Abstract
Venom proteins evolve rapidly, and as a trophic adaptation are excellent models for predator–prey evolutionary studies. The key to a deeper understanding of venom evolution is an integrated approach, combining prey assays with analysis of venom gene expression and venom phenotype. Here, we use such an approach to study venom evolution in the Amazon puffing snake, Spilotes sulphureus, a generalist feeder. We identify two novel three-finger toxins: sulditoxin and sulmotoxin 1. These new toxins are not only two of the most abundant venom proteins, but are also functionally intriguing, displaying distinct prey-specific toxicities. Sulditoxin is highly toxic towards lizard prey, but is non-toxic towards mammalian prey, even at greater than 22-fold higher dosage. By contrast, sulmotoxin 1 exhibits the reverse trend. Furthermore, evolutionary analysis and structural modelling show highest sequence variability in the central loop of these proteins, probably driving taxon-specific toxicity. This is, to our knowledge, the first case in which a bimodal and contrasting pattern of toxicity has been shown for proteins in the venom of a single snake in relation to diet. Our study is an example of how toxin gene neofunctionalization can result in a venom system dominated by one protein superfamily and still exhibit flexibility in prey capture efficacy.
Keywords: adaptation, gene duplication, proteome, structure-function, taxon-specific toxin, transcriptome
1. Introduction
Snake venoms are composed of numerous proteins and peptides as a result of extensive gene duplication and neofunctionalization events [1–4]. The evolutionary advantage of this can be twofold: individual toxin genes can evolve alternate functionalities through sequence substitutions without incurring negative selection [5], and duplicate gene copies may evolve and adapt transcriptionally, via differential expression of each copy [6,7]. However, venom proteins are also rapidly evolving on a molecular scale, with many toxin superfamilies experiencing positive selection [8,9] and commonly exhibiting nucleotide substitution rates at a higher frequency in the exons rather than the intron regions, especially in those exons that code for interactive surface residues [4,10–13]. This evolutionary process results in large venom protein superfamilies with toxins that could potentially and actually perform very different biological activities.
Three-finger toxins (3FTxs), a non-enzymatic superfamily found in many snake venoms, are a prime example of toxin functional diversification. Although 3FTxs share a common structural scaffold consisting of three β-stranded ‘fingers’ or loops stabilized by four conserved disulfide bridges, they exhibit a large diversity of biological activities, including neurotoxic, cardiotoxic, cytotoxic, haemorrhagic or necrotic effects [14]. In front-fanged Elapidae (cobras, mambas, kraits, etc.) venoms, prominent 3FTxs cause neurotoxicity and respiratory paralysis that can lead to rapid death [15]. 3FTxs have also been identified in several rear-fanged venomous snakes, but they have typically not been found to impart lethal mammalian toxicity [16–19]. For example, two Boiga sp. (cat snakes) and Oxybelis fulgidus (green vinesnake) secrete 3FTxs that are highly lethal to lizards and/or birds, their primary prey, but non-lethal towards mammals [20–22]. By contrast, the most abundant 3FTx (α-cobratoxin) in the venom of the front-fanged monocled cobra (Naja kaouthia) is equally toxic towards both endothermic and ectothermic prey [23]. To date, only the venoms of rear-fanged snakes are known to contain 3FTxs with prey-specific effects, but it is unknown how widely-occurring this phenomenon is among venom-producing snakes within Colubroidea. However, individual toxins that are prey-specific towards lizards/birds or towards mammals are not known to co-occur in the venom of a single species of rear-fanged or front-fanged snake. It is hypothesized that prey-specific toxins may co-occur, as some species use many prey types, commonly showing an ontogenetic shift in prey preference, so a two-pronged arsenal should be an adaptive trait in these instances.
Despite the fact that rear-fanged snake venoms may contain novel toxins, front-fanged snake families Elapidae and Viperidae have remained the primary focus of venom research, as they cause large-scale human morbidity and mortality [24,25]. While snakebite from several species of rear-fanged snakes (e.g. Dispholidus typus and Thelotornis sp.) have caused human fatalities [19,26,27], most rear-fanged venomous snakes rarely produce systemic envenomation in humans. This lack of serious effects explains to a large extent why venoms from most rear-fanged snake species remain unstudied [28–30]. Further, the presence of novel venom protein families in these snakes, coupled with the lack of relevant toxin sequences in public databases, present major hurdles for characterizing rear-fanged snake venoms using proteomic methods [30–33]. However, the combined use of venom gland transcriptomics and venom proteomic analysis now allows researchers to explore rear-fanged snake venom by generating a custom transcriptome database for venom peptide identification [27,34,35].
To determine if multiple novel prey-specific toxins exist within the venom of a single rear-fanged snake species, a combined transcriptomic and proteomic approach was used to characterize the currently unstudied venom from Spilotes sulphureus (formerly Pseustes sulphureus; Amazon puffing snake), a neotropical rear-fanged species [36]. Though considered by Taub (1967) to lack a Duvernoy's venom gland [37], it does in fact possess an elongate maxilla with three enlarged rear teeth, and a large Duvernoy's venom gland, distinct from the supralabial salivary gland (see the electronic supplementary material, figure S7a–e), with a size and location very similar to that of the well-characterized brown treesnake, Boiga irregularis ([38]; see also the electronic supplementary material, figure S7f). These snakes are semi-arboreal and feed on mammals, birds and lizards, and they do not appear to use constriction, either in the laboratory or the field [39]. They are among the largest of the advanced colubroid snakes, reaching lengths of up to 2.7 m [40]. This species has become popular in the herpetoculture trade, but nothing is known about their venom composition or production; because of their large size, they have the potential for producing high venom yields that could result in serious envenomations. We describe here the results of transcriptomic, proteomic and functional assays of S. sulphureus venom, and we uncover a venom rich in 3FTxs, several of which show differential toxicity towards specific prey types. Our study shows that S. sulphureus venom production represents a venom system of toxin gene duplication and neofunctionalization which, while dominated by one venom protein superfamily (3FTxs), exhibits an unparalleled capacity to target different prey specifically using taxon-specific chemical weapons. As a trophic adaptation, venom has the potential to provide insights into unique predator and prey evolutionary dynamics, and the venom of S. sulphureus provides a prime example of minimizing overall compositional complexity while retaining broad predatory efficacy.
2. Material and methods
(a). Rear-fanged snake venom extraction
Venom was manually extracted from all rear-fanged snakes used in this study. Snakes were maintained in the University of Northern Colorado Animal Resource Facility in accordance with UNC-IACUC protocol no. 9204 (refer to the electronic supplementary material, table S1 for snake localities). Three Amazon puffing snakes (S. sulphureus) originating from Suriname, South America were purchased from Bushmaster Reptiles. All specimens were adults, weighing over 500 g and measuring over 1500 mm snout-to-vent length. Venom was manually extracted from each individual after subcutaneous injections of ketamine-HCl (20–30 mg kg−1) followed by pilocarpine-HCl (6 mg kg−1) [41]. Venom was centrifuged at 9500 x g for 5 min, frozen at −80°C, lyophilized, and stored at −20°C until use.
(b). Toxin gene expression
Venom glands were dissected from one S. sulphureus within several hours of death from natural causes and added to TRIzol reagent (Thermo Fisher Scientific, USA) for immediate RNA isolation. To characterize toxin gene expression in the venom gland of S. sulphureus, RNA-seq was performed and the S. sulphureus venom gland transcriptome assembled de novo. Toxin sequences were identified and individual toxin transcript abundances determined (see the electronic supplementary material for RNA-seq, transcriptome assembly, annotation and quantification details).
(c). Venom phenotype
A complete venom compositional profile was generated for S. sulphureus with one-dimensional sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), size exclusion high performance liquid chromatography (SEC HPLC), and matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed on trypsin digested venom and individual purified toxins for protein identification using the translated venom gland transcriptome dataset as a custom reference, in addition to vertebrate protein databases. Proteomic analyses were completed to both characterize S. sulphureus venom and individual toxins, including determining protein subunit composition (see the electronic supplementary material for complete proteomic methodology). Biochemical assays for common venom enzymes were also performed (see the electronic supplementary material).
(d). Characterization of prey-specific toxins
Crude S. sulphureus venom toxicity and the toxicity of the three most abundant toxins observed within S. sulphureus venom were evaluated with LD50 assays using a non-model species, house gecko (Hemidactylus frenatus), and a standard model species, NSA mouse (Mus musculus). House geckos were obtained from Bushmaster Reptiles and NSA mice were bred in the UNC Animal Resource Facility (UNC IACUC protocol no. 9401). All doses (in sterile 0.9% saline) were injected intraperitoneally and three animals per dose were used. Three mice and three lizards were injected with only 0.9% saline as controls. Doses were adjusted to individual animal body masses, with lethality expressed as µg venom or toxin per gram body mass producing 50% mortality after 24 h [42]. The nonparametric Spearman-Kärber method was used for LD50 value estimations and determination of 95% confidence intervals [43]. All procedures were reviewed and approved by the UNC IACUC (protocol 1504D-SM-SMLBirds-18).
(e). Prey-specific toxin evolution in rear-fanged venomous snakes
Prey-specific monomeric 3FTxs from O. fulgidus (C0HJD3), Boiga dendrophila (Q06ZW0) and sulmotoxin 1 from S. sulphureus (this study) were submitted to the Phyre2 server [44] for modelling based on homology detection methods using server default parameters. To determine 3FTx residues available for binding, modelling was restricted to monomeric 3FTxs because the dimerization between the sulditoxin 3FTx subunits might result in different modes of receptor binding. These three models were overlain in UCSF Chimera (http://www.rbvi.ucsf.edu/chimera), and amino acids forming the central structural loop were visualized.
Nucleotide and amino acid sequences belonging to non-conventional 3FTxs were aligned, and model selection performed for tree construction (see the electronic supplementary material for additional details). The K80 model [45] was selected by Akaike information criterion (AIC). A maximum-likelihood tree incorporating the model was constructed in PhyML 3.1 [46], with 1000 bootstrap replicates for node support evaluation.
3. Results
(a). Toxin gene expression
Among the top 1000 expressed contigs, many highly expressed contigs were found to be venom toxins (figure 1a). The venom gland transcriptome assemblies produced 37 full-length coding DNA sequences (CDS) that were identified as venom proteins (figure 1b). These were from thirteen toxin superfamilies: 3FTxs, a cysteine rich secretory protein (CRiSP), C-type lectins (CTLs), l-amino acid oxidases (LAAOs), ficolins, a snake venom metalloproteinase (SVMP), a cobra venom factor (VF), a phospholipase B (PLB), a 5′ nucleotidase (NUC), a venom endothelial growth factor (VEGF), a Kunitz-type protease inhibitor (KUN), phospholipase inhibitors, and metalloproteinase inhibitors.
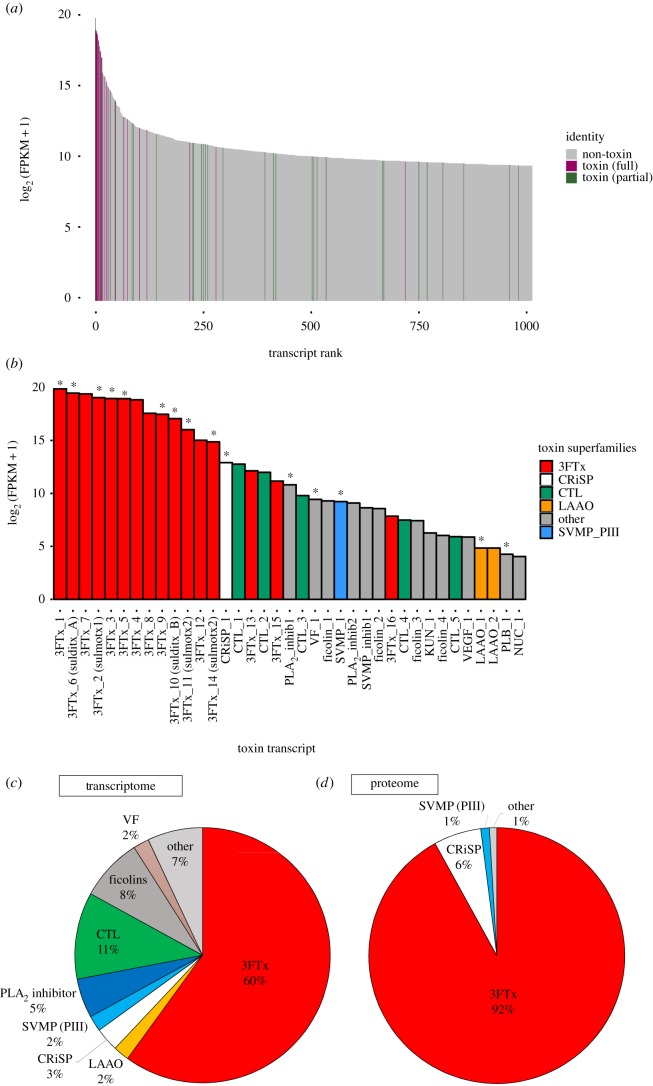
Figure 1.
Venom gland transcriptome and venom proteome abundance profiles for Spilotes sulphureus. Toxin contigs are expressed at high levels (a), with three-finger toxins exhibiting the highest levels of toxin gene expression (b). Toxin transcripts for venom proteins detected within the venom via LC-MS/MS are indicated with an asterisk (*). Abundances for transcripts belonging to each venom protein superfamily are shown (c) with superfamilies making up less than 1% combined to form the ‘other’ category. Relative abundances of each superfamily within the venom proteome are also shown (d). Abbreviations: 3FTx = three-finger toxin, CRiSP = cysteine-rich secretory protein, CTL = C-type lectin, KUN = Kunitz-type protease inhibitor, LAAO = l-amino acid oxidase, PLA2_inhib = phospholipase A2 inhibitor, PLB = phospholipase B, SVMP = snake venom metalloproteinase (PIII), SVMP_inhib = snake venom metalloproteinase inhibitor, NUC = 5′ nucleotidase, VEGF = venom endothelial growth factor, and VF = cobra venom factor. (Online version in colour.)
Of the full-length venom protein sequences, the most abundantly expressed toxin superfamily in the S. sulphureus venom gland was 3FTx (60% of total toxin reads), followed by CTLs (11%) and ficolins (8%) (figure 1c). In addition to being highly expressed, these three superfamilies also displayed the greatest diversity of isoforms, with 16 isoforms found among 3FTxs, 5 CTL isoforms and four ficolin-like isoforms (electronic supplementary material, table S2). The 3FTx isoforms were found to be highly diverse, with sequence similarities ranging from 20.7% to 91.9%.
All toxin transcript sequences were submitted to GenBank and are available at NCBI under Bioproject PRJNA448354 and accession numbers MH232964–MH233012.
(b). Venom phenotype
A venom proteome profile was constructed using a combination of SDS-PAGE, MALDI-TOF MS, SEC HPLC, and LC-MS/MS in order to determine the actively secreted venom phenotype of S. sulphureus. Reduced SDS-PAGE and MALDI-TOF MS demonstrated a high abundance of low molecular weight proteins (6000–9000 Da), within the mass range of 3FTxs observed in rear-fanged venomous snakes [20–22,34,35] (see the electronic supplementary material, results and figures S1 and S2). When using the S. sulphureus venom gland toxin translated transcriptome as a custom reference, seven venom protein families were identified with LC-MS/MS: the most abundant included 3FTxs (92%), CRiSPs (6%) and PIII SVMPs (1%), and all other toxins (cobra venom factor, LAAO, phospholipase A2 (PLA2) inhibitor and PLB) comprised less than 1% of the total toxin normalized spectra (figure 1d). Of the 37 full-length venom proteins that were derived from the venom gland transcriptome, peptides from LC-MS/MS mapped to 15 of these toxins (figure 1b, noted by an asterisk).
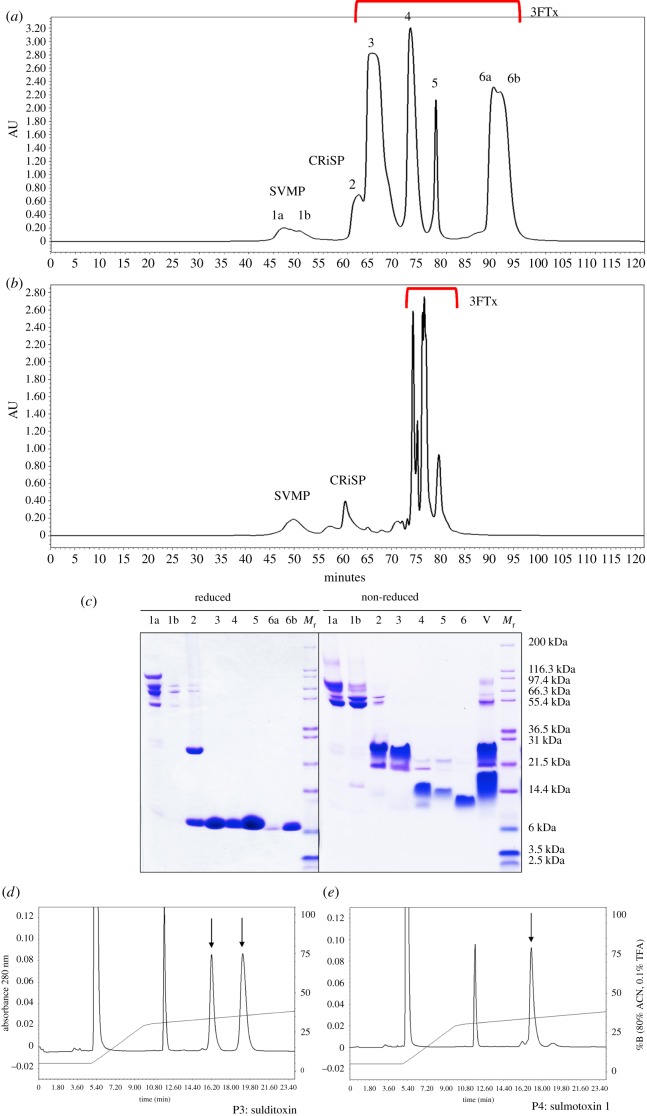
Size exclusion chromatography was used to profile crude S. sulphureus venom and to purify venom proteins for identification and biological assays, resulting in the resolution of six peaks (figure 2a). A size exclusion profile for B. irregularis venom, known to contain an abundance of 3FTxs and reduced amounts of larger toxins [47], is shown for comparison (figure 2b), and reduced and non-reduced SDS-PAGE profiles are shown for S. sulphureus peaks (figure 2c). The most abundant peaks (3–6) comprised approximately 91% of the total peak area and only contained proteins of 6000–8000 Da when reduced (figure 2c).
Figure 2.
Size exclusion venom profiles for Spilotes sulphureus and Boiga irregularis, with SDS-PAGE results of S. sulphureus peaks. Crude venoms (3 mg) from S. sulphureus (a) and B. irregularis (b) were fractionated using size exclusion HPLC; protein identities are based on SDS-PAGE results, N-terminal sequencing, LC-MS/MS, and enzymatic assays. Reduced and non-reduced SDS-PAGE gels are shown for the S. sulphureus chromatogram (c); crude venom (V) was also run under non-reduced conditions. HPLC chromatograms for reduced sulditoxin (d, arrows) and reduced sulmotoxin 1 (e, arrow) show the presence of two and one peaks, respectively; other peaks (approx. 5.4 & 12 min, unlabelled) are reduction chemistry peaks. Abbreviations: Mr = relative molecular masses, 3FTx = three-finger toxin, CRiSP = cysteine-rich secretory protein, and SVMP = snake venom metalloproteinase. (Online version in colour.)
Peaks 3, 4 and 6 (figure 2a) were subjected to reversed phase high pressure liquid chromatography (RP-HPLC), and each produced a single purified protein product (electronic supplementary material, figure S3). Peak 3 had a MALDI-TOF mass of 17 800 Da, peak 4 a mass of 8179.1, and peak 6a, the most abundant isoform of peak 6, a mass of 7579.9 (electronic supplementary material, figure S4). Purified peaks were also digested with trypsin and subjected to LC-MS/MS analysis. Transcripts from the S. sulphureus venom gland transcriptome were used as a reference to identify these venom proteins, and peptide spectra from peaks 3, 4, and 6 all matched with high probability to 3FTx sequences (greater than 99.9%). Using LC-MS/MS spectra in combination with the S. sulphureus transcriptome reference and MALDI-TOF masses of each purified 3FTxs (electronic supplementary material, figure S4), transcripts corresponding to each 3FTx were positively identified, although two transcripts (3FTx11 and 14) were possible for peak 6 (differing by one residue) (figure 3).
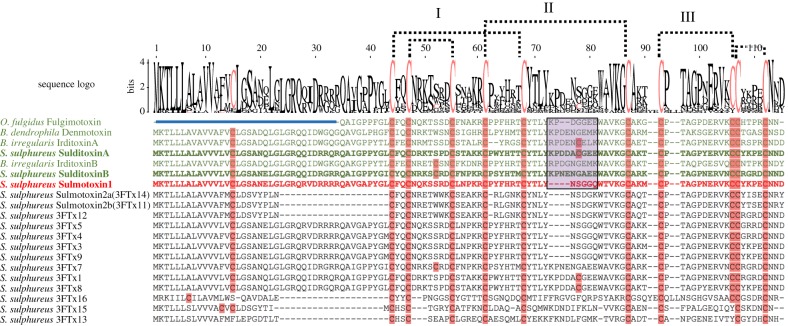
Figure 3.
Amino acid sequence alignment of all Spilotes sulphureus three-finger toxin (3FTx) isoforms and other known prey-specific 3FTxs. Characterized lizard-specific 3FTxs are shown in green, and sulmotoxin 1, a mammal-specific 3FTx, is in red. All prey-specific 3FTx sequences from this study are bolded. Sulmotoxin 2 is labelled twice because of the presence of either of these isoforms within RP-HPLC peak 6. Cysteines are highlighted in pink. The five disulfides characteristic of non-conventional 3FTxs are shown with dotted lines. The numbers above the alignments note the arrangement of the three 3FTx structural loops. Amino acid variation observed within the end of the central structural loop is highlighted in purple for characterized toxins. The signal peptides are indicated by the blue bar above all sequences except fulgimotoxin. (Online version in colour.)
Peaks 3 and 4 were reduced, alkylated and then subjected to RP-HPLC to evaluate potential subunit composition (figure 2d,e). Peak 3 showed two peaks for the reduced and alkylated protein and peak 4 showed only one, demonstrating that peak 3 is a 3FTx heterodimer complex and peak 4 is a monomer. For peak 3, the 3FTx complex, reduced and alkylated subunits A and B had masses of 9338.7 Da and 9222.6 Da, respectively (electronic supplementary material, figure S5). The protein from peak 3 is only the second heterodimeric 3FTx complex that has been described from a rear-fanged snake venom and we have named it sulditoxin, derived from S. sulphureus dimeric toxin. The monomers (peaks 4 and 6) are named sulmotoxin 1 and 2, from S. sulphureus monomeric toxins. Two transcripts have been labelled sulmotoxin 2 because both are equally likely to belong to the peak 6 protein; they differ by only one amino acid residue in loop 3.
For crude S. sulphureus venom, no detectable enzyme activity was present for many common venom enzymatic families: LAAO, acetylcholinesterase, phosphodiesterase (PDE), PLA2, thrombin-like or kallikrein-like snake venom serine proteinases. SVMP activity was detectable, but at relativity low levels (0.150 ± 0.014 units; U = ΔA342 nm min−1 mg−1 venom protein). These results demonstrate that S. sulphureus crude venom largely lacks enzymatic activity and larger proteolytic proteins that are observed in many other snake venoms [38]. Instead this venom is heavily dominated by 3FTxs.
Despite the low levels of enzymatic activity, crude S. sulphureus venom was lethally toxic towards both H. frenatus (house geckos) and M. musculus (NSA mice), with considerably greater toxicity towards geckos than mice (table 1). The murine LD50 value for S. sulphureus venom (2.56 µg g−1) was similar to values reported for moderately toxic rear-fanged venomous snakes, such as Alsophis portoricensis [48], although A. portoricensis venom was not nearly as toxic towards lizards as S. sulphureus venom. The higher toxicity of S. sulphureus venom towards geckos (1.01 µg g−1) strongly suggests the presence of prey-specific toxins, as observed in several other rear-fanged snake venoms [20–22].
Table 1.
Intraperitoneal lethal toxicity (LD50) of Spilotes sulphureus venom and purified three-finger toxins. (95% confidence intervals are listed in parentheses (lower, upper), determined by Spearman-Kärber methodology.)
| mice (Mus musculus) | house geckos (Hemidactylus frenatus) | |
|---|---|---|
| crude venom (Spilotes sulphureus) | 2.56 (2.51, 2.61) µg g−1 | 1.01 (0.78, 1.24) µg g−1 |
| sulditoxin | nontoxic >5 µg g−1 |
0.22 (−0.09, 0.53) µg g−1 |
| sulmotoxin 1 | 4.0 (3.7, 4.3) µg g−1 | nontoxic >5 µg g−1 |
| sulmotoxin 2 | nontoxic >5 µg g−1 |
nontoxic >5 µg g−1 |
(c). Characterization of prey-specific toxins
Lethal toxicity (LD50) assays were performed with sulditoxin and with sulmotoxins 1 and 2 using house geckos and NSA mice (table 1). Sulditoxin was highly toxic to lizards (LD50 = 0.22 µg g−1 in house geckos) and was non-toxic in mammals (LD50 >> 5 µg g−1 for mice). Sulmotoxin 1 exhibited the opposite trend, with a higher LD50 for mice (approx. 4 µg g−1) and no toxicity towards house geckos (greater than 5 µg g−1) (table 1). Sulmotoxin 2 was non-toxic (greater than 5 µg g−1) towards both geckos and mice. These results demonstrate that the two most abundant 3FTxs in S. sulphureus venom display prey-specific toxicity, with sulditoxin exhibiting lizard-specific toxicity and sulmotoxin 1 showing mammal-specific toxicity.
(d). Prey-specific toxin evolution in rear-fanged venomous snakes
Sulditoxin and sulmotoxins exhibit similar 3FTx structural scaffolds, and the majority of S. sulphureus 3FTx isoforms also contain 10 conserved cysteines, a characteristic feature of both non-conventional and other prey-specific 3FTxs (figure 3) [4,20–22,49]. The greatest amino acid residue variability between 3FTxs occurs in the central/second loop (figure 3 and electronic supplementary material, figure S6). However, sulditoxin A shares only 66% sequence identity with irditoxin A, and sulditoxin B shares only 64% identity with irditoxin B. Interestingly, sulditoxin shares the same additional cysteine residues in the first and central loops (figure 3) that are involved in the intersubunit disulfide bond for irditoxin, and these conserved cysteines probably also link the sulditoxin subunits, forming the heterodimeric complex (electronic supplementary material, figure S6A).
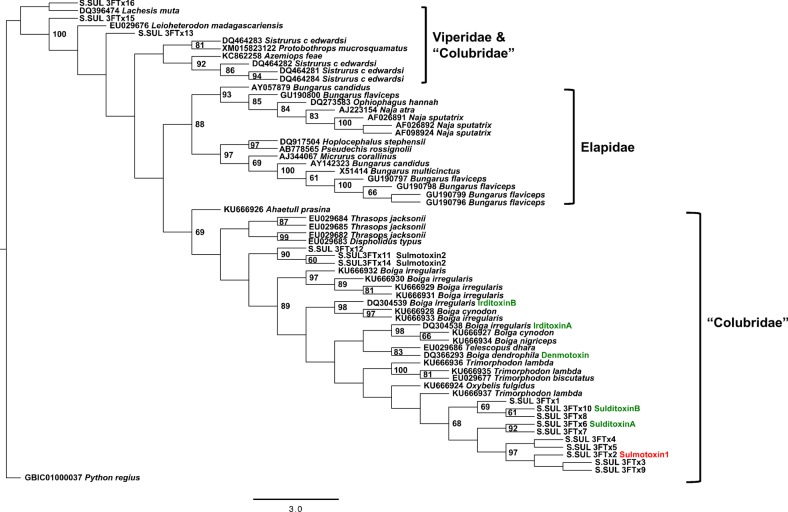
A maximum-likelihood tree constructed with non-conventional 3FTx nucleotide sequences from rear-fanged venomous snakes and 3FTxs from the front-fanged families Viperidae and Elapidae revealed extensive diversification of 3FTxs within rear-fanged venomous snakes (figure 4). The S. sulphureus 3FTx genes demonstrated species-level neofunctionalization, with both lizard- and mammal-specific 3FTxs clustering together, separate from the lizard-specific 3FTxs of Boiga, in spite of shared features such as the conserved intersubunit disulfide linkage forming the heterodimeric complex for both irditoxin and sulditoxin.
Figure 4.
Non-conventional three-finger toxin (3FTx) maximum-likelihood tree showing transcript sequence divergence between the three venomous snake families (Viperidae, Elapidae and ‘Colubridae’). Prey-specific 3FTxs are coloured green for lizard-specific and red for mammal-specific. Only bootstrap node support ≥60 are displayed. (Online version in colour.)
4. Discussion
Venoms are a complex trophic adaptation used by many snakes to facilitate feeding by causing prey quiescence, immobilization, death and/or tissue degradation [38]. Spilotes sulphureus, like many rear-fanged snakes, produces a venom with homologues of the toxins from venoms of front-fanged snakes, and its venom is clearly lethal to both lizards and rodents. Unexplored venoms and venom gland transcriptomes from rear-fanged snakes are probably rich sources in which to discover functional diversification of toxin proteins. We find that the venom of S. sulphureus is unique, as two distinct toxins that target very different prey taxa have evolved in this species. This is, to our knowledge, the first instance in which this dual phenomenon has been discovered in any snake species.
Venom gene expression within the venom gland of S. sulphureus is similar to the 3FTx-dominated elapid-like venom gland transcriptome of the rear-fanged brown treesnake B. irregularis [34,35]. However, the presence of many fewer 3FTx transcript isoforms in S. sulphureus (16) compared to B. irregularis (65) [35] indicates that gene duplication is much less extensive and that S. sulphureus venom composition is considerably less complex, and this is also corroborated by the presence of only five 3FTxs highly expressed in the venom. Fifteen of the 37 full-length venom proteins in S. sulphureus were detected via LC-MS/MS as translated proteins in crude venom, demonstrating that post-transcriptional regulation of specific isoforms within a single venom protein superfamily is occurring, especially in the case of 3FTxs.
Of these 3FTxs, only two showed moderate to high protein abundance: sulditoxin, a lizard-specific heterodimeric 3FTx complex, and sulmotoxin 1, a mammal-specific monomeric 3FTx. Sulditoxin is a lizard-specific toxin that is highly toxic towards house geckos, with a low LD50 (0.22 µg g−1). Strikingly however, we found that even at a greater than 22-fold increase in dosage, it is non-toxic towards inbred NSA mice. On the other hand, sulmotoxin 1 exhibits the opposite trend, showing higher toxicity towards mice, whereas it is non-toxic towards house geckos. These results represent a classic example of adaptation via venom gene duplication and neofunctionalization within a toxin superfamily, allowing for broader prey use.
In a previous study focused on the monomeric lizard-specific 3FTx fulgimotoxin, the amino acid sequence motifs WAVK and CYTLY in the 3FTx loop II were found to be conserved in all lizard/bird-specific 3FTxs and were identified as possible receptor binding sites involved in prey-specific toxicity [22]. Sulditoxin, which we found to be lizard-specific in this study, also shares these WAVK and CYTLY motifs in the 3FTx loop II of both subunits, supporting this hypothesis. In contrast to this, the mammal-specific sulmotoxin 1 has the sequence WTVK (A → T substitution) and CYNLY (T → N substitution). These findings are a classic illustration of sequence-based adaptive evolution of a venom gene family (3FTx) which has occurred directly in the genome of the snake; previously, adaptive venom evolution has been reported to occur via post-genomic mechanisms such as differential regulation of expression of venom genes and venom proteins [6,7,50]. Here it is observed that amino acid substitutions, products from variations in gene sequences, have resulted in the evolution of 3FTxs that display distinct toxicities. It is interesting to note that all lizard-specific 3FTxs also have a longer second loop owing to the presence of at least two (fulgimotoxin) or four (denmotoxin, irditoxin, and sulditoxin) additional amino acids. This longer central loop, combined with an additional proline followed by a negatively charged amino acid (either aspartic acid or glutamic acid, or both), could also result in an altered structure that facilitates binding with lizard/bird nicotinic acetylcholine receptors (electronic supplementary material, figure S6b).
It appears that the mammalian-specific toxicity is a more recent state, based on gene tree topology (figure 4) and multiple sequence indels. Deletions in 3FTx second loop result in a reduced number of amino acids in this region (14 amino acids), and this is only seen for sulmotoxin 1 (mammalian specific), sulmotoxin 2A and 2b, and several other isoforms (transcript-based). Having at least 16 amino acids, and usually 18, in this central loop, as is seen in the lizard-specific 3FTxs, is a shared feature with all other nonconventional 3FTxs from rear-fanged snake species, and probably it is more similar to the ancestral state. In addition, the mammal-specific toxin is not as potent as lizard-specific 3FTxs, supporting the idea that perhaps a 3FTx with more moderate toxicity has not been under selection as long as the development of lizard-specific toxicity, which also appears to have evolved convergently in Boiga [21]. However, it is very difficult to perform an ancestral state reconstruction or determine time scales confidently because critical data are missing for rear-fanged snakes. It is also difficult to perform proper selection analyses because with multiple gene duplications in species, it is problematic to identify orthologous genes.
The gene tree composed of non-conventional 3FTxs from Viperidae, Elapidae and ‘Colubridae’ demonstrated diversification of 3FTxs within rear-fanged venomous snakes and neofunctionalization of these genes within species in which prey-specific toxins have evolved. It is likely that in the genus Boiga, lizard/bird-specific heterodimeric 3FTxs evolved once, and these novel toxins were then conserved across numerous species of Boiga. The lack of toxic venom components specifically targeting mammalian prey, as demonstrated by previous studies [47], does not restrict the trophic ecology of B. irregularis owing to the use of differing predatory strategies when subduing prey: envenomation without constriction to subdue bird and lizard prey (primarily via irditoxin), and constriction alone to subdue mammalian prey [47]. Spilotes sulphureus, on the other hand, produces venom containing both lizard- and mammal-specific toxins, and adult S. sulphureus do not constrict mammalian prey, but use venom and body pinning to subdue both lizard and mammal prey (C.M. Modahl 2014, personal observation). The efficacy of this mode of feeding is greatly facilitated by the presence of different taxon-specific toxins that affect both prey types. The occurrence of prey-specific 3FTxs in the venom of S. sulphureus, a snake species native to the western hemisphere and evolutionarily distinct from Asian Boiga, emphasizes the significance of diet and predatory strategies in snake toxin evolution: both species preferentially use bird and lizard prey, but both can also take mammalian prey using different strategies (chemical versus mechanical subjugation). Sulditoxin and irditoxin therefore probably represent convergence upon a common structural solution (heterodimerism) to produce a lizard/bird-specific toxin, and the lack of close sequence identity between them supports the interpretation of functional convergence.
Relatively simple venoms with a small number of highly abundant toxins have been suggested to be characteristic of snakes with simple diets, such as sea snakes feeding on teleost fishes [51]. The sea snake Hydrophis cyanocinctus has only six major venom components, five 3FTxs and one PLA2 [51], and another sea snake species, Aipysurus laevis, has only one primary 3FTx and five PLA2s that make up the large majority of its venom [52]. Other elapids exhibit more complex venoms; even though they are often dominated by one or two venom protein superfamilies (typically 3FTxs and PLA2s), they have a greater number of individual protein isoforms within each of these families, with at least 12 3FTx isoforms, and usually more, present in venoms. This includes the diverse genera Micrurus, Naja, Ophiophagus, Dendroaspis and Bungarus [53–58]. Even the rear-fanged snake B. irregularis produces a venom with at least 25 different 3FTx isoforms, 10 of which are abundant [36]. The venom of S. sulphureus is therefore simple, similar to a sea snake, in that only five 3FTx isoforms make up 92% of the venom (based upon the HPLC chromatogram). The identification of prey-specific 3FTxs targeting lizards or mammals within the venom of S. sulphureus indicates that snakes with relatively simple venoms, such as many rear-fanged snake species, can still target a diversity of prey types. Among the 3FTx protein superfamily, a wide variety of disparate pharmacologies has evolved on a highly conserved structural scaffold [14], and within this one colubrid snake species, the venom has capitalized upon this conserved yet versatile platform to produce a simple but effective trophic adaptation. This diversified activity is in contrast to observations for the front-fanged N. kaouthia (monocled cobra), where a single abundant 3FTx (α-cobratoxin) was the only 3FTx lethal towards both lizards and mammals [23]. This study of S. sulphureus venom toxins is, to our knowledge, the first time that 3FTxs targeting different prey items have been characterized in a single venom and demonstrates the significance of venom protein gene neofunctionalization as an important and versatile trophic adaptation.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Colorado HistoPrep (Ft. Collins, CO, USA) for providing histological preparations used in this work.
Ethics
All experiments involving animals were subjected to prior review by the University of Northern Colorado Institutional Animal Care and Use Committee (UNC-IACUC), and protocols 9204 and 1504D-SM-SMLBirds-18 were approved before any work was initiated. All work conforms to the ethics guidelines of the University of Northern Colorado, the International Society on Toxinology and the Society for the Study of Amphibians and Reptiles.
Data accessibility
Additional details of the analyses can be found in the electronic supplementary material, appendices S1 and S2. All toxin transcript sequences were submitted to GenBank and are available at NCBI under Bioproject PRJNA448354 and accession nos MH232964–MH233012.
Authors' contributions
C.M.M. and S.P.M. jointly conceived of and designed the study. C.M.M. and S.F. conducted all transcriptome experiments and C.M.M. and S.P.M. jointly conducted proteome and biochemical/biological characterization experiments. C.M.M., M. and S.F. conducted all bioinformatic analyses. C.M.M. and S.P.M. jointly wrote the original draft of the manuscript, and all authors edited the final manuscript and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
The University of Northern Colorado Graduate Student Association provided a grant (C.M.M.) for research materials and the UNC Office of Research provided additional financial support (S.P.M., S.F.). The National University of Singapore (South East Asian Biodiversity Genomics grant no. R-154-000-648-646) also provided resources and support (M.). Bioinformatic research was conducted using equipment provided by NSF grant no. MRI-1429003 to S.F.
References
- 1.Vonk FJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl Acad. Sci. USA 110, 20 651–20 656. ( 10.1073/pnas.1314702110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whittington AC, Mason AJ, Rokyta DR. 2018. A single mutation unlocks cascading exaptations in the origin of a potent pitviper neurotoxin. Mol. Biol. Evol. 35, 887–898. ( 10.1093/molbev/msx334) [DOI] [PubMed] [Google Scholar]
- 3.Brust A, et al. 2013. Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol. Cell. Proteomics 12, 651–663. ( 10.1074/mcp.M112.023135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs HL, Rossiter W. 2008. Rapid evolution by positive selection and gene gain and loss: PLA2 venom genes in closely related Sistrurus rattlesnakes with divergent diets. J. Mol. Evol. 66, 151–166. ( 10.1007/s00239-008-9067-7) [DOI] [PubMed] [Google Scholar]
- 5.Aird SD, Arora J, Barua A, Qiu L, Terada K, Mikheyev AS. 2017. Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genome Biol. Evol. 9, 2640–2649. ( 10.1093/gbe/evx199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margres MJ, Bigelow AT, Lemmon EM, Lemmon AR, Rokyta DR. 2017. Selection to increase expression, not sequence diversity, precedes gene family origin and expansion in rattlesnake venom. Genetics 206, 1569–1580. ( 10.1534/genetics.117.202655) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margres MJ, Wray KP, Hassinger ATB, Ward MJ, McGivern JJ, Lemmon EM, Lemmon AR, Rokyta DR. 2017. Quantity, not quality: rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol. Biol. Evol. 34, 3099–3110. ( 10.1093/molbev/msx231) [DOI] [PubMed] [Google Scholar]
- 8.Aird SD, Aggarwal S, Villar-Briones A, Tin MM, Terada K, Mikheyev AS. 2015. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genomics 16, 647 ( 10.1186/s12864-015-1832-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rokyta DR, Wray KP, Lemmon AR, Lemmon EM, Caudle BS. 2011. A high-throughput venom-gland transcriptome for the eastern diamondback rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon 57, 657–671. ( 10.1016/j.toxicon.2011.01.008) [DOI] [PubMed] [Google Scholar]
- 10.Nakashima K, Ogawa T, Oda N, Hattori M, Sakaki Y, Kihara H, Ohno M. 1993. Accelerated evolution of Trimeresurus flavoviridis venom gland phospholipase A2 isozymes. Proc. Natl Acad. Sci. USA 90, 5964–5968. ( 10.1073/pnas.90.13.5964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kini RM, Chan YM. 1999. Accelerated evolution and molecular surface of venom phospholipase A2 enzymes. J. Mol. Evol. 48, 125–132. ( 10.1007/PL00006450) [DOI] [PubMed] [Google Scholar]
- 12.Doley R, Mackessy SP, Kini RM. 2009. Role of accelerated segment switch in exons to alter targeting (ASSET) in the molecular evolution of snake venom proteins. BMC Evol. Biol. 9, 146 ( 10.1186/1471-2148-9-146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa T, Oda N, Nakashima K, Sasaki H, Hattori M, Sakaki Y, Kihara H, Ohno M. 1992. Unusually high conservation of untranslated sequence in cDNA for Trimeresurus flavoviridis phospholipase A2 isozymes. Proc. Natl Acad. Sci. USA 89, 8557–8561. ( 10.1073/pnas.89.18.8557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kini RM, Doley R. 2010. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon 56, 855–867. ( 10.1016/j.toxicon.2010.07.010) [DOI] [PubMed] [Google Scholar]
- 15.Karlsson E, Eaker D. 1972. Isolation of the principal neurotoxins of Naja naja subspecies from the Asian mainland. Toxicon 10, 217–225. ( 10.1016/0041-0101(72)90006-2) [DOI] [PubMed] [Google Scholar]
- 16.Weinstein SA, Warrell DA, White J, Keyler DE. 2011. ‘Venomous’ bites from non-venomous snakes. Amsterdam, The Netherlands: Elsevier Inc. [Google Scholar]
- 17.Weinstein SA, White J, Keyler DE, Warrell DA. 2013. Non-front-fanged colubroid snakes: a current evidence-based analysis of medical significance. Toxicon 69, 103–113. ( 10.1016/j.toxicon.2013.02.003) [DOI] [PubMed] [Google Scholar]
- 18.Fry BG, Wüster W, Ramjan RSF, Jackson T, Martelli P, Kini RM. 2003. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: evolutionary and toxinological implications. Rapid Commun. Mass Spectrom. 17, 2047–2062. ( 10.1002/rcm.1148) [DOI] [PubMed] [Google Scholar]
- 19.Mackessy SP. 2002. Biochemistry and pharmacology of colubrid snake venoms. J. Toxicol. Toxin Rev. 21, 43–83. ( 10.1081/TXR-120004741) [DOI] [Google Scholar]
- 20.Pawlak J, et al. 2006. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (mangrove catsnake) with bird-specific activity. J. Biol. Chem. 281, 29 030–29 041. ( 10.1074/jbc.M605850200) [DOI] [PubMed] [Google Scholar]
- 21.Pawlak J, et al. 2009. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 23, 534–545. ( 10.1096/fj.08-113555) [DOI] [PubMed] [Google Scholar]
- 22.Heyborne WH, Mackessy SP. 2013. Identification and characterization of a taxon-specific three-finger toxin from the venom of the green vinesnake (Oxybelis fulgidus; family Colubridae). Biochimie 95, 1923–1932. ( 10.1016/j.biochi.2013.06.025) [DOI] [PubMed] [Google Scholar]
- 23.Modahl CM, Mukherjee AK, Mackessy SP. 2016. An analysis of venom ontogeny and prey-specific toxicity in the monocled cobra (Naja kaouthia). Toxicon 119, 8–20. ( 10.1016/j.toxicon.2016.04.049) [DOI] [PubMed] [Google Scholar]
- 24.Gutiérrez JM, Williams D, Fan HW, Warrell DA. 2010. Snakebite envenoming from a global perspective: towards an integrated approach. Toxicon 56, 1223–1235. ( 10.1016/j.toxicon.2009.11.020) [DOI] [PubMed] [Google Scholar]
- 25.Harrison RA, Cook D, Renjifo C, Casewell NR, Currier R, Wagstaff SC. 2011. Research strategies to improve snakebite treatment: challenges and progress. J. Proteomics 74, 1768–1780. ( 10.1016/j.jprot.2011.06.019) [DOI] [PubMed] [Google Scholar]
- 26.Kuch U, Mebs D. 2002. Envenomations by colubrid snakes in Africa, Europe, and the Middle East. J. Toxicol. Toxin Rev. 21, 159–179. ( 10.1081/TXR-120004745) [DOI] [Google Scholar]
- 27.Pla D, Sanz L, Whiteley G, Wagstaff SC, Harrison RA, Casewell NR, Calvete JJ. 2017. What killed Karl Patterson Schmidt? Combined venom gland transcriptomic, venomic and antivenomic analysis of the South African green tree snake (the boomslang), Dispholidus typus. Biochim. Biophys. Acta 1861, 814–823. ( 10.1016/j.bbagen.2017.01.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saviola AJ, Peichoto ME, Mackessy SP. 2014. Rear-fanged snake venoms: an untapped source of novel compounds and potential drug leads. Toxin Rev. 33, 1–17. ( 10.3109/15569543.2014.942040) [DOI] [Google Scholar]
- 29.Mackessy SP, Saviola AJ. 2016. Understanding biological roles of venoms among the Caenophidia: the importance of rear-fanged snakes. Integr. Comp. Biol. 56, 1004–1021. ( 10.1093/icb/icw110) [DOI] [PubMed] [Google Scholar]
- 30.Junqueira-de-Azevedo IL, Campos PF, Ching AT, Mackessy SP. 2016. Colubrid venom composition: an -omics perspective. Toxins 8, 230 ( 10.3390/toxins8080230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumsden NG, Banerjee Y, Kini RM, Kuruppu S, Hodgson WC. 2007. Isolation and characterization of rufoxin, a novel protein exhibiting neurotoxicity from venom of the psammophiine, Rhamphiophis oxyrhynchus (Rufous beaked snake). Neuropharmacology 52, 1065–1070. ( 10.1016/j.neuropharm.2006.11.002) [DOI] [PubMed] [Google Scholar]
- 32.OmPraba G, Chapeaurouge A, Doley R, Devi KR, Padmanaban P, Venkatraman C, Velmurugan D, Lin Q, Kini RM. 2010. Identification of a novel family of snake venom proteins veficolins from Cerberus rynchops using a venom gland transcriptomics and proteomics approach. J. Proteome Res. 9, 1882–1893. ( 10.1021/pr901044x) [DOI] [PubMed] [Google Scholar]
- 33.Ching AT, et al. 2012. Venomics profiling of Thamnodynastes strigatus unveils matrix metalloproteinases and other novel proteins recruited to the toxin arsenal of rear-fanged snakes. J. Proteome Res. 11, 1152–1162. ( 10.1021/pr200876c) [DOI] [PubMed] [Google Scholar]
- 34.McGivern JJ, Wray KP, Margres MJ, Couch ME, Mackessy SP, Rokyta DR. 2014. RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genomics 15, 1061 ( 10.1186/1471-2164-15-1061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pla D, et al. 2017. Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged brown treesnake, Boiga irregularis, from Guam. J. Proteomics 174, 71–84. ( 10.1016/j.jprot.2017.12.020) [DOI] [PubMed] [Google Scholar]
- 36.Jadin RC, Burbrink FT, Rivas GA, Vitt LJ, Barrio-Amorós CL, Guralnick RP. 2013. Finding arboreal snakes in an evolutionary tree: phylogenetic placement and systematic revision of the Neotropical birdsnakes. J. Zool. Syst. Evol. Res. 52: 257–264. ( 10.1111/jzs.12055) [DOI] [Google Scholar]
- 37.Taub AM. 1967. Comparative histological studies on Duvernoy's gland of colubrid snakes. Bull. Mus. Nat. Hist. 138, 1–50. [Google Scholar]
- 38.Mackessy SP. 2010. The field of reptile toxinology: snakes, lizards and their venoms. In Handbook of venoms and toxins of reptiles (ed. Mackessy SP.), pp. 2–23. Boca Raton, FL: CRC Press/Taylor & Francis Group. [Google Scholar]
- 39.Boos HEA. 2001. The snakes of Trinidad and Tobago. College station, TX: Texas A&M University Press. [Google Scholar]
- 40.Lopez TJ, Maxson LR. 1995. Mitochondrial DNA sequence variation and genetic differentiation among colubrine snakes (Reptilia: Colubridae: Colubrinae). Biochem. Syst. Ecol. 23, 487–505. ( 10.1016/0305-1978(95)00034-R) [DOI] [Google Scholar]
- 41.Hill RE, Mackessy SP. 1997. Venom yields from several species of colubrid snakes and differential effects of ketamine. Toxicon 35, 671–678. ( 10.1016/S0041-0101(96)00174-2) [DOI] [PubMed] [Google Scholar]
- 42.Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Trop. Med. Hyg. 27, 493–497. (doi: 10.12691/jfnr-4-9-6) [Google Scholar]
- 43.Finney D. 1978. Statistical method in biological assay. London and High Wycombe, UK: Charles Griffin & Co. [Google Scholar]
- 44.Kelley LA, Mezulis S, Yates C, Wass M, Sternberg M. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. ( 10.1038/nprot.2015.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. ( 10.1007/BF01731581) [DOI] [PubMed] [Google Scholar]
- 46.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137. ( 10.1007/978-1-59745-251-9_6) [DOI] [PubMed] [Google Scholar]
- 47.Mackessy SP, Sixberry NM, Heyborne WH, Fritts T. 2006. Venom of the brown treesnake, Boiga irregularis: ontogenetic shifts and taxa-specific toxicity. Toxicon 47, 537–548. ( 10.1016/j.toxicon.2006.01.007) [DOI] [PubMed] [Google Scholar]
- 48.Weldon CL, Mackessy SP. 2010. Biological and proteomic analysis of venom from the Puerto Rican racer (Alsophis portoricensis: Dipsadidae). Toxicon 55, 558–569. ( 10.1016/j.toxicon.2009.10.010) [DOI] [PubMed] [Google Scholar]
- 49.Nirthanan S, Gopalakrishnakone P, Gwee MCE, Khoo HE, Kini RM. 2003. Non-conventional toxins from elapid venoms. Toxicon 41, 397–407. ( 10.1016/s0041-0101(02)00388-4) [DOI] [PubMed] [Google Scholar]
- 50.Casewell NR, et al. 2014. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl Acad. Sci. USA 111, 9205–9210. ( 10.1073/pnas.1405484111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calvete JJ, Ghezellou P, Paiva O, Matainaho T, Ghassempour A, Goudarzi H, Kraus F, Sanz L, Williams DJ. 2012. Snake venomics of two poorly known Hydrophiinae: comparative proteomics of the venoms of terrestrial Toxicocalamus longissimus and marine Hydrophis cyanocinctus. J. Proteomics 75, 4091–4101. ( 10.1016/j.jprot.2012.05.026) [DOI] [PubMed] [Google Scholar]
- 52.Laustsen AH, Gutiérrez JM, Rasmussen AR, Engmark M, Gravlund P, Sanders KL, Lohse B, Lomonte B. 2015. Danger in the reef: proteome, toxicity, and neutralization of the venom of the olive sea snake, Aipysurus laevis. Toxicon 107, 187–196. ( 10.1016/j.toxicon.2015.07.008) [DOI] [PubMed] [Google Scholar]
- 53.Sanz L, Pla D, Perez A, Rodríguez Y, Zavaleta A, Salas M, Lomonte B, Calvete JJ. 2016. Venomic analysis of the poorly studied desert coral snake, Micrurus tschudii tschudii, supports the 3FTx/PLA2 dichotomy across Micrurus venoms. Toxins 8, 178 ( 10.3390/toxins8060178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang HW, Liu BS, Chien KY, Chiang LC, Huang SY, Sung WC, Wu WG. 2015. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteomics 128, 92–104. ( 10.1016/j.jprot.2015.07.015) [DOI] [PubMed] [Google Scholar]
- 55.Dutta S, Chanda A, Kalita B, Islam T, Patra A, Mukherjee AK. 2017. Proteomic analysis to unravel the complex venom proteome of eastern India Naja naja: correlation of venom composition with its biochemical and pharmacological properties. J. Proteomics 156, 29–39. ( 10.1016/j.jprot.2016.12.018) [DOI] [PubMed] [Google Scholar]
- 56.Chang HC, Tsai TS, Tsai IH. 2013. Functional proteomic approach to discover geographic variations of king cobra venoms from Southeast Asia and China. J. Proteomics 89, 141–153. ( 10.1016/j.jprot.2013.06.012) [DOI] [PubMed] [Google Scholar]
- 57.Ainsworth S, et al. 2018. The medical threat of mamba envenoming in sub-Saharan Africa revealed by genus-wide analysis of venom composition, toxicity and antivenomics profiling of available antivenoms. J. Proteomics 172, 173–189. ( 10.1016/j.jprot.2017.08.016) [DOI] [PubMed] [Google Scholar]
- 58.Rusmili MR, Yee TT, Mustafa MR, Hodgson WC, Othman I. 2014. Proteomic characterization and comparison of Malaysian Bungarus candidus and Bungarus fasciatus venoms. J. Proteomics 110, 129–144. ( 10.1016/j.jprot.2014.08.001) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional details of the analyses can be found in the electronic supplementary material, appendices S1 and S2. All toxin transcript sequences were submitted to GenBank and are available at NCBI under Bioproject PRJNA448354 and accession nos MH232964–MH233012.