FST < 0.001, p > 0.95)), data from all locations were combined for subsequent analyses. Within Lake Chala, F-statistics were calculated for each morph-pair, between groups with distinct mitochondrial haplotypes, and between groups of distinct male nuptial coloration. To test for significance for FSTs between morph pairs, we used a permutation test (electronic supplementary material, appendix S2).
(g). Estimation of fitness surfaces
Fitness variation is ideally measured as variation in individual lifetime reproductive success, which is almost impossible to obtain for wild fishes in large populations. Growth rate has been successfully used as a fitness proxy in fishes, including cichlids [43–45]. We inferred growth rate for individuals by relating their standard length to their age measured from scale circuli, and used this as a fitness proxy, an approach that we had experimentally validated previously ([45], see also the electronic supplementary material, appendix S8).
Growth rate variation was assessed against variation in phenotypic traits to assess evidence of phenotypic selection using Lande & Arnold's [46] ordinary least-squares regression approach (electronic supplementary material, appendix S9) and canonical analyses with subsequent Eigen-analyses [47,48]. Additionally, we performed projection pursuit regression analyses (PPR) that reduce multivariate phenotypic data to two axes that explain most fitness variation and allow for more complex fitness surfaces than canonical analyses [48,49]. Both axes of the PPR were checked for their relationship to our raw growth rate estimate, using Akaike information criterion (ΔAICc)-thresholds (electronic supplementary material, appendix S9), allowing a maximum of three fitness optima in phenotype space. Fitness surfaces were visualized using the Tps function in the R-package fields [50].
To test for effects of phenotype-environment interactions on fitness, we tested how the relationship between fitness and the same trait combinations change along water depth and whether the trait combinations that best predict fitness variation differ between different depths. For this, we created fitness surfaces for five 10 m depth bands using the same morphological trait axes estimated by the PPR with all fish from all depths combined (a1 and a2), and additionally created fitness surfaces for three different depth categories by calculating depth-specific eigenvalues/projections (electronic supplementary material, appendix S10). To test whether the differences among the fitness surfaces for the different depth bands were significant, we performed paired Wilcoxon signed-rank tests. To test whether including depth as a variable increases the model fit, we applied generalized additive models (for details, see the electronic supplementary material, appendix S11).
To test for evolutionary response to the selection that might result from the inferred fitness surfaces, we tested whether individuals clustered around local fitness optima in phenotype space, and whether the relative occurrence of one morph on one side of a fitness valley is more frequent than expected (electronic supplementary material, appendix S12).
3. Results
(a). Colonization history of Lake Chala
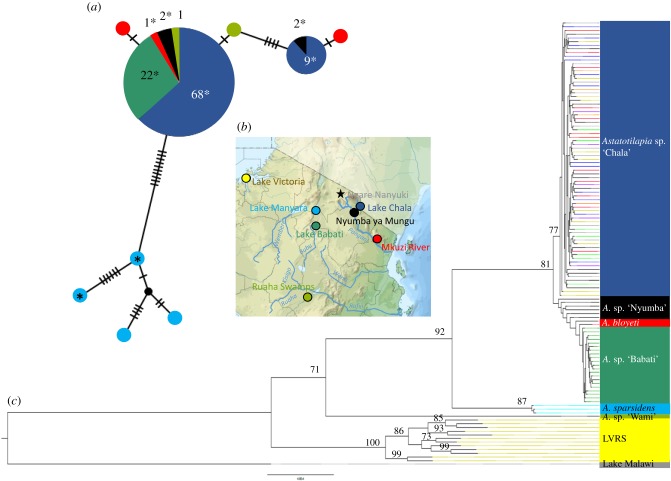
Sequences of the mitochondrial D-loop of Astatotilapia from Lake Chala revealed two clearly distinct haplotypes (separated by five mutations in 830 bp). Both haplotypes are shared with A. cf. bloyeti from the nearby Pangani River reservoir (figure 1a, for more details, see the electronic supplementary material, appendix S13).
Figure 1.
(a) TCS haplotype network (D-loop, 830 bp) for Astatotilapia of Lake Chala (dark blue), Astatotilapia cf. bloyeti of the lower Pangani (red) and of Nyumba ya Mungu (black), Astatotilapia sparsidens of Lake Manyara (light blue), Astatotilapia sp. of Lake Babati (green) and Astatotilapia sp. of the Ruaha Swamps (olive). Numbers at the pie charts state number of individuals per sampling location. Asterisks indicate individuals that were included in the phylogenetic tree. (b) Map indicating sampling sites. (c) Phylogenetic tree based on approximately 2 million sites indicating monophyly of Astatotilapia sp. ‘Chala’ with its closest relative being A. cf. bloyeti from the close-by Pangani River catchment (red and black). Colour code as in (a,b), with representatives of the Lake Victoria Region Super-flock in yellow (Pundamilia spp. and Enterochromis spp.), and Metriaclima zebra of Lake Malawi in grey. Coloured tip extensions within the Lake Chala clade represent the morph assignments (blue, BBE; black, GAL; green, LEA; light green, LEO; brown, OSH; purple, PLB; violet, PLR; yellow, YBE). Bootstrap supports greater than 70% are given. (Online version in colour.)
In our phylogenomic analyses based on approximately 2 million sites, Astatotilapia sp. ‘Chala’ form a monophyletic group (bootstrap = 77%), sister to Astatotilapia from the Pangani drainage and from Lake Babati (figure 1c). Introgression tests (ABBA-BABA) revealed no evidence for introgression either from A. sparsidens of the Lake Manyara basin (which was connected to the Pangani River system in recent geological history), from cichlids of the Lake Victoria region (of the nearest large adaptive radiations), or from Astatotilapia sp. from the Ruaha swamps (which share one of the mitochondrial haplotypes with Astatotilapia sp. ‘Chala’; electronic supplementary material, table S5).
Both mitochondrial haplotypes present in the haplochromines of Lake Chala, and all mitochondrial haplotypes of both other introduced cichlid species of Lake Chala (O. sp. ‘blue head’ and C. rendalli), were shared with members of the same taxa from the upper Pangani River (electronic supplementary material, appendix S13 and figure S4). Therefore, an accidental introduction of Astatotilapia as a bycatch when Oreochromis and Coptodon were introduced from the Pangani River during the early 1970s [22,51] seems the most plausible explanation for the arrival of Astatotilapia in Lake Chala. Supportive evidence for the recent arrival of Astatotilapia in Lake Chala comes from our demographic modelling. Our best models suggest that the Chala population diverged from the Pangani River population approximately 40 generations (approx. 80 years) ago (electronic supplementary material, appendix S14 and figure S3).
(b). Phenotypic and ecological expansion
Comparing the Lake Chala Astatotilapia to its closest known relatives from outside the lake revealed significant differences in eight morphometric traits. While the range of trait values for HW, BD, SnL, IOW, EyD and POW had expanded (albeit not significantly in any one of them alone) and their mean had shifted (p < 0.0015), LJW and EyL showed a significant shift of the mean without a change in variance (electronic supplementary material, figure S5 and table S7). Generally, Astatotilapia in Lake Chala are more narrow (smaller width measurements), less deep bodied, and have larger eyes than their relatives in the Pangani reservoir. Comparisons of multivariate morphospace occupancy suggest a significant increase in morphospace occupation in Lake Chala (electronic supplementary material, figure S6). However, this result, albeit statistically corrected for uneven sample size, should be taken with a note of caution given the low sample size from the Pangani reservoir.
Within Lake Chala, we found several traits with significant phenotype-environment correlations. HW and SnW were significantly correlated with water depth (wider in the shallow water, electronic supplementary material, table S8). LJL was significantly correlated with δ13C (fish with shorter lower jaws relied more on terrestrial carbon sources). EyL and EyD were significantly correlated with δ15N (fish feeding higher in the food web had smaller eyes, electronic supplementary material, table S8).
The PCA on colour traits revealed three distinct clusters in male colour space (figure 2b; electronic supplementary material, figure S10A). Stomach content analyses of 59 individuals revealed considerable individual variation in diet. Most abundant food items were insect larvae (25% of the overall volume), followed by fish fry (21%), detritus (20%), annelids (8%), zooplankton (8%) and fish eggs (6%). The most common prey items in the stomachs of A. cf. bloyeti from the Pangani reservoir were small fishes and insects (electronic supplementary material, figure S9).
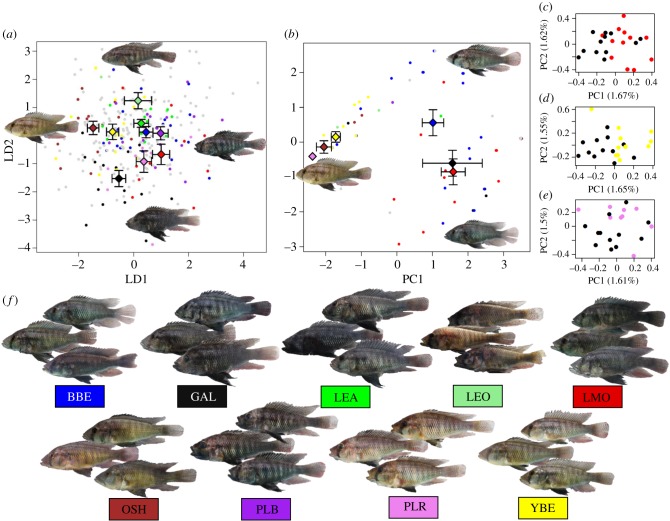
Figure 2.
Individual phenotypic variation within Astatotilapia sp. ‘Chala’ and the visually recognized phenotype groups (morphs). (a) LDA based on 14 linear morphometric traits, (b) PCA based on male nuptial coloration, which revealed three rather distinct clusters (morph means were only plotted for the morphs with more than four males in the colour analyses), (c–e) pairwise genomic PCA for the morph pairs with significant genomic differentiation. Dots represent individuals, whereas diamonds represent morph centroids with standard errors for morphs with more than three individuals for the corresponding analyses (blue, BBE; black, GAL; green, LEA; light green, LEO; brown, OSH; purple, PLB; violet, PLR; yellow, YBE; grey, not assigned). (f ) Images of three individual males representative of the variation within each of the visually recognized phenotype groups. (Online version in colour.)
Stable isotopes revealed large between-individual variation in both δ13C and δ15N within Lake Chala (electronic supplementary material, figure S10). The δ13C values changed along water depth (deep caught fish had more negative δ13C, F = 28.04, p < 0.001, electronic supplementary material, figure S11). The strong correlation between δ13C and water depth suggests that most fish were caught at the depth at which they had been predominantly foraging for the last several months prior to capture (the turnover rate of muscle tissue). The δ15N values ranged from 7.7 to 11.8. They were not correlated with water depth but differed between morphs (F = 2.47, p = 0.013; electronic supplementary material, table S12).
(c). Phenotypic and ecological differentiation
LDA on morphometric traits resulted in significant differences (after correcting for multiple testing) between the different morphs. While in the global LD1-LD2 morphospace spanned by LD1 (HW, LJL) and LD2 (BD, SnW; figure 2a; electronic supplementary material, tables S9, S10, figures 2A, S7A, S8) 15 of 36 pairwise comparisons between morphs were significant (electronic supplementary material, table S9), all pairwise LD-comparisons were highly significant (p < 0.0011, electronic supplementary material, figure S7). The most distinct morph was GAL, which differed significantly from every other morph except PLR even in the global morphospace (electronic supplementary material, table S9).
In the colour space, most morphs fell entirely within one of the three colour clusters (figure 2; electronic supplementary material, appendix S15).
Morphs were non-randomly distributed along the depth gradient (ANOVA: F = 8.47, p < 0.0001, electronic supplementary material, figure S9). The most distinct prey composition was found for PLR and GAL (electronic supplementary material, table S11), the morphs with the most evasive (chironomids and fishes) and the least evasive (diatoms, bryozoans and detritus) prey, respectively (electronic supplementary material, figure S9). Both stable isotopic measures (δ13C and δ15N) differed significantly between the morphs (δ13C: F = 3.17, p = 0.002 and δ15N: F = 2.47, p = 0.013, electronic supplementary material, table S12). ANCOVA analyses further revealed that three morphs (GAL: p = 0.003, LMO: p = 0.005 and PLB: p = 0.006) had δ13C signatures significantly different from the other morphs living in the same depth habitat as any of those three (electronic supplementary material, figure S11), indicating ecological divergence between morphs even within the same depth habitat.
(d). Genomic differentiation
The Astatotilapia of Lake Chala are significantly differentiated genomically from their close relatives in the Pangani reservoir (FST = 0.140). But, neither F-statistics nor Structure analyses detected significant genomic differentiation between morphs (best supported K = 1, electronic supplementary material, figure S12), permutation tests and pairwise PCA revealed significant genomic differentiation between the morph GAL and several other morphs (figure 2c,d; electronic supplementary material, figures S13–S16, table S13). GAL is genomically most distinct among the morphs, albeit pairwise FST are small (FST > 0.006 against 5/7 morphs (electronic supplementary material, table S13)). Neither groups made by the two distinct mitochondrial haplotypes, nor by the three major colour types were genomically differentiated from one another.
(e). Fitness surfaces change along the water depth gradient
We could detect evidence for disruptive selection either on any of the morphological traits using the Lande & Arnold-approach (electronic supplementary material, table S14) or on morphological trait combinations using the canonical analyses (electronic supplementary material, appendix S16 and table S15).
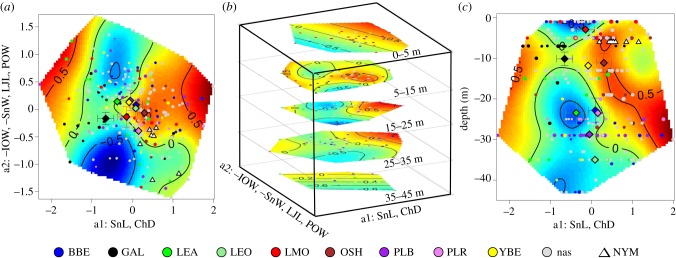
Using the PPR to find the multivariate trait axes explaining most fitness variation, we detected significantly multimodal fitness surfaces on a1 (SnL, ChD; electronic supplementary material, table S16) with two alternative regions of high fitness (subsequently called fitness optima, γ = 0.211, figure 3a; electronic supplementary material, figure S19A and table S17), consistent with disruptive selection. A single significant fitness optimum consistent with stabilizing selection was found on a2 (IOW, SnW, LJL and POW, γ = −0.216, electronic supplementary material, figure S19B). While most individuals cluster around the optimum with positive a1-values, the other optimum is only sparsely occupied (figure 3a). Projecting the A. cf. bloyeti from the Pangani reservoir into the same fitness surface reveals that they share the morphospace around the optimum at positive a1-values with the majority of the Lake Chala individuals (figure 3a). This may hence represent the ancestral fitness optimum.
Figure 3.
Fitness surfaces visualizing (a) the projections from the PPR, showing evidence of two local fitness optima along a1, (b) the differences in the fitness surface at different depths (in 10 m depth intervals), and (c) the change of the fitness surface along the smoothed depth gradient. In the depth range between 5 and 35 m, two fitness optima exist. The position of Astatotilapia from the Pangani Reservoir (=NYM; triangles) indicates that the ancestral Chala phenotype was near the optimum with positive a1 values. The two optima converge at about 10 m depth. At this depth, GAL (black) occupies the otherwise sparsely occupied, derived optimum at negative a1. Dots represent individuals from Lake Chala and diamonds represent morph means with standard errors. The size of the dots represents individual raw growth rate estimates; surface colour reflects smoothed overall fitness estimates. Fish from NYM were not used for the inference of the fitness landscape, but were projected into the phenotype space afterwards. (Online version in colour.)
Calculating fitness surfaces for fish occurring within a certain water depth band revealed that selection on the same morphological trait combinations changes significantly along the depth gradient (figure 3b; electronic supplementary material, figures S20–S22). We found evidence for the two syntopic fitness optima on a1 along most of the depth gradient (5–35 m; individuals with extreme negative a1-values are missing for shallower and deeper parts, figure 3c). These optima were connected by a ridge of rather high fitness (>0.1) at intermediate depth (5–15 m, figure 3b). Interestingly, this coincides with the depth, where the derived optimum at negative a1-values (short snout and small cheek depth) is most densely occupied. Interestingly, most of the fish occupying the optimum at negative a1-values were assigned to the most distinct morph (GAL). A similar pattern, with evidence for relaxed disruptive selection in the intermediate habitat, was revealed if the phenotypic axes were calculated for three different depth categories separately (electronic supplementary material, appendix S17, figure S25 and table S18).
Individuals clustered significantly in high fitness regions in deep water, and tended to cluster in shallow water, but neither in the intermediate depth range where the inferred fitness valley was shallow and narrow, nor in an overall analysis where we pooled fish across depths (electronic supplementary material, figure S23 and table S18).
4. Discussion
Even though many adaptive radiations have been extensively studied, empirical data about the very beginnings of the process are scarce [2,5,12–14]. In this study, we investigated phenotypic, ecological and genomic diversification within a population of Astatotilapia cichlid fish that colonized a deep clear water crater lake in very recent times. We find that within quite likely less than 50 generations after colonization, phenotypic trait means have significantly changed and trait diversity has increased relative to the riverine ancestors, as predicted by theory for the onset of adaptive radiation [2,5]. Most of the observed phenotypic changes are consistent with ecological expansion into the lacustrine deep-water habitat, and with adaptations to new prey items. Moreover, we find evidence for ecological and morphological differentiation between phenotypic groups (morphs) within the lake (figure 2; electronic supplementary material, video S1), at least one of which appears to be genomically differentiated from most others, suggesting that the crater lake population is in an early phase of sympatric differentiation. We observe evidence for multimodal fitness surfaces that change with water depth. Below we discuss each of these aspects in more detail.
(a). Phenotypic change and ecological niche expansion in Lake Chala
In line with the theoretical prediction of phenotypic expansion at the onset of the adaptive radiation process [2,4,5], we found evidence for significant phenotypic change and increased morphological variation within the Astatotilapia of Lake Chala, compared to their relatives in the Pangani reservoir (electronic supplementary material, appendix S18) which most likely represent their ancestral condition. Such phenotypic expansion might occur owing to relaxed stabilizing selection associated with the release from interspecific competition in a new, less species-rich environment and/or owing to the availability of new niches, free of competitors (reviewed in [52]). Both scenarios are likely to apply to Lake Chala. Interspecific competition is likely less severe in Lake Chala than in the Pangani River, because when Astatotilapia were introduced into the lake, at most three other fish species were present, none of which overlaps with Astatotilapia in either morphospace (electronic supplementary material, figure S26) or trophic ecology [20]. Only one of these (the endemic cichlid O. hunteri) was present for sufficiently long time to adapt to general conditions or specific niches in Lake Chala [51]. The other two were introduced probably at the same time as Astatotilapia. In sharp contrast, Astatotilapia in the Pangani River coexist with another ecologically and morphologically similar haplochromine cichlid (Ctenochromis pectoralis), and with a variety of cyprinid species, characids and catfish besides a set of Oreochromis and Coptodon species similar to that in Lake Chala. Because deep, clear and well-oxygenated lakes provide a larger range of ecological niches for cichlid fishes than rivers do [19], Astatotilapia in Lake Chala would simultaneously have gained access to new niches that were not available to the ancestors in the Pangani River.
The most likely direct progenitors of today's haplochromines of Lake Chala, riverine cichlids from the Pangani, were dietary generalists that fed mainly on insect larvae and fish fry. Most of the morphological differences between Astatotilapia from the Pangani versus Lake Chala are consistent with increased ecological opportunity in Lake Chala (see the electronic supplementary material, appendix S19). Within Lake Chala, some adaptation to lacustrine habitats and lacustrine dietary resources (e.g. zooplankton feeding) seems to have occurred rapidly. Phenotypic plasticity, rapid evolution or both may explain this. On the other hand, several feeding modes, such as algivory and molluscivory, which are widespread in full-fledged cichlid radiations are entirely lacking. Similarly, in another case of very recent crater lake colonization by cichlid fishes, in a few hundred years young morph pair of Nicaraguan crater lake cichlids [53], where herbivory (algae, Chara and biofilm) most likely represents the ancestral state, some new items (fishes and insects) are regularly encountered in the stomachs, whereas others (zooplankton and snails) are rare [53]. Whereas we did not see large amounts of filamentous algae in Lake Chala, perhaps owing to the presence of algae grazing Oreochromis [54], gastropods (Melanoides sp.) were observed in high abundance (electronic supplementary material, video S1). This indicates that Astatotilapia of Lake Chala may have difficulties feeding on hard-shelled prey potentially owing to adaptational constraints, e.g. in the ability to express hypertrophied pharyngeal bones and molariform pharyngeal teeth required to crush these snails, or procumbent oral teeth needed to grab the snail by their foot [55,56]. Similarly, the pelagic zone of Lake Chala remained unoccupied by Astatotilapia, even though full-fledged haplochromine cichlid radiations typically have pelagic species. Finally, while the haplochromines of Lake Chala vary in their male nuptial coloration from blue to yellowish with traces of orange colour, red dorsum or red chest morphs common among Lake Victoria cichlids are absent.
(b). Phenotypic and ecological differentiation within Lake Chala
We observed strongest phenotypic divergence along the water depth gradient, a major predictor of within-lake cichlid speciation in older radiations [18,27]: morphology (electronic supplementary material, table S8) as well as trophic ecology (electronic supplementary material, figure S11) varied along the depth gradient, and most morphs occupied restricted depth ranges (electronic supplementary material, figure S8). Such depth-associated morphological variation has also been reported in older crater lake cichlid radiations [27,57]. However, besides depth-related differentiation, we found evidence for significant differentiation between morphs within the shallow habitat in morphological traits that are often associated with different feeding modes (electronic supplementary material, appendix S20), and significant trophic differentiation between morphs within several depth habitats (electronic supplementary material, figure S11). Some of these differences indicate divergence along some well-known ecological axes for pairwise divergence in fishes, such as the littoral/profundal or the benthic/limnetic axis (see the electronic supplementary material, appendix S19, and [27,57–60]) but contrary to ecological speciation in cases of postglacial Northern Hemisphere fishes, we do not observe discrete non-overlapping ecotypes in Lake Chala haplochromines. Rather more continuous but multidimensional differentiation along several ecological axes may signal the nearly simultaneous beginnings of divergence into several eco-phenotypic groups, and perhaps eventually nascent species in haplochromines. Similar to the pattern observed in a slightly older (few hundred generations) crater lake cichlid morph pair in Nicaragua [53], the ecological and phenotypic divergence is not associated with strong genomic differentiation, indicating that phenotypic and ecological divergence can happen faster than the accumulation of genomic differentiation. Interestingly, this is different from cases of similarly rapid recent speciation from hybrid populations demonstrated in Lake Victoria cichlids [61].
(c). Rapid speciation in sympatry?
Potential mechanisms underlying morphological and ecological differentiation of morphs with significant phenotype-environment associations include phenotypic plasticity, adaptive polymorphisms with habitat matching and speciation. Habitat matching and phenotypic plasticity seem plausible explanations for the significant phenotype-environment associations, as we did not detect a direct pattern of disruptive selection along the depth axis. Nevertheless, as we found evidence for phenotypic and dietary differentiation and disruptive selection within the same depth habitat, mechanisms other than plasticity and habitat matching are most likely involved in the evolution of the phenotypic diversity that we observed.
The rapid phenotypic expansion after the invasion of a new habitat supports that successful invasions of new habitats are often associated with ecological versatility and partially plastic responses rather than the usually slower evolutionary changes [62,63]. However, despite the likely contribution of phenotypic plasticity, the fact that the phenotypically and ecologically most distinct morph GAL is also genomically the most distinct morph, points to a genetic basis of at least some of the phenotypic diversification observed in Lake Chala.
(d). Changing fitness surfaces along the depth gradient
Individual fitness variation is often mediated by interactions between phenotype and environment [2,64], but few studies of selection in the wild have investigated how fitness surfaces may change along environmental gradients. The same morphological trait combination that may be beneficial in one habitat can be disadvantageous in another one [65,66]. In this study, we have provided evidence for such changes in fitness along water depth in two ways. First, we showed that fitness optima at different places of the ecological gradient (water depth) are associated with different morphological trait combinations (electronic supplementary material, figure S25). Second, we show that the fitness effects of the same trait combination change along the depth gradient (figure 3; electronic supplementary material, figures S20–S22). Even though the general pattern of two distinct fitness optima persists along most of the depth gradient, the width and the depth of the intervening fitness valley changes. Interestingly, the novel fitness optimum is best occupied in the depth habitat where the fitness valley between the ancestral and the derived morphological optima is the narrowest and shallowest. This is consistent with the hypothesis that variation in the ruggedness of a fitness landscape along a spatial ecological gradient may facilitate the colonization of new peaks [19,67]. This implies that not just parapatric speciation along a habitat gradient, but also sympatric speciation, may be facilitated by occupation of wide sectors of a habitat gradient.
5. Conclusion
Haplochromine cichlids of the riverine species A. cf. bloyeti have colonized the entire, habitable water depth range of Lake Chala probably within only about 50 generations. Compared to the putatively ancestral Pangani River population, the lake population increased its morphospace occupation, most likely driven by the ecological expansion to deeper habitats and additional prey items. Besides phenotypic divergence along the depth gradient, we find evidence for rugged fitness surfaces within depth habitats, potentially facilitating the coexistence of morphologically differentiated morphs in narrow sense sympatry within the same depth habitat. Divergence along the depth gradient in some trait combinations together with differentiation within depth habitats in other trait combinations results in several phenotypic groups that differ in morphology and ecology.
Although, rapid and contemporary incipient speciation into two sympatric ecotypes within a lake has now been demonstrated in a few cases [17,60,68], the rapid diversification along multiple orthogonal ecological and phenotypic axes simultaneously in contemporary time has not been shown before, but is consistent with patterns observed in several thousand years old radiations of Amphilophus crater lake cichlids [53] and in the larger but also very young radiations in the Lake Victoria region [19,26].
Supplementary Material
Supplementary Material
Acknowledgements
We are thankful to the Tanzanian Fisheries Research Institute (TAFIRI), COSTECH and the Lake Chala Safari Lodge for their support during the fieldwork. We thank Mhoja Kayeba, Mohamed Haluna and Jonathan Makoye for their help in the field, Jonas Walker, Petra Nobs, Guy Schnidrig and Ariane LeGros for preparing stable isotope samples, stomach contents and conducting morphological measurements, respectively. We thank the Lausanne Genomic Technologies Facility for conducting the RAD-sequencing, the Genetic Diversity Centre of the ETH Zürich for providing the bioinformatics facilities, the Seehausen group for intensive discussions and two anonymous reviewers for their critical and constructive reviews.
Ethics
This research was done under research permits no. 2013-251-ER-2014-177 (F.N.M.), 2013-256-NA-2014-177 (J.C.v.R.) and 2013-251-NA-2014-177 (O.S.) from the Tanzania Commission for Science and Technology (COSTECH).
Data accessibility
Mitochondrial DNA sequences are available at GenBank (MH644430-MH64538). RADseq reads are available at the NCBI Sequence Read Archive (SAMN09692994-SAMN09693110). Phenotypic data are available in the Dryad Digital Repository: http://doi.org/10.5061/dryad.67v66m9 [69].
Authors' contribution
F.N.M. carried out fieldwork, participated in the design of the study, generated stable isotope, coloration and genomic data, analysed the data, performed demographic modelling and wrote the manuscript together with O.S. J.C.v.R carried out fieldwork, participated in the design of the study and helped drafting the manuscript. S.M. generated mitochondrial sequences and helped in the laboratory. J.I.M. helped analysing the genomic data and demographic models and drafting the manuscript. B.N. carried out fieldwork. O.S. designed and coordinated the study, made the sampling design and wrote the manuscript together with F.M.
Competing interests
We have no competing interests.
Funding
This research was supported by Swiss National Science Foundation grant no. 31003A_144046 to O.S.
References
- 1.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 2.Schluter D. 2000. The ecology of adaptive radiation.
- 3.Stroud JT, Losos JB. 2016. Ecological opportunity and adaptive radiation. Annu. Rev. Ecol. Evol. Syst. 47, 507–532. ( 10.1146/annurev-ecolsys-121415-032254) [DOI] [Google Scholar]
- 4.Simpson GG. 1953. A general discussion of evolutionary radiations that integrates population genetics with paleontology. The major features of evolution New York, NY: Columbia University Press. [Google Scholar]
- 5.Yoder JB, et al. 2010. Ecological opportunity and the origin of adaptive radiations. J. Evol. Biol. 23, 1581–1596. ( 10.1111/j.1420-9101.2010.02029.x) [DOI] [PubMed] [Google Scholar]
- 6.Grant PR, Grant BR. 2008. How and why species multiply: the radiation of Darwin's finches. Princeton, NJ: Princeton University Press. [Google Scholar]
- 7.Carlquist S. 2003. Tarweeds and silverswords: evolution of the Madiinae (Asteraceae). St Louis, MO: Missouri Botanical Garden Press. [Google Scholar]
- 8.Givnish TJ, et al. 2009. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc. R. Soc. B 276, 407–416. ( 10.1098/rspb.2008.1204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seehausen O. 2006. African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B 273, 1987–1998. ( 10.1098/rspb.2006.3539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lerner HRL, Meyer M, James HF, Hofreiter M, Fleischer RC. 2011. Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Curr. Biol. 21, 1838–1844. ( 10.1016/j.cub.2011.09.039) [DOI] [PubMed] [Google Scholar]
- 11.Losos JB. 2009. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Berkeley, CA: University of California Press. [Google Scholar]
- 12.Schluter D, Price T, Mooers AO, Ludwig D. 1997. Likelihood of ancestor states in adaptive radiation. Evolution 51, 1699–1711. ( 10.1111/j.1558-5646.1997.tb05095.x) [DOI] [PubMed] [Google Scholar]
- 13.Rainey PB, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72. ( 10.1038/27900) [DOI] [PubMed] [Google Scholar]
- 14.Kassen R. 2009. Toward a general theory of adaptive radiation insights from microbial experimental evolution. Ann. NY Acad. Sci. 1168, 3–22. ( 10.1111/j.1749-6632.2009.04574.x) [DOI] [PubMed] [Google Scholar]
- 15.Schluter D. 1996. Ecological speciation in postglacial fishes. Phil. Trans. R. Soc. Lond. B 351, 807–814. ( 10.1098/rstb.1996.0075) [DOI] [Google Scholar]
- 16.Hendry AP, Nosil P, Rieseberg LH. 2007. The speed of ecological speciation. Funct. Ecol. 21, 455–464. ( 10.1111/j.1365-2435.2007.01240.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques DA, Lucek K, Meier JI, Mwaiko S, Wagner CE, Excoffier L, Seehausen O. 2016. Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genet. 12, 1005887 ( 10.1371/journal.pgen.1005887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner CE, Harmon LJ, Seehausen O. 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487, 366–369. ( 10.1038/nature11144) [DOI] [PubMed] [Google Scholar]
- 19.Seehausen O. 2015. Process and pattern in cichlid radiations: inferences for understanding unusually high rates of evolutionary diversification. New Phytologist 207, 304–312. ( 10.1111/nph.13450) [DOI] [PubMed] [Google Scholar]
- 20.Moser FN, Van Rijssel JC, Mwaiko S, Ngatunga BP, Seehausen O. Submitted. The origin and future of an endangered crater lake endemic; phylogeography and ecology of Oreochromis hunteri and its invasive relatives. Hydrobiologia. [Google Scholar]
- 21.Lowe RH. 1955. New species of Tilapia (Pisces, Cichlidae) from Lake Jipe and the Pangani River in East Africa. Bull. Br. Mus. 2, 19. [Google Scholar]
- 22.Dadzie S, Haller RD, Trwavas E. 1988. A note on the fishes of Lake Jipe and Lake Chale on the Kenya-Tanzania border J. East Afr. Nat. Hist. Soc. Nat. Mus. Kenya 192, 46–42. [Google Scholar]
- 23.Damste JSS, Ossebaar J, Abbas B, Schouten S, Verschuren D. 2009. Fluxes and distribution of tetraether lipids in an equatorial African lake: constraints on the application of the TEX86 palaeothermometer and BIT index in lacustrine settings. Geochim. Cosmochim. Ac. 73, 4232–4249. ( 10.1016/j.gca.2009.04.022) [DOI] [Google Scholar]
- 24.Genner MJ, Ngatunga BP, Mzighani S, Smith A, Turner GF. 2015. Geographical ancestry of Lake Malawi's cichlid fish diversity. Biol. Lett. 11, 20150232 ( 10.1098/rsbl.2015.0232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzburger W, Mack T, Verheyen E, Meyer A. 2005. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol. Biol. 5, 17 ( 10.1186/1471-2148-5-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier JI, Marques DA, Mwaiko S, Wagner CE, Excoffier L, Seehausen O. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 14363 ( 10.1038/ncomms14363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malinsky M, et al. 2015. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science 350, 1493–1498. ( 10.1126/science.aac9927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 29.Baird NA, et al. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 ( 10.1371/journal.pone.0003376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. ( 10.1093/oxfordjournals.molbev.a026036) [DOI] [PubMed] [Google Scholar]
- 31.Nagl S, Tichy H, Mayer WE, Samonte IE, McAndrew BJ, Klein J. 2001. Classification and phylogenetic relationships of African tilapiine fishes inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 20, 361–374. ( 10.1006/mpev.2001.0979) [DOI] [PubMed] [Google Scholar]
- 32.Kinaro ZO, Xue LY, Volatiana JA. 2016. Complete mitochondrial DNA sequences of the Victoria tilapia (Oreochromis variabilis) and redbelly tilapia (Tilapia zilli): genome characterization and phylogeny analysis. Mitochondr. DNA 27, 2455–2457. ( 10.3109/19401736.2015.1033695) [DOI] [PubMed] [Google Scholar]
- 33.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Excoffier L. 2011. Foll M. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27, 1332–1334. ( 10.1093/bioinformatics/btr124) [DOI] [PubMed] [Google Scholar]
- 35.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. ( 10.1177/117693430500100003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson N, Moorjani P, Luo YT, Mallick S, Rohland N, Zhan YP, Genschoreck T, Webster T, Reich D. 2012. Ancient admixture in human history. Genetics 192, 1065–1093. ( 10.1534/genetics.112.145037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barel CDN, Van Oijen MJP, Witte F, Wittemaas ELM. 1977. Introduction to taxonomy and morphology of haplochromine cichlidae from Lake Victoria: manual to Greenwoods Revision Papers. Neth. J. Zool. 27, 333–380. ( 10.1163/002829677X00199) [DOI] [Google Scholar]
- 38.Witte F. 1984. Ecological differentiation in Lake Victoria haplochromines: comparison of cichlid species flocks in African lakes Evol. Fish Spec. Flocks 155–167.
- 39.R-Core-Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
- 40.Skulason S, Smith TB. 1995. Resource polymorphisms in vertebrates. Trends Ecol. Evol. 10, 366–370. ( 10.1016/S0169-5347(00)89135-1) [DOI] [PubMed] [Google Scholar]
- 41.Besnier F, Glover KA. 2013. ParallelStructure: a R package to distribute parallel runs of the population genetics program STRUCTURE on multi-core computers. PLoS ONE 8, e70651 ( 10.1371/journal.pone.0070651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng XW, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. ( 10.1093/bioinformatics/bts606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rundle HD, Vamosi SM, Schluter D. 2003. Experimental test of predation's effect on divergent selection during character displacement in sticklebacks. Proc. Natl Acad. Sci. USA 100, 14 943–14 948. ( 10.1073/pnas.2036360100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin CH. 2012. Weak disruptive selection and incomplete phenotypic divergence in two classic examples of sympatric speciation: Cameroon crater lake cichlids. Am. Nat. 180, E90–E109. ( 10.1086/667586) [DOI] [PubMed] [Google Scholar]
- 45.Van Rijssel JC, Moser FN, DFrei D, Seehausen O. 2018. Prevalence of disruptive selection predicts extent of species differentiation in Lake Victoria cichlids. Proc. R. Soc. B 285, 20172630 ( 10.1098/rspb.2017.2630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lande R, Arnold SJ. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226. ( 10.1111/j.1558-5646.1983.tb00236.x) [DOI] [PubMed] [Google Scholar]
- 47.Phillips PC, Arnold SJ. 1989. Visualizing multivariate selection. Evolution 43, 1209–1222. ( 10.1111/j.1558-5646.1989.tb02569.x) [DOI] [PubMed] [Google Scholar]
- 48.Cole GL, Endler JA. 2015. Variable environmental effects on a multicomponent sexually selected trait. Am. Nat. 185, 452–468. ( 10.1086/680022) [DOI] [PubMed] [Google Scholar]
- 49.Friedman JH, Stuetzle W. 1981. Projection pursuit regression. J. Am. Stat. Assoc. 76, 817–823. ( 10.1080/01621459.1981.10477729) [DOI] [Google Scholar]
- 50.Nychka D, Furrer R, Sain S. 2015. Fields: tools for spatial data. See http://www.image.ucar.edu/fields.
- 51.Seegers L, De Vos L, Okeyo DO. 2003. Annotated checklist of the freshwater fishes of Kenya (excluding the lacustrine haplochromines from Lake Victoria). J. East Afr. Nat. Hist. 92, 11–47. ( 10.2982/0012-8317(2003)92%5B11:ACOTFF%5D2.0.CO;2) [DOI] [Google Scholar]
- 52.Grant PR. 1971. Evolution on islands. Oxford, UK: Oxford University Press. [Google Scholar]
- 53.Elmer KR, Lehtonen TK, Kautt AF, Harrod C, Meyer A. 2010. Rapid sympatric ecological differentiation of crater lake cichlid fishes within historic times. BMC Biol. 8, 60 ( 10.1186/1741-7007-8-60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trewavas E. 1983. Tilapiine fishes of the genera Sarotherodon, Oreochromis and Danakilia. London, UK: Brit. Mus. (Nat. Hist.). [Google Scholar]
- 55.Greenwood PH. 1979. Towards a phyletic classification of the ‘genus’ Haplocromis (Pisces, Cichlidae) and related taxa. Bull. Br. Mus. 35, 265–322. ( 10.5962/bhl.part.20455) [DOI] [Google Scholar]
- 56.Witte F, Van Oijen MJP. 1990. Taxonomy, ecology and fishery of Lake Victoria haplochromine trophic groups. Zoologische Verhandelingen. 262, 20–24. [Google Scholar]
- 57.Recknagel H, Elmer KR, Meyer A. 2014. Crater lake habitat predicts morphological diversity in adaptive radiations of cichlid fishes. Evolution 68, 2145–2155. ( 10.1111/evo.12412) [DOI] [PubMed] [Google Scholar]
- 58.Mcphail JD. 1984. Ecology and evolution of sympatric sticklebacks (gasterosteus): morphological and genetic-evidence for a species pair in Enos Lake, British Columbia. Can. J. Zool. 62, 1402–1408. ( 10.1139/z84-201) [DOI] [Google Scholar]
- 59.Ostbye K, Amundsen PA, Bernatchez L, Klemetsen A, Knudsen R, Kristoffersen R, Naesje TF, Hindar K. 2006. Parallel evolution of ecomorphological traits in the European whitefish Coregonus lavaretus (L.) species complex during postglacial times. Mol. Ecol. 15, 3983–4001. ( 10.1111/j.1365-294X.2006.03062.x) [DOI] [PubMed] [Google Scholar]
- 60.Kautt AF, Machado-Schiaffino G, Torres-Dowdall J, Meyer A. 2016. Incipient sympatric speciation in Midas cichlid fish from the youngest and one of the smallest crater lakes in Nicaragua due to differential use of the benthic and limnetic habitats? Ecol. Evol. 6, 5342–5357. ( 10.1002/ece3.2287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meier JI, Marques DA, Wagner CE, Excoffier L, Seehausen O. 2018. Genomics of parallel ecological speciation in Lake Victoria cichlids. Mol. Biol. Evol. 35, 1489–1506. ( 10.1093/molbev/msy051) [DOI] [PubMed] [Google Scholar]
- 62.Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP. 2010. Phenotypic plasticity's impacts on diversification and speciation. Trends Ecol. Evol. 25, 459–467. ( 10.1016/j.tree.2010.05.006) [DOI] [PubMed] [Google Scholar]
- 63.Baldwin MJ. 1896. A new factor in evolution. Amer. Soc. Nat. 30, 10. [Google Scholar]
- 64.Laughlin DC, Messier J. 2015. Fitness of multidimensional phenotypes in dynamic adaptive landscapes. Trends Ecol. Evol. 30, 487–496. ( 10.1016/j.tree.2015.06.003) [DOI] [PubMed] [Google Scholar]
- 65.Karino K, Orita K, Sato A. 2006. Long tails affect swimming performance and habitat choice in the male guppy. Zool. Sci. 23, 255–260. ( 10.2108/zsj.23.255) [DOI] [PubMed] [Google Scholar]
- 66.Calsbeek R, Irschick DJ. 2007. The quick and the dead: correlational selection on morphology, performance, and habitat use in island lizards. Evolution 61, 2493–2503. ( 10.1111/j.1558-5646.2007.00206.x) [DOI] [PubMed] [Google Scholar]
- 67.Steinberg B, Ostermeier M. 2016. Environmental changes bridge evolutionary valleys. Sci. Adv. 2, e1500921 ( 10.1126/sciadv.1500921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meier JI, Sousa VC, Marques DA, Selz OM, Wagner CE, Excoffier L, Seehausen O. 2017. Demographic modelling with whole-genome data reveals parallel origin of similar Pundamilia cichlid species after hybridization. Mol. Ecol. 26, 123–141. ( 10.1111/mec.13838) [DOI] [PubMed] [Google Scholar]
- 69.Moser FN, van Rijssel JC, Mwaiko S, Meier JI, Ngatunga B, Sechausen O. 2018. Data from: The onset of ecological diversification 50 years after colonization of a crater lake by haplochromine cichlid fishes Dryad Digital Repository. ( 10.5061/dryad.67v66m9) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Moser FN, van Rijssel JC, Mwaiko S, Meier JI, Ngatunga B, Sechausen O. 2018. Data from: The onset of ecological diversification 50 years after colonization of a crater lake by haplochromine cichlid fishes Dryad Digital Repository. ( 10.5061/dryad.67v66m9) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Mitochondrial DNA sequences are available at GenBank (MH644430-MH64538). RADseq reads are available at the NCBI Sequence Read Archive (SAMN09692994-SAMN09693110). Phenotypic data are available in the Dryad Digital Repository: http://doi.org/10.5061/dryad.67v66m9 [69].