Abstract
An understanding of the balance of interspecific competition and the physical environment in structuring organismal communities is crucial because those communities structured primarily by their physical environment typically exhibit greater sensitivity to environmental change than those structured predominantly by competitive interactions. Here, using detailed phylogenetic and functional information, we investigate this question in macrofaunal assemblages from Northwest Atlantic Ocean continental slopes, a high seas region projected to experience substantial environmental change through the current century. We demonstrate assemblages to be both phylogenetically and functionally under-dispersed, and thus conclude that the physical environment, not competition, may dominate in structuring deep-ocean communities. Further, we find temperature and bottom trawling intensity to be among the environmental factors significantly related to assemblage diversity. These results hint that deep-ocean communities are highly sensitive to their physical environment and vulnerable to environmental perturbation, including by direct disturbance through fishing, and indirectly through the changes brought about by climate change.
Keywords: supertree, functional traits, community phylogenetics, climate change, bottom trawling, deep sea
1. Introduction
Competition between species has long been recognized as an important factor determining the ecological diversity and structure of organismal communities [1–4]. Intense interspecific competition for scarce resources can result in the exclusion of certain taxa [2,4], shaping species' realized niches and distributions, and influencing ecosystem functioning [5,6]. Numerous studies have investigated the role of competition in structuring terrestrial (such as plant [7]) and shallow-water (such as coral reef [8]) assemblages, but the importance of interspecific competition in structuring the expansive and functionally important communities of the deep ocean has been a matter of debate because the discovery of high alpha diversity in deep-water sediments [9–11]. Some researchers have emphasized an important role of biological interactions in structuring deep-seafloor communities, theorizing a dynamic balance between competitive forces and predation [12,13]. Others have argued that extensive niche differentiation, coupled with typically low organismal densities and the availability of space, mean that competitive interactions are unlikely to be significant in structuring modern-day deep-ocean communities [9,10,14,15].
Empirical evidence in support of either of these viewpoints is limited. Studies that have examined the morphological or trophic characteristics of deep-sea assemblages present some evidence for the displacement of ecologically similar taxa by their competitive dominants [16–18]. Conversely, studies that have investigated the taxonomic or phylogenetic structure of assemblages provide some evidence that variation in physical environmental conditions may be of greater influence than competitive interactions in structuring deep-ocean communities [19,20]. However, investigations to date have been limited in their analytical scope by the challenges associated with sampling and/or conducting experiments in the deep ocean.
Knowing whether interspecific competition or the physical environment dominates in structuring natural communities is important because it further enhances our understanding of the sensitivity of ecosystems to environmental change. Communities that are structured predominantly by interspecific competition are typically more stable under environmental stress than those whose structure is governed by the physical environment [21]. Understanding the sensitivity of deep-sea ecosystems to environmental change is of pressing concern because they are predicted to experience increasing direct and indirect anthropogenic pressure over this century [22–24]. For example, fishing fleets are operating at ever-increasing depths [25], there is growing commercial interest in the mining of seabed minerals [26], and greenhouse gas emissions are increasing oceanic temperature, reducing pH and dissolved oxygen concentrations, and altering food supply to the deep ocean [23,24,27].
In this study, we perform ‘community phylogenetic’ analyses to investigate the importance of interspecific competition versus the physical environment in shaping the composition of deep-seafloor assemblages in the Northwest Atlantic Ocean—a region predicted to experience particularly rapid environmental change over this century [28]. Application of a community phylogenetic approach avoids many of the problems associated with conducting experiments in deep-ocean environments, enabling an investigation of previously unprecedented scale. Under this approach, the dispersion of taxa within samples across a phylogeny or functional trait dendrogram is compared with that which would be expected by chance [29]. If taxa within samples are found, on average, to be less similar to one another than would be expected by random draw from the available taxa pool, assemblages are described as ‘over-dispersed’; this is typically considered evidence of a dominance of competitive exclusion in shaping assemblage structure, because phylogenetically/functionally similar taxa are assumed to be ecologically similar [3,29,30]. Conversely, if taxa within samples are found, on average, to be more similar to one another than would be expected by chance, assemblages are described as ‘under-dispersed’; this is typically considered evidence of a dominance of the physical environment in determining assemblage structure, because phylogenetically/functionally similar taxa are assumed to share the particular traits that are necessary for survival under the prevailing environmental conditions [3,29,30] (although see [31]).
Our results provide evidence that deep-seafloor communities may typically be both phylogenetically and functionally under-dispersed. We therefore also investigate and discuss a number of physical environmental parameters which may be of importance in structuring the enigmatic but widespread communities of deep-ocean sediments.
2. Material and methods
(a). Sampling of deep-seafloor assemblages
We analysed 312 sediment samples, forming the largest macrofaunal sample set yet collected from the deep ocean. Samples were collected with a box corer (area 0.25 m2) from the continental slopes of the Northwest Atlantic Ocean (depth range: 582–2294 m) (figure 1) between May-August 2009 and June–August 2010, and form part of the international ‘NEREIDA’ programme (https://www.nafo.int/About-us/International-Cooperation), a project instigated by the Northwest Atlantic Fisheries Organisation (NAFO) in order to investigate the impacts of high seas fisheries on Vulnerable Marine Ecosystems. Sediment subsamples were taken for geochemical and particle size analyses and remaining sediment was washed over a 1 mm mesh sieve for faunal analyses; 20 245 specimens of peracarid crustacean were identified to the genus level (177 genera within 74 families, in total). Peracarid crustaceans were chosen for analysis because of their low dispersal potential, extremely high taxonomic diversity, superabundance in marine sediments and ecological importance as prey, predators and ecosystem engineers [32–34].
Figure 1.
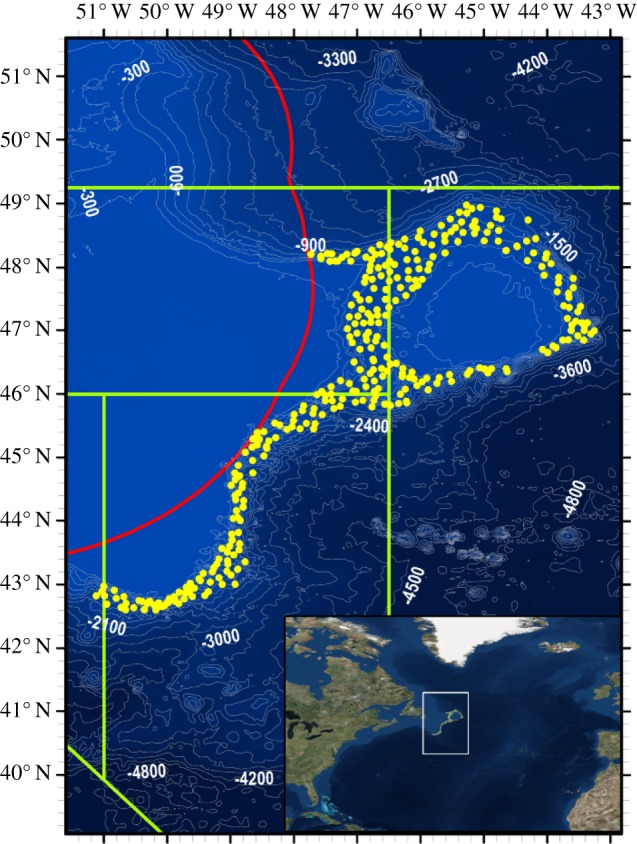
Box corer deployment locations (yellow dots, n = 312) and bathymetry (darker areas = greater water depth) of the sampling area in the Northwest Atlantic Ocean (300 m depth contours; SRTM30 bathymetric data). Red line shows the extent of the Canadian Exclusive Economic Zone. Green boxes show locations of Northwest Atlantic Fisheries Organization subarea divisions. Inset map places the sampling area (white box) in a global context (satellite imagery courtesy of ESRI World Imagery). (Online version in colour.)
(b). Supertree phylogeny construction
To investigate the phylogenetic structure of the sampled peracarid assemblages, we constructed a ‘supertree’ [35] (figure 2a). We used Google Scholar to identify 59 studies containing suitable evolutionary source trees. From each study, only unique source trees were retained for analysis to ensure that there was no duplication of source tree topology (an emergent characteristic of evolutionary trees as phylogenetic hypotheses, and the information directly used during supertree construction) that would otherwise unfairly weight the analysis as a result of pseudo-replication [35]. One hundred and twenty-seven evolutionary trees were retained for analysis (electronic supplementary material, table S2), and monophyletic taxonomic groups were labelled following World Register of Marine Species (WoRMS) systematic nomenclature.
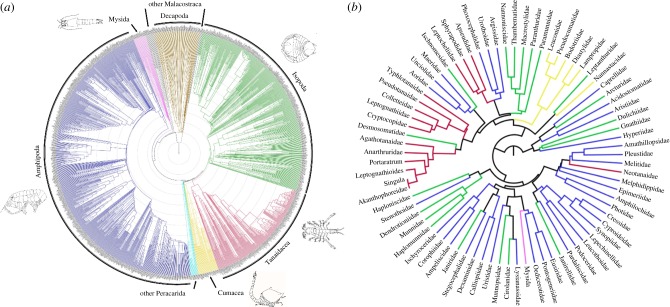
Figure 2.
(a) Supertree chronogram of Peracarida (Crustacea), including other malacostracan taxa as outgroups; 1487 terminal taxa; produced from 129 source trees using MultiLevelSupertree 1.0 [36]. Root at 600 Ma, concentric circles representing 50 Ma steps to present day (tips). Examples of taxa—Amphipoda (blue branches): Leucothoe rudicola (modified from [37]); Cumacea (yellow branches): Procampylaspis chathamensis (image © Sarah Gerken); Tanaidacea (red branches): Pseudosphyrapus anomalus (image © Graham J. Bird); Isopoda (green branches): Atlantoserolis vemae (modified from [38]); Mysida (pink branches): Heteromysis modlini (modified from [39]). (b) Functional dendrogram of 77 peracarid taxa sampled by box corer from the NW Atlantic Ocean. Produced by the clustering (UPGMA, Euclidean distance) of a database of 38 functional traits in 10 trait groupings. Branches coloured by higher taxonomic identity of terminal taxa using same palette as the supertree. ‘Newick’ format files for both the supertree and functional dendrogram are available in the electronic supplementary material to enable detailed examination of their topology.
Supertrees were constructed using MultiLevelSupertree (MLS) v. 1.0 [36] run on the Oxford University Advanced Research Computing supercomputer ARCUS (Phase B) (http://www.arc.ox.ac.uk/content/home). Because of prohibitive run times, the program was run individually for the peracarid orders Amphipoda, Isopoda, Tanaidacea and Cumacea. For each run, a taxonomy tree was used to guide the program.
To provide reference branch length information for the supertree, we constructed two further phylogenetic trees based on 18S SSU rDNA, 16S rDNA, cytochrome c oxidase 1 (COI) and histone H3 gene sequences downloaded from GenBank (electronic supplementary material, appendix S1; the topologies of these trees are available upon request). Genes were aligned individually using MAFFT v. 7.273 [40] running on the MAFFT online server (http://mafft.cbrc.jp/alignment/server/). Alignments were scrutinized using trimAl v. 1.2 [41]. The final alignment was concatenated using SequenceMatrix v. 1.8 [42] and consisted of 285 taxa and 2586 bp. PartitionFinder v. 1.1.1 [43] was used to select the most appropriate model of evolution and partitioning scheme. ML and Bayesian topologies were estimated using RAxML v. 8.2.8 and MrBayes v. 3.2.6, respectively, on the CIPRES Science Gateway v. 3.3 online server [44].
During a fifth round of supertree construction, we used MLS 1.0 to combine the output trees from the four previous MLS runs and the ML and Bayesian analyses with all source trees focusing on order-level relationships within Peracarida and Malacostraca to produce a final supertree with 1487 terminal taxa. This supertree topology was then trimmed to include only those taxa present in the GenBank sequence concatenated alignment. We estimated maximum-likelihood branch lengths for this topology using RAxML v. 8.2.8. Common nodes between the supertree and ML branch length tree were labelled using PhyloCom v. 4.2 [45] and the labelled ML branch length tree was used as an input for the program R8s v. 1.8 [46] in order to obtain a dated phylogeny. Twenty-two nodes were constrained with age estimates based on fossil data (electronic supplementary material, table S1). Non-parametric rate smoothing (NPRS) with Powell optimization was selected as the analysis method. The BLADJ function of PhyloCom [45] was then used to obtain a fully dated supertree (figure 2a; see electronic supplementary material for a ‘Newick’ format representation of the supertree to enable detailed examination of its topology).
(c). Functional dendrogram construction
To characterize the functional structure of the sampled peracarid assemblages, we constructed a dendrogram (figure 2b) describing the functional similarity of sampled families based on their scoring for a selection of traits (electronic supplementary material, table S3). Trait groupings and traits were chosen based on ecological relevance and data availability. We used a fuzzy coding [47] approach to enable the coding of variability in trait scores within a family/individual. Based on available literature and the expert opinion of the authors T.H., A.B., G.J.B., S.G. and O.S.A., 77 taxa were scored for 38 traits in 10 trait groupings. The trait database was converted into a dendrogram via hierarchical clustering (figure 2b; see electronic supplementary material for a ‘Newick’ format representation of the functional dendrogram). We used cophenetic correlation coefficient values [48] to select the most appropriate distance metric and clustering method as Euclidean distance and unweighted pair group method using arithmetic averages (UPGMA) clustering. Analyses were conducted in R v. 3.0.2 [49].
(d). Testing for phylogenetic and functional assemblage structure
We investigated phylogenetic assemblage structure using the phylostruct function of the R package ‘picante 1.6–2’ [50] based on the constructed supertree (figure 2a) and complete genus-level peracarid assemblage matrix. To investigate assemblage structure at smaller spatial scales, we used ESRI ArcGIS v. 10.1 to produce seven data subsets, each consisting of 30 box cores chosen at random from within a set radius (50, 100, 200, 300, 400, 500 and 600 km) of the central-most sampling point. To quantify the phylogenetic dispersion (diversity) of peracarid genera within each sample, we calculated the metric ‘phylogenetic species variability’ (PSV) [51]. We employed permutation tests (100 000 permutations) to determine whether the average PSV of all samples, and sample subsets, was significantly different from that expected under two null hypotheses—‘Null 1’ and ‘Null 2’ [51]. Under Null 1, phylogenetic structure was removed from both taxon prevalence and sample composition by the randomization of taxon presence within samples. Under Null 2, phylogenetic structure was removed from sample composition, but not from taxon prevalence, by the randomization of taxon occurrence between samples.
Since functional dendrograms are analogous to phylogenies in form, we employed the same methods as outlined above to quantify the functional dispersion (referred to herein as ‘functional species variability’ (FSV)) of peracarids contained within each sample based on the constructed functional dendrogram (figure 2b) and compared this to the expected outcome under the two null hypotheses stated above.
(e). Testing for phylogenetic signal
As the interpretation of community phylogenetic patterns relies on knowledge of the evolution of taxon traits [29], we tested for phylogenetic signal across the peracarid trait matrix by applying a Mantel test [52]. Based on the constructed supertree (figure 2a), we calculated the square root of patristic distance as a measure of phylogenetic distance between taxa using the R package ‘ape 4.1’ [53]. Euclidean distance was calculated as a measure of trait similarity between taxa based on the peracarid trait matrix. The Mantel test was performed using the R package ‘vegan 2.0-9’ [54] (100 000 permutations). To assess the strength of phylogenetic signal in individual traits, based on the supertree of Peracarida (figure 2a), we calculated Pagel's λ [55] for each trait in the peracarid functional trait table using the R package ‘phylosignal 1.2’ [56] (100 000 permutations).
(f). Characterization of the deep-sea physical environment
To investigate relationships between the PSV/FSV of sampled peracarid assemblages and the prevailing environmental conditions, we examined the following environmental parameters: bathymetry (depth, slope, aspect, seafloor rugosity, bathymetric position index (BPI)); fishing intensity (vessel monitoring system (VMS) signal density and total trawl length per km2); geological context; seafloor sediment particle size (percentage clay/silt/sand); carbon availability (percentage inorganic, organic and total carbon, surface chlorophyll a and particulate organic carbon (POC) concentrations, modelled transport of POC to depth); physical oceanographic variables (temperature, salinity and current speed); and month and year of sample collection.
Water depth at each sampling location was extracted using ArcGIS 10.1 based on multibeam bathymetric surveys (5625 m2 cell size). Slope, eastness and northness, roughness (225 × 225 m analysis window) and standard deviation of multibeam bathymetry values (225 × 225 m analysis window) were calculated using the Spatial Analyst extension of ArcGIS 10.1. Benthic Terrain Modeler [57] was used to calculate BPI over a range of radii as well as seafloor rugosity (375 × 375 m and 1875 × 1875 m analysis windows).
We quantified bottom trawling intensity using VMS signal locations. Individual trawl paths were identified based on boat identity, speed, location, date and time using ArcGIS 10.1, and the Line Density Tool (Spatial Analyst extension) was used to measure the total length of trawls per km2 within a set radius (1, 3 or 5 km) from each box core.
The sediments of the study area were classified into 12 discreet geological categories based on their acoustic characteristics, depth and slope [58]. We extracted the relevant geological category for each sampling location using ArcGIS 10.1.
Sediment per cent clay/silt/sand was calculated for each core subsample based the following particle size categories consistent with the ‘Phi’ (Φ) scale. We calculated particle size diversity following the methodology of Etter & Grassle [59] and Leduc et al. [60].
Sediment total carbon and organic carbon content were determined using a Leco TruSpec CHN analyser. Inorganic carbon was determined by the difference between the total carbon and organic carbon measurements for each sample [61].
We obtained surface chlorophyll a and POC concentrations from the Giovanni ocean colour radiometry online data system (https://giovanni.gsfc.nasa.gov/giovanni/). MODIS AQUA 4 km resolution data were downloaded for the years 2008–2010. These data were interpolated to 2500 × 2500 pixels (525 m resolution) in QGIS 2.2. We estimated POC delivery to the seafloor from surface POC concentrations following region-specific equations [62].
Seafloor temperature, salinity, and meridional and zonal current speed values were extracted from a modelled monthly average data layer for the study area [63] and averaged both across the year prior to sample collection and across the year of sample collection. These values were interpolated to 3000 × 3000 pixels (578 m resolution) in QGIS 2.2. Absolute current speed was calculated using Pythagoras's theorem; 10-year minimum/maximum values for temperature and current speed, respectively, and 10-year average values for both variables were calculated to capture longer term variability.
(g). Statistical analyses
We removed highly correlated environmental variables following consideration of variance inflation factors (VIFs). Variables were removed from the analysis in a stepwise manner (those with highest VIF first). The resulting dataset contained 19 variables (electronic supplementary material, table S4). VIF calculations were undertaken in R 3.0.2 [49] using the package ‘HH 3.1-32’ [64].
We constructed generalized additive models (GAMs) using the R package ‘mgcv 1.7-26’ [65] to determine which combination of environmental variables most effectively explained variability in PSV and FSV between samples. Initial GAMs consisted of all variables contained within electronic supplementary material, table S4, with smoothers (penalized thin-plate regression spline) added to all continuous variables. Appropriate error distributions and link functions were selected based on model diagnostics and the Akaike information criterion (AIC). Acceptable satisfaction of model assumptions was confirmed using the gam.check function. Smoothing parameters were optimized automatically on the basis of the generalized cross-validation criterion [65]. Explanatory terms included in each GAM were refined by backwards stepwise selection considering variable p-values and model AIC until a minimum AIC value was reached (see the electronic supplementary materials for additional detail relating to the methodology employed by this study).
3. Results
(a). Phylogenetic and functional structure of deep-seafloor assemblages
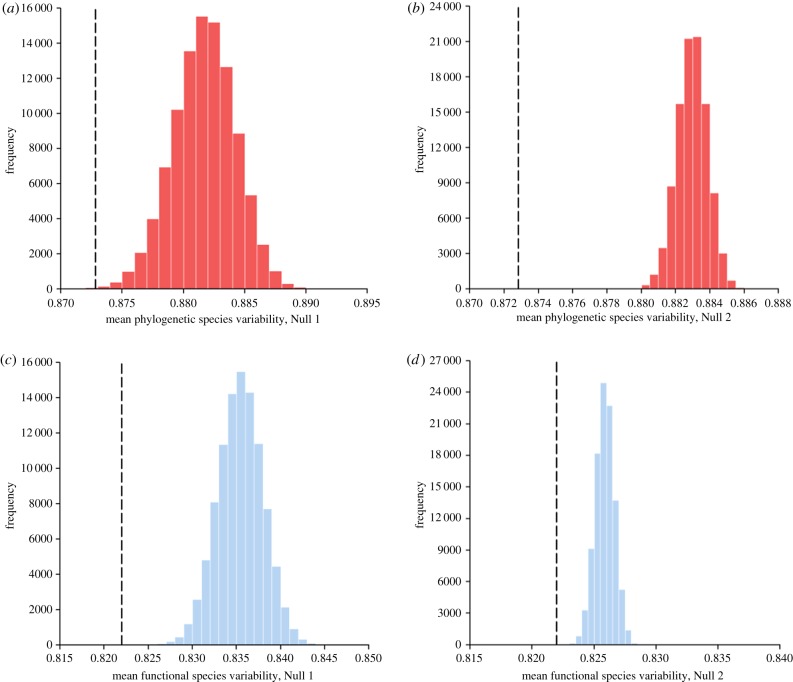
We found the average PSV value of the deep-sea assemblages analysed to be significantly smaller than that which would be expected under both null hypotheses (mean PSVobserved = 0.8728; mean PSVNull 1 = 0.8817, mean PSVNull 2 = 0.8830; probability mean PSVobserved taken from Null 1 distribution < 0.001; figure 3a; probability mean PSVobserved taken from the Null 2 distribution < 0.0001; figure 3b). Similar results were obtained for all data subsets analysed. Based on these results, we conclude that the deep-sea assemblages analysed are phylogenetically ‘under-dispersed’ (i.e. that the peracarid taxa within a sample are on average more closely related to one another than would be expected by chance).
Figure 3.
Contrast of observed (black dashed lines; n = 299) and null distribution (histograms) of average (a,b) ‘phylogenetic species variability’ (red) and (c,d) ‘functional species variability’ (blue) values for peracarid assemblages sampled from the NW Atlantic Ocean given phylogenetic relationships specified by the supertree of Peracarida (figure 2a) and functional similarity specified by the functional dendrogram of Peracarida (figure 2b). (Online version in colour.)
Further, we found the average FSV value of the assemblages analysed also to be significantly smaller than that which would be expected under both null hypotheses (mean FSVobserved = 0.8220; mean FSVNull 1 = 0.8354, mean FSVNull 2 = 0.8259; probability mean FSVobserved taken from Null 1 distribution < 0.0001; figure 3c; probability mean FSVobserved taken from the Null 2 distribution < 0.0001; figure 3d). Similar results were obtained for all data subsets analysed. We therefore conclude that the deep-sea assemblages analysed are functionally ‘under-dispersed’ (i.e. that the peracarid taxa within a sample share, on average, more functional traits with each other than would be expected by chance).
(b). Phylogenetic signal
Significant phylogenetic signal was identified in the peracarid trait matrix (Mantel test: ρ = 0.3583, p < 0.001; Pagel's λ: 34 of 38 traits exhibit significant phylogenetic signal; electronic supplementary material, table S5). We thus conclude that, on average, phylogenetically similar taxa tend also to be functionally similar.
(c). Environmental drivers of phylogenetic and functional structure
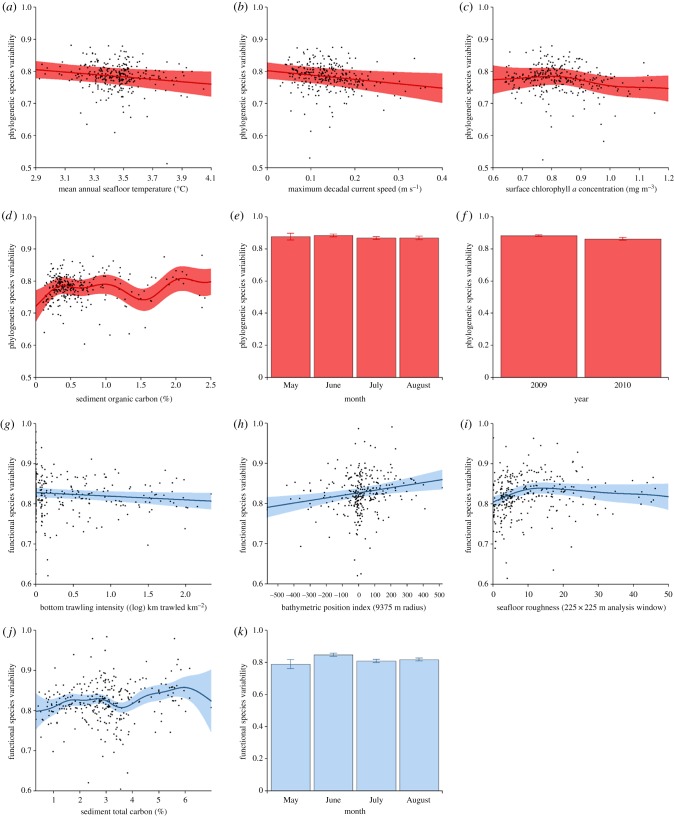
We found assemblage PSV to relate negatively with average seafloor temperature for the year of sample collection (p = 0.0349; figure 4a), and maximal current speed values for 10 years prior to sample collection (p = 0.0214; figure 4b). However, we found PSV to vary unimodally with average surface chlorophyll a concentration for the year of sample collection (p = 0.0011; figure 4c), and in a complex but weakly positive manner with sediment organic carbon content (p = 0.0001; figure 4d). We also found assemblage PSV to be significantly related to the month (p < 0.0001; figure 4e) and year (p < 0.0001; figure 4f) of sample collection.
Figure 4.
Relationships between the phylogenetic (red)/functional variability (blue) of sampled peracarid assemblages (n = 299) and environmental parameters. (a) PSV/mean annual seafloor temperature, (b) PSV/maximal decadal current speed, (c) PSV/surface chlorophyll a concentration, (d) PSV/sediment organic carbon content, (e) PSV/month, (f) PSV/year, (g) FSV/bottom trawling intensity, (h) FSV/BPI, (i) FSV/seafloor roughness, (j) FSV/sediment total carbon content, (k) FSV/month. Error around best-fit lines/bar values = 95% confidence intervals. (Online version in colour.)
We found assemblage FSV to be negatively related to bottom trawling intensity (p = 0.0452; figure 4g), but positively related to BPI (p = 0.0037; figure 4h), while FSV varied unimodally with seafloor roughness (p = 0.0472; figure 4i). Sediment total carbon content was found to relate in a complex but weakly positive manner with FSV (p = 0.0185; figure 4j). We also found assemblage FSV to be significantly related to the month of sample collection (p < 0.0001; figure 4k).
4. Discussion
The complementary phylogenetic and functional analyses performed here provide evidence for a compositional under-dispersion of the focal deep-sea assemblages at the spatial scales investigated. This under-dispersion may reflect the selection of favourable phenotypic traits that are shared between similar taxa [29]. Typically, the selecting agent in question is the physical environment, and, as a result, this process is known as ‘environmental filtering’ [29]. Although not global in extent, and focusing only on continental slope depths, the results of our study provide evidence that the physical environment may be more important than interspecific competition in shaping the composition of deep-water communities, emphasizing a potentially high sensitivity of deep-sea ecosystems to environmental perturbation [21].
Our findings challenge those studies that hypothesize an importance of competition and character displacement as a significant ecological structuring agent in the deep sea [12,13], and conflict with investigations that have examined the morphological or trophic characteristics of deep-sea assemblages [16–18]. Instead, they substantiate the hypothesis that competitive interactions between species in the deep ocean are weak and unlikely to be significant in structuring communities at the spatial scales investigated [14,15]. Further, they support the results of previous analyses of lesser spatial scope that have investigated the taxonomic and phylogenetic structure of deep-sea assemblages [19,20].
Comparison of our results with those of other studies employing the PSV metric suggests that the phylogenetic signal observed here is comparably strong or stronger than that reported for many non-marine assemblages, including temperate lake fish assemblages [30,51], tropical plant assemblages [66], archaea assemblages [67] and tropical bird assemblages [68].
Although significant phylogenetic signal was apparent in the functional trait matrix, our results provide some evidence for the convergence of functional traits between relatively distantly related crustacean taxa. For example, in figure 3, mean FSVNull 2 is closer to mean FSVobserved than mean FSVNull 1 is (figure 3c,d), suggesting that a portion of the observed under-dispersion reflects elevated functional similarity of the most prevalent taxa (Null 2 removes phylogenetic/functional structure only from assemblage composition, maintaining any structure in relative taxon prevalence, while Null 1 removes phylogenetic/functional structure from both assemblage composition and taxon prevalence). However, while the more prevalent taxa are more functionally similar to each other than would be expected by chance, they are not correspondingly phylogenetically similar; mean PSVNull 2 is not closer to mean PSVobserved than mean PSVNull 1 is (figure 3a,b), suggesting that their functional similarity is convergent to an extent. Examples of apparent functional convergence can be identified in figure 2b—the amphipod family Lysianassidae, and the isopod family Cirolanidae, for example. Our results suggest that traits related to fecundity and armament exhibit greatest propensity for convergent evolution among the peracarid taxa analysed (electronic supplementary material, table S5).
Our investigation into the possible environmental drivers of assemblage variability demonstrates that both natural and anthropogenic factors may influence the structure of the deep-sea assemblages (figure 4). The negative relationship between bottom trawling intensity and assemblage FSV (figure 4g) suggests that physical disturbance by bottom trawling reduces soft-sediment functional diversity, with the resulting assemblages exhibiting a reduced subset of the functional traits that would otherwise be present in an undisturbed assemblage. While generally concordant with the small number of studies that have investigated trawling impacts on deep-sea macrofauna and meiofauna [69–73], this finding adds a new facet to our understanding of the impacts of bottom trawling in the deep ocean.
Our analyses demonstrate a negative relationship between seafloor temperature and the phylogenetic diversity of the sampled peracarid assemblages (figure 4a), indicating that the physiological tolerances of peracarid taxa to temperature change are preserved within evolutionary lineages. That this relationship is apparent across a temperature range of only approximately 1.2°C (figure 4a) suggests that even the superficially small increases in deep-ocean temperature that are predicted to occur over this century as a result of climate change [74,75], particularly in the high seas of the North Atlantic [28], will significantly reduce the phylogenetic diversity of the communities found there, potentially impacting deep-ocean ecosystem functioning [76]. An altered phylogenetic profile of deep-sea ecosystems may eventually lead to a change in the cycling, storage and sequestration pathways of nutrients and chemicals, such as carbon.
Under current climate change scenarios, global patterns of the export of surface production to the deep ocean are expected to change in a complex manner [23,24,27,77]. Food supply to the deep ocean may dwindle in some regions, such as the North and South Atlantic Oceans [24], while being enhanced in others, such as the Arctic and Southern Oceans [24]. Our analyses suggest that changes in food availability in the deep ocean may affect both the phylogenetic and functional variability of communities, but in a complex manner (figure 4c,d,j). This, in turn, may affect the availability and variety of food for demersal and pelagic organisms that feed on sediment-dwelling prey. This multifaceted relationship is complex and still poorly understood.
Overall, the results of our analyses suggest that deep-water soft-sediment ecosystems, which constitute the majority of global seafloor area, may be particularly sensitive to environmental change. Such ecosystems are central to a number of important ecosystem services, including carbon sequestration [78], and are predicted to come under increasing direct and indirect anthropogenic pressures [22–24,27]. Even superficially small changes in natural and anthropogenic disturbance regimes, temperature, food availability and bathymetry may significantly alter the phylogenetic and functional variability of deep-seafloor communities (figure 4), and this may alter ecosystem functioning and the provision of ecosystem services by the deep ocean [76]. Our findings are therefore relevant to the understanding of anthropogenic pressures on deep-sea ecosystems, including, for example, the prediction of possible mining impacts on deep-sea fauna. We advocate that the precautionary principle be exercised in all circumstances where anthropogenic actions may disrupt the natural ecology of deep-water ecosystems.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Dr Maud Mouchet (Muséum National d'Histoire Naturelle, Paris, France) for contributing towards the production of the functional dendrogram of Peracarida. We thank Oxford University's Advanced Research Computing team for their computational support. We thank the crew and scientists aboard the Spanish research vessel Miguel Oliver who collected the box core samples analysed in this study. We are grateful to Ms Anne Downie (Cefas, Lowestoft, UK) for providing access to bathymetric and oceanographic data, Dr Jesse van der Grient (University of Oxford, UK) for aiding in the analysis of surface productivity and oceanographic data, and Dr Zeliang Wang, Dr Blair Greenan, Dr Owen Brown and Dr William Leblanc and colleagues (Bedford Institute of Oceanography, Halifax, Canada) for providing Oceanographic, sediment particle size and carbon content data. We thank the Institute of Estuarine and Coastal Studies (University of Hull, UK) for initial box core processing. We are grateful to Prof. Lisa A. Levin and two anonymous reviewers for providing insightful comments on this manuscript.
Ethics
This research was performed in accordance with all applicable international, national and/or institutional laws, guidelines and ethical standards.
Data accessibility
All data used by this study are freely available in electronic supplementary material.
Authors' contributions
O.S.A., M.B.B. and A.D.R. conceived the study. A.D.R., A.J.K. and C.R.S.B.F. provided biological specimens, access to environmental data and secured funding. O.S.A. and T.H. identified the biological specimens, with additional input from G.J.B. O.S.A., T.H., A.B., G.J.B. and S.G. scored the functional traits. O.S.A. and C.R.S.B.F. processed the environmental data. O.S.A. produced the supertree and functional dendrogram and undertook all analyses. O.S.A. wrote the manuscript, which was contributed to and edited by A.J.K., C.R.S.B.F., M.B.B., T.H., A.B., G.J.B., S.G. and A.D.R.
Competing interests
The authors declare no competing interests.
Funding
This research was supported a Natural Environment Research Council (NERC) Collaborative Awards in Science and Engineering (CASE) studentship (NE/K006886/1), by Merton College, University of Oxford, UK and through the European Union's Horizon 2020 research and innovation programme under grant agreement no. 678760 (ATLAS). This output reflects only the authors' views and the European Union cannot be held responsible for any use that may be made of the information contained therein. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. A.D.R. also acknowledges the Oxford Martin School in the funding of the Oxford Martin School Sustainable Oceans Project, which has also contributed to the current paper.
References
- 1.Lotka AH. 1932. The growth of mixed populations: two species competing for a common food supply. J. Wash. Acad. Sci. 22, 461–469. [Google Scholar]
- 2.Gause GF. 1934. The struggle for existence. Baltimore, MD: The Williams and Wilkins Company. [Google Scholar]
- 3.Elton C. 1946. Competition and the structure of ecological communities. J. Anim. Ecol. 15, 54–68. ( 10.2307/1625) [DOI] [Google Scholar]
- 4.Hardin G. 1960. The competitive exclusion principle. Science 131, 1292–1297. ( 10.1126/science.131.3409.1292) [DOI] [PubMed] [Google Scholar]
- 5.Riginos C. 2009. Grass competition suppresses savanna tree growth across multiple demographic stages. Ecology 90, 335–340. ( 10.1890/08-0462.1) [DOI] [PubMed] [Google Scholar]
- 6.Leduc AOHC, da Silva EM, Rosenfeld JS. 2015. Effects of species vs. functional diversity: understanding the roles of complementarity and competition on ecosystem function in a tropical stream fish assemblage. Ecol. Indic. 48, 627–635. ( 10.1016/j.ecolind.2014.09.027) [DOI] [Google Scholar]
- 7.Goldberg DE, Barton AM. 1992. Patterns and consequences of interspecific competition in natural communities: a review of field experiments with plants. Am. Nat. 139, 771–801. ( 10.1086/285357) [DOI] [Google Scholar]
- 8.McCook L, Jompa J, Diaz-Pulido G. 2001. Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs 19, 400–417. ( 10.1007/s003380000129) [DOI] [Google Scholar]
- 9.Hessler RR, Sanders HL. 1967. Faunal diversity in the deep-sea. Deep Sea Res. Oceanogr. Abstr. 14, 65–78. ( 10.1016/0011-7471(67)90029-0) [DOI] [Google Scholar]
- 10.Sanders HL. 1968. Marine benthic diversity: a comparative study. Am. Nat. 102, 243–282. ( 10.1086/282541) [DOI] [Google Scholar]
- 11.McClain CR, Schlacher TA. 2015. On some hypotheses of diversity of animal life at great depths on the sea floor. Mar. Ecol. 36, 849–872. [Google Scholar]
- 12.Rex MA. 1976. Biological accomodation in the deep-sea benthos: comparative evidence on the importance of predation and productivity. Deep Sea Res. Oceanogr. Abstr. 23, 975–987. ( 10.1016/0011-7471(76)90827-5) [DOI] [Google Scholar]
- 13.Rex MA. 1981. Community structure in the deep-sea benthos. Annu. Rev. Ecol. Syst. 12, 331–353. ( 10.1146/annurev.es.12.110181.001555) [DOI] [Google Scholar]
- 14.Wilson WH. 1990. Competition and predation in marine soft-sediment communities. Annu. Rev. Ecol. Syst. 21, 221–241. ( 10.1146/annurev.es.21.110190.001253) [DOI] [Google Scholar]
- 15.Grant A. 2000. Deep-sea diversity: overlooked messages from shallow-water sediments. Mar. Ecol. 21, 97–112. ( 10.1046/j.1439-0485.2000.00713.x) [DOI] [Google Scholar]
- 16.McClain CR. 2005. Bathymetric patterns of morphological disparity in deep-sea gastropods from the western north Atlantic basin. Evolution 59, 1492–1499. ( 10.1111/j.0014-3820.2005.tb01798.x) [DOI] [PubMed] [Google Scholar]
- 17.Laakmann S, Kochzius M, Auel H. 2009. Ecological niches of Arctic deep-sea copepods: vertical partitioning, dietary preferences and different trophic levels minimize inter-specific competition. Deep Sea Res. I 56, 741–756. ( 10.1016/j.dsr.2008.12.017) [DOI] [Google Scholar]
- 18.Levin LA, Ziebis W, Mendoza GF, Bertics VJ, Washington T, Gonzalez JP, Thurber AR, Ebbe B, Lee RW. 2013. Ecological release and niche partitioning under stress: lessons from dorvilleid polychaetes in sulfidic sediments at methane seeps. Deep Sea Res. II 92, 214–233. ( 10.1016/j.dsr2.2013.02.006) [DOI] [Google Scholar]
- 19.Rex MA, Waren A (eds) 1981. Evolution in the deep sea: taxonomic diversity of gastropod assemblages In Proc. of the XIV Pacific Science Congress, 1981 Vladivostock, Russia: Academy of Sciences of the USSR, Far East Science Center, Institute of Marine Biology. [Google Scholar]
- 20.Quattrini AM, Gomez CE, Cordes EE. 2017. Environmental filtering and neutral processes shape octocoral communities in the deep sea. Oecologia 183, 221–236. ( 10.1007/s00442-016-3765-4) [DOI] [PubMed] [Google Scholar]
- 21.Didham RK, Watts CH, Norton DA. 2005. Are systems with strong underlying abiotic regimes more likely to exhibit alternative stable states? Oikos 110, 409–416. ( 10.1111/j.0030-1299.2005.13883.x) [DOI] [Google Scholar]
- 22.Ramirez-Llodra E, et al. 2011. Man and the last great wilderness: human impact on the deep sea. PLoS ONE 6, e22588 ( 10.1371/journal.pone.0022588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers AD. 2015. Environmental change in the deep ocean. Annu. Rev. Environ. Resour. 40, 1–38. ( 10.1146/annurev-environ-102014-021415) [DOI] [Google Scholar]
- 24.Sweetman AK, et al. 2017. Major impacts of climate change on deep-sea benthic ecosystems. Elem. Sci. Anthropocene 5, article 4. [Google Scholar]
- 25.Watson RA, Morato T. 2013. Fishing down the deep: accounting for within-species changes in depth of fishing. Fish. Res. 140, 63–65. ( 10.1016/j.fishres.2012.12.004) [DOI] [Google Scholar]
- 26.Rogers AD, et al. 2015. Delving deeper: critical challenges for 21st century deep-sea research. Ostend, Belgium: European Marine Board.
- 27.Levin LA, Le Bris N. 2015. The deep ocean under climate change. Science 350, 766–768. ( 10.1126/science.aad0126) [DOI] [PubMed] [Google Scholar]
- 28.Saba VS, et al. 2016. Enhanced warming of the Northwest Atlantic Ocean under climate change. J. Geophys. Res. Oceans 121, 118–132. ( 10.1002/2015JC011346) [DOI] [Google Scholar]
- 29.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. ( 10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 30.Helmus MR, Savage K, Diebel MW, Maxted JT, Ives AR. 2007. Separating the determinants of phylogenetic community structure. Ecol. Lett. 10, 917–925. ( 10.1111/j.1461-0248.2007.01083.x) [DOI] [PubMed] [Google Scholar]
- 31.Mayfield MM, Levine JM. 2010. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 13, 1085–1093. ( 10.1111/j.1461-0248.2010.01509.x) [DOI] [PubMed] [Google Scholar]
- 32.Snelgrove PVR. 1997. The importance of marine sediment biodiversity in ecosystem processes. Ambio 26, 578–583. [Google Scholar]
- 33.Poore GCB. 2005. Peracarida: monophyly, relationships and evolutionary success. Nauplius 13, 1–27. [Google Scholar]
- 34.Larsen K, Tuya F, Froufe E. 2014. Genetic divergence of tanaidaceans (Crustacea: Peracarida) with low dispersal ability. Sci. Mar. 78, 81–90. ( 10.3989/scimar.03878.19A) [DOI] [Google Scholar]
- 35.Bininda-Emonds ORP. 2004. The evolution of supertrees. Trends Ecol. Evol. 19, 315–322. ( 10.1016/j.tree.2004.03.015) [DOI] [PubMed] [Google Scholar]
- 36.Berry V, Bininda-Emonds ORP, Semple C. 2013. Amalgamating source trees with different taxonomic levels. Syst. Biol. 62, 231–249. ( 10.1093/sysbio/sys090) [DOI] [PubMed] [Google Scholar]
- 37.White KN, Thomas JD. 2009. Leucothoidae. Zootaxa. 2260, 494–555. [Google Scholar]
- 38.Brandt A, Brix S, Held C, Kihara TC. 2014. Molecular differentiation in sympatry despite morphological stasis: deep-sea Atlantoserolis Wägele, 1994 and Glabroserolis Menzies, 1962 from the south-west Atlantic (Crustacea: Isopoda: Serolidae). Zool. J. Linn. Soc. 172, 318–359. ( 10.1111/zoj.12178) [DOI] [Google Scholar]
- 39.Price WW, Heard RW. 2011. Two new species of Heteromysis (Olivemysis) (Mysida, Mysidae, Heteromysinae) from the tropical northwest Atlantic with diagnostics on the subgenus Olivemysis Băcescu, 1968. Zootaxa 2823, 32–46. [Google Scholar]
- 40.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. ( 10.1111/j.1096-0031.2010.00329.x) [DOI] [PubMed] [Google Scholar]
- 43.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 44.Miller MA, Pfeiffer W, Schwartz T (eds). 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees In Gateway Computing Environments Workshop (GCE), 2010 New Orleans, LA: IEEE. [Google Scholar]
- 45.Webb CO, Ackerly DD, Kembel SW. 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100. ( 10.1093/bioinformatics/btn358) [DOI] [PubMed] [Google Scholar]
- 46.Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302. ( 10.1093/bioinformatics/19.2.301) [DOI] [PubMed] [Google Scholar]
- 47.Chevenet F, Doledec S, Chessel D. 1994. A fuzzy coding approach for the analysis of long-term ecological data. Freshw. Biol. 31, 295–309. ( 10.1111/j.1365-2427.1994.tb01742.x) [DOI] [Google Scholar]
- 48.Sokal RR, Rohlf FJ. 1962. The comparison of dendrograms by objective methods. Taxon 11, 33–40. ( 10.2307/1217208) [DOI] [Google Scholar]
- 49.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 50.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2014. ‘picante’ R tools for integrating phylogenies and ecology, 1.6-2 ed. CRAN repository. [DOI] [PubMed]
- 51.Helmus MR, Bland TJ, Williams CK, Ives AR. 2007. Phylogenetic measures of biodiversity. Am. Nat. 169, E68–E83. ( 10.1086/511334) [DOI] [PubMed] [Google Scholar]
- 52.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27(2 Part 1), 209–220. [PubMed] [Google Scholar]
- 53.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language (version 3.0-11). Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 54.Oksanen J, et al. 2013. Vegan: community ecology package. R package version 2 0-92013.
- 55.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 56.Keck F. 2018. Phylosignal: exploring the phylogenetic signal in continuous traits. R package version 1.2.
- 57.Wright DJ, Pendleton M, Boulware J, Walbridge S, Gerlt B, Eslinger D, Sampson D, Huntley E. 2012. ArcGIS Benthic Terrain Modeler (BTM). 3.0 ed. Redlands, CA: Environmental Systems Research Institute, NOAA Coastal Services Center, Massachusetts Office of Coastal Zone Management.
- 58.Anon. 2011. Vulnerable marine ecosystems (VMEs) in the NAFO regulatory area (NRA). EVID 4 Evidence Project Final Report (Rev. 06/11). London, UK: DEFRA.
- 59.Etter RJ, Grassle JF. 1992. Patterns of species diversity in the deep sea as a function of sediment particle size diversity. Nature 360, 576–578. ( 10.1038/360576a0) [DOI] [Google Scholar]
- 60.Leduc D, Rowden AA, Probert PK, Pilditch CA, Nodder SD, Vanreusel A, Duineveld GC, Witbaard R. 2012. Further evidence for the effect of particle-size diversity on deep-sea benthic biodiversity. Deep Sea Res. I 63, 164–169. ( 10.1016/j.dsr.2011.10.009) [DOI] [Google Scholar]
- 61.Weitzman J, Ledger S, Stacey CD, Strathdee G, Piper DJW, Jarrett KA, Higgins J. 2014. Logs of short push cores, deep-water margin of Flemish Cap and the eastern Grand Banks of Newfoundland. Ottawa, Canada: Geological Survey of Canada. [Google Scholar]
- 62.Lutz M, Dunbar R, Caldeira K. 2002. Regional variability in the vertical flux of particulate organic carbon in the ocean interior. Glob. Biogeochem. Cycles 16, 11-1–11-18. ( 10.1029/2000GB001383) [DOI] [Google Scholar]
- 63.Wang Z, Greenan BGW. 2014. Physical oceanographic conditions on Newfoundland Shelf/Flemish Cap from a model perspective (1990–2012). Serial no. N6298. Dartmouth, Canada: Northwest Atlantic Fisheries Organization.
- 64.Heiberger RM. 2016. HH statistical analysis data display. 3.1-32 ed. CRAN repository.
- 65.Wood SN. 2013. Mixed GAM computation vehicle with GCV/AIC/REML smoothness estimation. 1.7-26 ed. CRAN repository.
- 66.Umaña MN, Norden N, Cano Á, Stevenson PR. 2012. Determinants of plant community assembly in a mosaic of landscape units in Central Amazonia: ecological and phylogenetic perspectives. PLoS ONE 7, e45199 ( 10.1371/journal.pone.0045199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Auguet J-C, Barberan A, Casamayor EO. 2009. Global ecological patterns in uncultured Archaea. ISME J. 4, 182–190. ( 10.1038/ismej.2009.109) [DOI] [PubMed] [Google Scholar]
- 68.Gómez JP, Bravo GA, Brumfield RT, Tello JG, Cadena CD. 2010. A phylogenetic approach to disentangling the role of competition and habitat filtering in community assembly of Neotropical forest birds. J. Anim. Ecol. 79, 1181–1192. ( 10.1111/j.1365-2656.2010.01725.x) [DOI] [PubMed] [Google Scholar]
- 69.Koslow JA, Gowlett-Holmes K, Lowry JK, O'Hara T, Poore GCB, Williams A. 2001. Seamount benthic macrofauna off southern Tasmania: community structure and impacts of trawling. Mar. Ecol. Prog. Ser. 213, 111–125. ( 10.3354/meps213111) [DOI] [Google Scholar]
- 70.Clark MR, Rowden AA. 2009. Effect of deepwater trawling on the macro-invertebrate assemblages of seamounts on the Chatham Rise, New Zealand. Deep Sea Res. I 56, 1540–1554. ( 10.1016/j.dsr.2009.04.015) [DOI] [Google Scholar]
- 71.Pusceddu A, Bianchelli S, Martín J, Puig P, Palanques A, Masqué P, Danovaro R. 2014. Chronic and intensive bottom trawling impairs deep-sea biodiversity and ecosystem functioning. Proc. Natl Acad. Sci. USA 111, 8861–8866. ( 10.1073/pnas.1405454111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clark MR, Althaus F, Schlacher TA, Williams A, Bowden DA, Rowden AA. 2016. The impacts of deep-sea fisheries on benthic communities: a review. ICES J. Mar. Sci. 73, i51–i69. ( 10.1093/icesjms/fsv123) [DOI] [Google Scholar]
- 73.Román S, Vanreusel A, Romano C, Ingels J, Puig P, Company JB, Martin D. 2016. High spatiotemporal variability in meiofaunal assemblages in Blanes Canyon (NW Mediterranean) subject to anthropogenic and natural disturbances. Deep Sea Res. I 117, 70–83. ( 10.1016/j.dsr.2016.10.004) [DOI] [Google Scholar]
- 74.Balmaseda MA, Trenberth KE, Kallen E. 2013. Distinctive climate signals in reanalysis of global ocean heat content. Geophys. Res. Lett. 40, 1754–1759. ( 10.1002/grl.50382) [DOI] [Google Scholar]
- 75.Somavilla R, Schauer U, Budéus G. 2013. Increasing amount of Arctic Ocean deep waters in the Greenland Sea. Geophys. Res. Lett. 40, 4361–4366. ( 10.1002/grl.50775) [DOI] [Google Scholar]
- 76.Danovaro R, Gambi C, Dell'Anno A, Corinaldesi C, Fraschetti S, Vanreusel A, Vincx M, Gooday AJ. 2008. Exponential decline of deep-sea ecosystem functioning linked to benthic biodiversity loss. Curr. Biol. 18, 1–8. ( 10.1016/j.cub.2007.11.056) [DOI] [PubMed] [Google Scholar]
- 77.Jones DOB, Yool A, Wei C-L, Henson SA, Ruhl HA, Watson RA, Gehlen M. 2014. Global reductions in seafloor biomass in response to climate change. Glob. Change Biol. 20, 1861–1872. ( 10.1111/gcb.12480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thurber AR, Sweetman AK, Narayanaswamy BE, Jones DOB, Ingels J, Hansman RL. 2014. Ecosystem function and services provided by the deep sea. Biogeosciences 11, 3941–3963. ( 10.5194/bg-11-3941-2014) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used by this study are freely available in electronic supplementary material.