Abstract
House sparrows (Passer domesticus) are a hugely successful anthrodependent species; occurring on nearly every continent. Yet, despite their ubiquity and familiarity to humans, surprisingly little is known about their origins. We sought to investigate the evolutionary history of the house sparrow and identify the processes involved in its transition to a human-commensal niche. We used a whole genome resequencing dataset of 120 individuals from three Eurasian species, including three populations of Bactrianus sparrows, a non-commensal, divergent house sparrow lineage occurring in the Near East. Coalescent modelling supports a split between house and Bactrianus sparrow 11 Kya and an expansion in the house sparrow at 6 Kya, consistent with the spread of agriculture following the Neolithic revolution. Commensal house sparrows therefore likely moved into Europe with the spread of agriculture following this period. Using the Bactrianus sparrow as a proxy for a pre-commensal, ancestral house population, we performed a comparative genome scan to identify genes potentially involved with adaptation to an anthropogenic niche. We identified potential signatures of recent, positive selection in the genome of the commensal house sparrow that are absent in Bactrianus populations. The strongest selected region encompasses two major candidate genes; COL11A—which regulates craniofacial and skull development and AMY2A, part of the amylase gene family which has previously been linked to adaptation to high-starch diets in humans and dogs. Our work examines human-commensalism in an evolutionary framework, identifies genomic regions likely involved in rapid adaptation to this new niche and ties the evolution of this species to the development of modern human civilization.
Keywords: Passer, anthrodependency, selective sweeps, amylase genes, Neolithic revolution
1. Introduction
Human activity has had a dramatic impact on life on earth, both negatively and positively with respect to biodiversity. With the advent of agriculture and establishment of more permanent settlements following the Neolithic revolution, came the creation of novel niches that a number of species have been able to utilise. Species that have adapted to a life in anthropogenic surroundings, ranging from pests such as bedbugs or head lice to the precursors to domesticated animals, have had a profound impact on our own societies. However, in many cases the ecological context and evolutionary dynamics of adaptation to a human niche are poorly understood [1].
Anthrodependent or human-commensal taxa differ from domesticated species in that humans do not play a direct role in their reproduction, i.e. they do not experience artificial selection [2,3]. Archetypal examples include our most common rodents such as the house mouse and black and brown rats that have spread with agriculture, colonialism and urbanization [4–6]. Such species were likely predisposed to use human resources as opportunistic scavengers and subsequently adapted to a dependent relationship with humans [7]. The evolutionary origins of several domesticated species—i.e. dogs, cats and horses—are now reasonably well understood [8,9]; however, relatively little is known about several charismatic and familiar species which may have had a long association with human civilization.
Anthrodependent species may act as bioproxies for our own history [10]. The distributions of human-commensals are largely linked to human activity and their evolutionary history should reflect large-scale human movements. House mice populations of the northern and western British Isles harbour an mtDNA lineage that is also present in Norway, suggesting mice were likely transported to the regions as stowaways on Norwegian Viking ships [5]. Analysis of some pest species has also shed light on human evolution; divergence in lice lineages reflect splits in the Homo genus [11] and genetic diversity in Helicobacter pylori reflects human migrations from prehistory to the modern era [12].
The house sparrow (Passer domesticus) is a ubiquitous human-commensal bird species occupying cities and farmland where it feeds on food waste and crops. Its native range covers Western and Central Eurasia; however, due to deliberate and accidental introductions by humans, its current distribution also encompasses Southern Africa, Australia, New Zealand and the Americas. It is strongly associated with human settlements with a clearly human associated ecology; the species is known to go locally extinct in abandoned settlements [13,14]. The house sparrow is a well-studied model for quantitative genetics [15,16] and also for adaptation to urban environments [17,18]. However, we know surprisingly little about the evolutionary history of this charismatic companion species.
House sparrow human-commensalism is thought to have arisen once in the Middle and Near East with the Neolithic revolution [19]. The species likely spread with the subsequent introduction of agriculture and establishment of fixed settlements in Europe [13]. This is also thought to have played an instrumental role in the hybrid origin of the Italian sparrow (P. italiae) in the Mediterranean region as a result of admixture with the Spanish sparrow (P. hispaniolensis), which was probably already present in Southern Europe ([20–24], figure 1a). Furthermore, genomic evidence suggesting multiple, independent hybrid speciation events is consistent with a stepwise introduction of agriculture via Mediterranean islands [25,26].
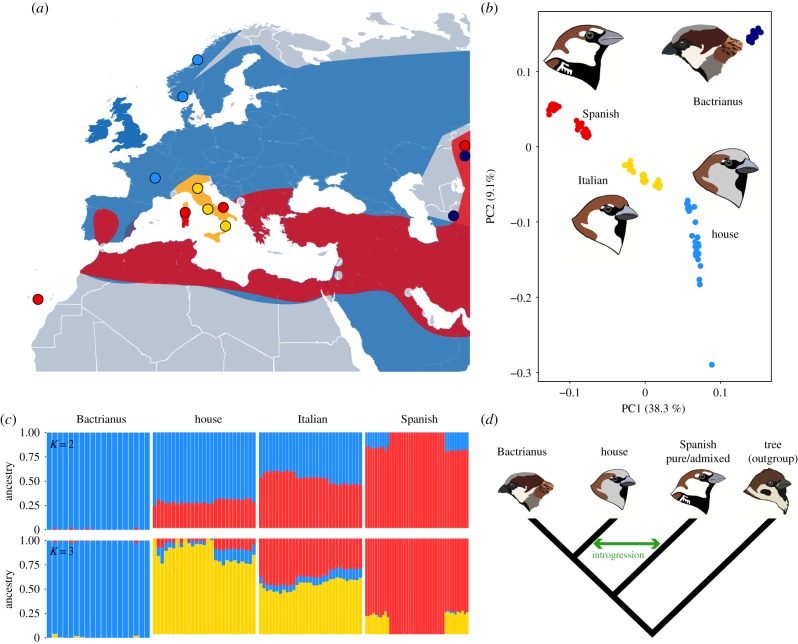
Figure 1.
(a) Sample sites for Eurasian sparrow species, point colours correspond to species shown in (b). Shading represents distribution of house (blue), Italian (yellow) and Spanish (red); darker red indicates house Spanish overlap. (b) Principal component analysis of high-quality, linkage pruned SNPs separate species along PC1. (c) Population clustering using ADMIXTURE for two best supported values of K. (d) Schematic of the four-population test used to calculate genome-window measures of house–Spanish sparrow introgression (fd).
Intriguingly, a house sparrow subspecies—P. d. bactrianus (hereafter Bactrianus sparrow)—occurs in the Middle East and Central Asian steppes (figure 1a). Bactrianus sparrows are ecologically unlike European house sparrows—they migrate, are less bold, and are not associated with human settlements [13,19]. Furthermore, Bactrianus skull and beak morphology are less robust compared to the more human-associated house sparrow [27]. This difference is consistent with divergent foraging—i.e. Bactrianus sparrows mainly feed on natural grass seeds, whereas the house sparrow is expected to have adapted to feeding on tougher seeds from domesticated crops [27]. Taken together, this suggests the Bactrianus sparrow represents a branch of the house sparrow lineage that diverged prior to the evolution of human-commensalism. If so, a comparative approach using both European house and Bactrianus sparrows may shed light on how the ubiquitous house sparrow became associated with humans.
Here we examine population genomic variation in three Eurasian Passer sparrow species, the Spanish, Italian and house sparrow; we also include non-commensal house sparrow lineage, the Bactrianus sparrow. We first investigate population genomic structuring and evolutionary relationships among species, testing for evidence of admixture across the European distribution. We then use coalescent modelling to infer demographic history, testing the split date between the house and Bactrianus sparrow and the timing of house sparrow population expansion. Finally, we perform a comparative genome scan between the house and Bactrianus sparrow, revealing strong signatures of divergent selection between these two P. domesticus lineages that points to several intriguing candidate genes that may have played a role in adaptation to a human niche.
2. Methods
(a). Study system
Three main species of Passer occur in Europe and Western Asia. The house sparrow is the most widely distributed and is found across Europe and the Middle East (figure 1a) [13]. The wild-like Bactrianus sparrow, one of these subspecies, occurs largely in Iran and Kazakhstan [19]. The Spanish sparrow is found across Southern Europe, Northern Africa and the Middle East and Central Asia where it co-occurs with Bactrianus (figure 1a). The hybrid Italian sparrow occurs in the Italian peninsula and also on some Mediterranean islands (figure 1a) [20,25]. A fourth, more divergent species, the tree sparrow (P. montanus) also occurs in Eurasia, in lower densities than the house sparrow [13].
(b). Sample collection
We collected house (n = 46), Spanish (n = 43), Italian (n = 31) and Bactrianus (n = 19) sparrows from across Eurasia (figure 1a) using mist nets at each sampling location. Blood samples were taken from captured individuals. See electronic supplementary material, table S1 for a breakdown of all samples and sampling locations. Blood was immediately stored in Queen's lysis buffer for preservation and individuals were released quickly, without harm, to minimize stress. In all cases, sparrows were collected under appropriate permissions and guidelines.
(c). DNA extraction and sequencing
We extracted 5–95 ng µl−1 of genomic DNA from blood samples using a Qiagen Blood & Tissue DNEasy kit (Qiagen, California, USA). Extracted DNA was prepared for sequencing using an Illumina TruSeq gDNA 180 bp kit (Illumina, California, USA). Sequencing was conducted on Illumina Hi-Seq 2000 and Illumina Hi-Seq X machines at Genome Quebec, McGill University, Canada.
(d). Read alignment, variant calling and filtering
Raw reads were trimmed and filtered for Illumina adapters using Trimmomatic 0.36 [28]. Base calls at the start and end of reads with a Phred quality score of less than 5 were removed (i.e. less than 70% base call accuracy) to account for errors with increasing read length. Reads with an average quality of less than 10 (i.e. less than 90% base call accuracy) across 5 bp step windows were removed to ensure high-confidence base calls. Trimmed and filtered reads were then aligned to the house sparrow reference genome [20] using bwa 0.7.10 [29]; both paired and unpaired reads were mapped separately and then merged to produce a final binary alignment map (bam) for each individual. Bams were then sorted, marked for duplicates and indexed using Picard 2.7.1 (https://github.com/broadinstitute/picard). Mapping sequence data from different species to a divergent genome are a potential source of bias for SNP calling. However, bird genomes are highly conserved, allowing efficient mapping across large evolutionary distances [30]. Indeed, mean total read mapping percentage was high (91.8% for all species), with a marginally lower mean mapping percentage in the Italian sparrow (90.3%; ANOVA, R2 = 0.13, F3, 125 = 7.63, p < 0.0001; see electronic supplementary material, figure S1).
Bams were realigned around insertion–deletion polymorphisms to prevent false positive variant detection; calls were then made for all sites (variant and invariant) using the GATK (2.7.1) HaplotypeCaller [31]. The raw vcf was filtered to remove all indels and annotated with filter thresholds. Additional filters were applied to create two, high-quality datasets for downstream analyses with different data requirements. The first dataset (hereafter ‘variant only’) included only polymorphic, biallelic SNPs occurring in at least 80% of individuals, with minimum site and genotype quality scores of 20, a mean site depth of between 10–40X. We additionally masked all genotypes with a depth below 5× and above 60×. We applied three different minor allele thresholds—no threshold (for demographic analyses using the SFS), 0.02 and 0.05. The second dataset was filtered for the same criteria but included calls at all sites—i.e. variant and invariant—and with no MAF thresholds (hereafter ‘all sites’). The invariant sites in this dataset were necessary to calculate sequence-based statistics such as dXY and fd (see Genome scans). All filtering was conducted using vcftools 0.1.13 [32] and bcftools 1.1 [33] with scripts available at www.github.com/markravinet and Dryad [34].
(e). Population structure analysis
To investigate population structure, we performed linkage pruning on the MAF 0.05 filtered variant only dataset using PLINK 1.9 [35], filtering for all loci within 100 Kb windows with an r2 exceeding 0.1, the baseline for genome wide linkage-disequilibrium (LD) [20]. PCA was then performed for allele frequencies of autosomal SNPs only using PLINK 1.9. For a model-based analysis of population structure on autosomal SNPs, we used admixture 1.3 [36], setting the number of assumed populations (K) between 1 and 6 and using leave-one-out cross-validation in order to determine the best supported value. Scripts for population structure analyses are available at www.github.com/markravinet and Dryad [34].
(f). Genome scans for signatures of selection and introgression
In order to identify genomic regions under selection, we first estimated FST, a relative measure of differentiation in 100 Kb sliding windows with a 25 Kb step using vcftools from our MAF 0.02 filtered variant only dataset. We additionally estimated dXY, an absolute measure of divergence for the same windows using popgenWindows.py (https://github.com/simonhmartin/genomics_general), [37] on our all sites dataset.
Since both FST and dXY are prone to biases, particularly with regard to genome-wide recombination rate variation, we also calculated long-range haplotype (LRH) estimates that incorporate information on linkage disequilibrium and recombination rate variation [38]. These tests are specifically designed to detect the increase in haplotype homozygosity around a target of selection during a selective sweep [38]. Since this requires haplotypes, we statistically phased the MAF = 0.02 threshold variant-only dataset. Bactrianus, house and Spanish sparrow individuals were all phased individually with ShapeIt2 [39]. For all autosomes, a previously published linkage map [20] was used to inform phasing—we did not include the Z chromosome. LRH and extended haplotype homozygosity statistics (specifically the integrated haplotype homozygosity score (iHS), cross-population extended haplotype homozygosity (xpEHH) and site specific extended haplotype homozygosity (EHHS)) were calculated on the phased variant only data using the R package rehh [40]. EHHS is a measure of the extent of haplotype homozygosity surrounding SNP potentially under selection, iHS is a derivation of EHHS, using the area under a curve of EHHS to detect positive selection; xpEHH compares haplotype lengths between populations and identifies regions where a selective sweep has proceeded to fixation in at least one lineage [38]. We note here that for visualization of genome-wide LRH statistics for greater than 1 million SNPs, figures are drawn using multiple randomly sampled subset (20%) of the full dataset in order to improve plotting efficiency. Scripts are available at www.github.com/markravinet and Dryad [34].
Our initial population structure analyses revealed evidence of admixture between house and Spanish lineages in Europe. We investigated fine-scale patterns of introgression within the house genome using a four population test to calculate fd—i.e. the proportion of introgressed sites within a genome window [37]. Using the test tree topology (((Bactrianus, house), Spanish), Tree), high levels of fd represent an enrichment of shared sites between house and Spanish sparrows (figure 1d). We calculated fd three separate times using all Spanish sparrows, using ‘pure’ Spanish sparrows and using Spanish sparrows showing evidence of admixture with European house sparrows. We note that our clustering of Spanish sparrows with admixture into a single group is arbitrary—i.e. they are instead likely to have experienced independent admixture events. Nonetheless, we used this grouping to demonstrate that these populations have undergone introgression and to test whether gene flow has occurred in secondary contact (see Demographic inference). Analyses were performed on 100 Kb sliding windows with a 25 Kb step using ABBABABAwindows.py (https://github.com/simonhmartin/genomics_general) on our all sites dataset [37].
(g). Demographic inference
To shed light on the evolutionary origins of Eurasian Passer sparrow lineages, we performed maximum likelihood based demographic inference using the site-frequency spectrum with fastsimcoal2 [41]. Because of the complexities of modelling hybrid origin and also the possibility that Italian sparrows have arisen from several independent hybridization events [25], we focused only on the house, Bactrianus and Spanish populations. Spanish sparrows were split into two further subsets—one with evidence of admixture with the house (Spanish admix) and one without (Spanish pure). This is an artificial split to simplify our demographic modelling approach; in reality, we expect that house–Spanish admixture is likely to have occurred independently in different locations. We derived the folded four-population multidimensional observed SFS using easySFS.py (https://github.com/isaacovercast/easySFS) from a set of high quality autosomal SNPs present in greater than 95% of individuals (from our variant only dataset) with no MAF filtering. Since sample sizes varied between each of the populations, we projected the SFS down to 30 chromosomes—i.e. 15 diploid individuals per population.
We tested three main models of divergence between sparrow species—isolation, migration and admixture (see electronic supplementary material, figure S7). For the isolation model, all species/populations diverged in the absence of gene flow (electronic supplementary material, figure S7). Under this model, contemporary evidence of admixture is due to incomplete lineage sorting. For the migration model, species/populations diverged in the face of interspecific gene flow throughout their divergence history (electronic supplementary material, figure S7). In the admixture model, interspecific gene flow occurs as a result of a pulse of admixture following divergence. Since more realistic models of gene flow can improve model performance [42], we also ran versions of the models with intraspecific gene flow included (electronic supplementary material, figure S7).
For all models, we drew priors from a loguniform distribution (see electronic supplementary material, table S2 for a full description of priors). Since we were not interested in divergence time between admixed and pure Spanish lineages, we fixed this to 10 000 generations, assuming population structuring as a result of postglacial range expansion; this also allowed us to constrain admixture and population expansion events to having occurred within this time. The split between house and Spanish sparrows was allowed to vary between 100 000 and 2 million generations. We also set priors so that house/Bactrianus divergence occurred 10 000 to 100 000 generations in the past (see electronic supplementary material, table S2).
For each model, we performed 100 independent runs of 100 000 coalescent simulations to estimate the maximum likelihood. We then performed model selection using the run with the highest likelihood for each of the models. Following Meier et al. [42], we used AIC for model selection and assessed the likelihood distribution for each model by calculating the likelihoods of 100 expected site frequency spectra, derived from 1 000 000 coalescent simulations. We derived 95% confidence intervals for parameters estimated from our models using non-parametric block bootstrapping [42]. To achieve this, we split the genome into 3204 1 Mb windows and resampled windows with replacement until the bootstrapped SFS was the same size as the observed. We created 100 bootstrapped site-frequency spectra and then performed 10 independent runs of likelihood estimation on them. Estimates from each of the best runs were then used to derive the 95% confidence intervals around all parameter estimates from our focal model.
(h). Candidate gene identification and gene ontology analysis
To identify potential candidate genes for adaptation to a human-commensal lifestyle, we looked for evidence of positive selective sweeps that have occurred in the house but not the Bactrianus sparrow. We set our threshold to identify outliers using the xpEHH statistic at a log10 p-value of 6. Using the Benjamani & Hochberg procedure for FDR correction, this is equivalent to a Q-value of 0.001—i.e. that in 1000 outliers, only one is expected to be a false positive. This threshold, although conservative, allowed us to identify SNPs exhibiting clear signatures of divergent selection. Using a custom R script (available at www.github.com/markravinet and Dryad [34]) we then clustered outlier SNPs occurring within 100 Kb of one another to produce a dataset of outlier regions occurring across the genome. We then identified all known genes within 250 Kb of the peaks of these outlier regions. Retaining only unique gene IDs we then performed gene ontology analysis using clueGO in Cytoscape 3.6.0 [43] using a human gene set, medium network specificity, a right-sided hypergeometric test, Benjamani & Hochberg FDR correction and a p < 0.05. In addition to GO analysis, we examined the identity of genes in outlier regions manually.
3. Results
(a). Population structure and differentiation
After mapping to the house sparrow reference genome, calling and filtering variants, we retained 21 930 880 SNPs. We used a subset of LD pruned 178 268 biallelic SNPs with an MAF > 0.05 for parametric (ADMIXTURE) and non-parametric (PCA) inference of population structure.
Species are clearly separated along the first principal component (38.3% PVE) with the Italian sparrow occurring intermediate between Spanish and house sparrows, consistent with hybrid origin (figure 1b). Interestingly, the Bactrianus sparrow is displaced on both PC1 and PC2 (9.1% PVE), forming a separate cluster from European house populations (figure 1b)—but remaining closer to the European house than the Spanish sparrow. For Spanish, house and Italian sparrows, within-species population structure is also apparent from the PCA (electronic supplementary material, figure S2).
ADMIXTURE analysis strongly supported K values of 2 and 3 (figure 1c; electronic supplementary material, figure S3). For K = 2, Bactrianus and Spanish sparrows form clear, separate ‘pure’ clusters and the Italian sparrow is clearly admixed, as expected for a hybrid species. However, European populations of both house and Spanish sparrows show evidence of introgression, which is also apparent from PC1 (electronic supplementary material, figure S1). For K = 3, house sparrows form a separate third cluster but retain a signature of Spanish and Bactrianus ancestry (figure 1c).
Mean genome-wide FST (±s.d.) estimates also support a Bactrianus lineage divergent from both the house (0.103 ± 0.075) and Spanish sparrow (0.326 ± 0.213; see also electronic supplementary material, figure S4). Pairwise mean FST between the Bactrianus and the European house is half that between the house and the Spanish sparrow. This is also apparent from the genome-wide distribution of FST between the house and Bactrianus—peaks of differentiation are apparent but in general differentiation is relatively low in comparison to either with the Spanish sparrow (electronic supplementary material, figure S5). Differentiation was also lower between European house and putatively admixed Spanish sparrows (0.166 ± 0.171) than pure Spanish populations from Italy and Kazakhstan (0.234 ± 0.208; p < 2.2 × 10−16, permutation test: electronic supplementary material, figure S4)—providing evidence of house–Spanish introgression upon secondary contact. Absolute genomic divergence between species, measured using dXY, was similarly higher between house/Bactrianus and the pure Spanish populations compared to the admixed (p < 2.2 × 10−16, permutation tests; see electronic supplementary material, figure S6). Finally, fd (±s.d.) was higher when the third population on the test phylogeny was set to the Spanish admixed populations (0.38 ± 0.17; figure 1d) compared to the Spanish pure (0.35 ± 0.16; see electronic supplementary material, figure S6)—again supporting house–Spanish admixture in Europe.
(b). Demographic inference
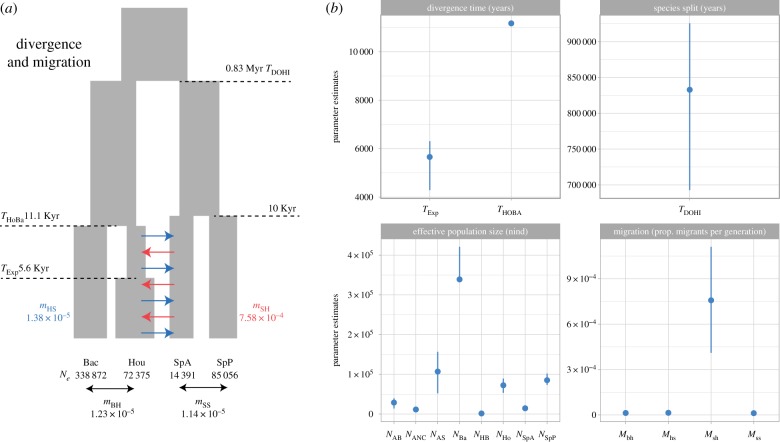
Our demographic analysis clearly rejected scenarios where no gene flow occurred between the house and Spanish sparrow (electronic supplementary material, figures S7 and S8). The best-supported model using log-likelihood and AIC was one of divergence with migration via secondary contact between the house and Spanish sparrow (figure 2a; electronic supplementary material, table S3 and figure S8). Under this model the two Passer species diverged 0.83 million years BP (0.69–0.93 mya 95% confidence intervals), assuming a single year generation time. Divergence between the Bactrianus and house sparrow was much more recent, occurring 11.1 Kya ago, whereas an expansion in the house lineage occurred 5657 generations (4292–6308 85% CI; figure 2a,b) in the past. Migration rate estimates also indicated introgression from the Spanish into the house was greater than gene flow in the opposite direction or within species (figure 2b).
Figure 2.
Site-frequency-spectrum based coalescent analyses supports a model of divergence in isolation with house–Spanish migration upon secondary contact (a). Parameter estimates of divergence time, the species split date between the house and Spanish sparrow, effective population sizes and migration rates and their associated 95% CI, derived from non-parametric block bootstraps (b). Parameter abbreviations are TDOHI = timing of house/Spanish divergence, THOBA = timing of house/Bactrianus divergence, TEXP = timing of population expansion in house lineage, mHS = migration house to Spanish, mSH = migration Spanish to house, mBH = migration Bactrianus to house, mSS migration between Spanish admixed (SpA) and Spanish pure (SpP) populations, NAB = ancestral Bactrianus Ne, NANC = ancestral Ne, NAS = ancestral Spanish Ne, NBa = Bactrianus Ne, NHB = house/Bactrianus ancestral Ne, NHO = house Ne, NSPA = Spanish admixed Ne, NSpP = Spanish pure Ne.
(c). Divergent selection between house and Bactrianus sparrows
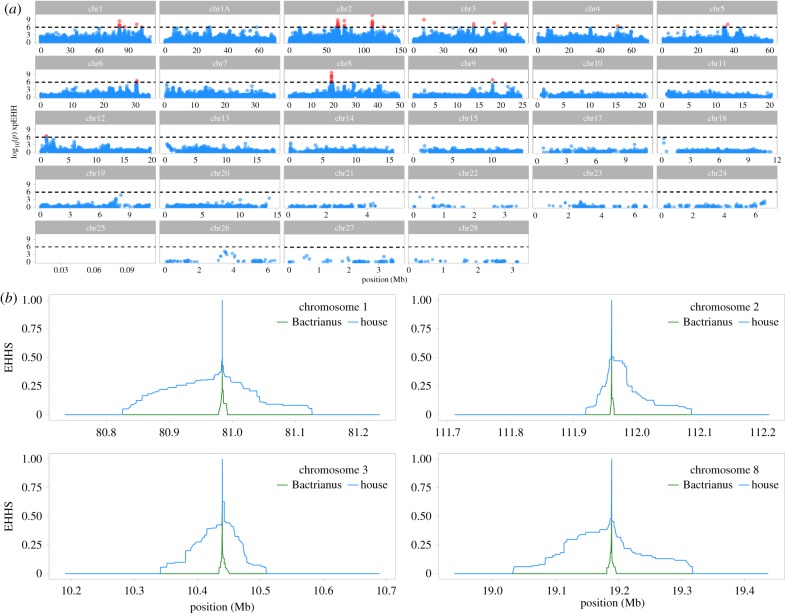
Divergence between house and Bactrianus sparrows is consistent with the onset of the Holocene; suggesting the latter is potentially a relict of the pre-human commensal house ancestor. A comparative genome scan between these species will help identify genome regions that may have played a role in adaptation to a human niche by the house sparrow. Examining only the house sparrow genome shows signatures of apparent strong selection on the majority of autosomes (electronic supplementary material, figure S9). However, cross-population long-range haplotype statistics also point to multiple regions throughout the genome exhibiting signatures of putative divergent selection between the house and Bactrianus sparrow, including the three highest peaks on chromosomes 1, 8 and 2 respectively (figures 3a,b and 4). Peaks are apparent on multiple autosomes and are not an artefact of increased baseline differentiation between the house and Bactrianus sparrows (electronic supplementary material, figure S5).
Figure 3.
Signatures of divergent selection between house and Bactrianus sparrows. (a) A genome-wide plot of xpEHH shows clear peaks of divergent haplotype homozygosity occurring throughout the genome, with particularly large peaks on chromosomes 1, 2, 3 and 8. Horizontal dashed line represents threshold of significance (log10 1 × 10−6) for outlier SNPs, red points indicate outlier SNPs. N.B: the full dataset has been randomly downsampled to 20% to aid plotting and visualization. (b) Examination of the four SNPs with highest xpEHH scores across on the chromosomes containing 63% of the xpEHH peaks shows a clear signature of site-specific extended haplotype homozygosity (EHHS) in the house (blue lines) but not the Bactrianus sparrow (green lines).
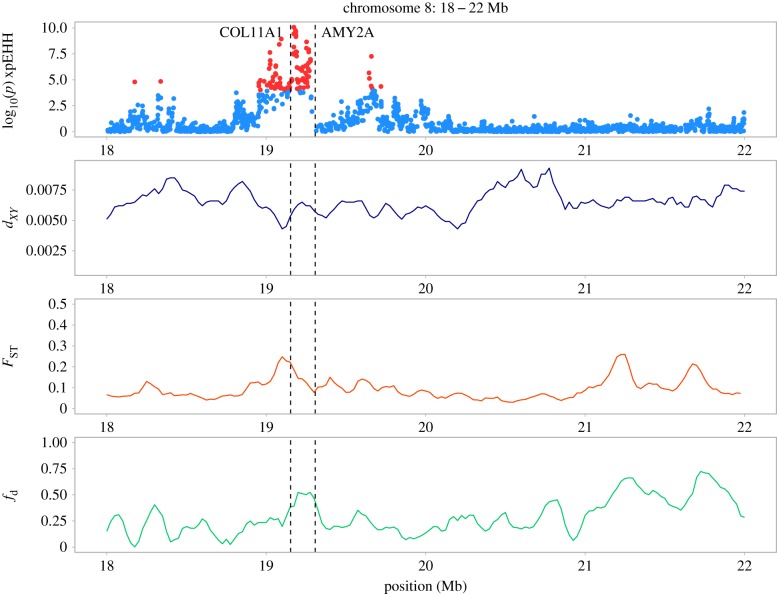
Figure 4.
A closer look at genomic divergence between house and Bactrianus sparrows along chromosome 8; top panel—log10 (p) xpEHH (blue = background, red = outliers where p < 0.0001); second panel—mean absolute nucleotide divergence (dXY) between house and Bactrianus; third panel—relative differentiation (FST) between house and Bactrianus; fourth panel—proportion of putatively introgressed sites per 100 Kb window (fd) between the house and Spanish sparrow.
To further investigate these putative signatures of selection, we identified 705 outlier SNPs (0.06% of 1 033 861 total SNPs analysed) where the log10 p-value of xpEHH between house and Bactrianus was greater than 6 (i.e. points coloured red in figure 4 and electronic supplementary material, figure S8). This cut-off, although conservative, represents a false discovery rate of 0.001 and only identifies SNPs with the strongest signatures of divergent selection. We grouped outlier SNPs occurring within 100 Kb of one another to identify 59 peak outlier regions across the genome (i.e. figure 4). The majority (63%) of these 59 outlier peaks fell on chromosomes 1, 2, 3 and 8; chromosomes 1 and 8 also harboured the SNPs with the highest signatures of divergent selection (see electronic supplementary material, tables S4 and S5). Using the highest outlier SNPs in each of the major peaks on these chromosomes, EHHS values show a clear pattern of increased extended haplotype homozygosity in the house but not the Bactrianus sparrow (figure 3b). In some cases, regions of increased FST corresponded to extended haplotype homozygosity and slight peaks of fd (figure 4). Overall this suggests positive selective sweeps have occurred at these regions in the house sparrow genome only and may point towards potential candidate genes involved in commensalism.
(d). Candidate gene and gene ontology analysis
We identified 153 unique genes falling within 250 Kb of the 59 xpEHH outlier peaks from across the genome. GO analysis identified 20 gene pathways with evidence of enrichment among the outlier gene set (see electronic supplementary material, table S6). These included cartilage condensation (p = 0.006) and regulation of circadian rhythm (p = 0.023 after FDR correction). A gene of interest from the cartilage condensation pathway is wnt7a on chromosome 12; this has previously been linked to feather development and melanogenesis in birds [44,45]. Additionally, a PARL transcript (presenilin associated rhomboid-like) on chromosome 9 is upregulated during migration in white-crowned sparrows (Zonotrichia leucophyrs) [46].
One of the highest xpEHH peaks in the genome occurred on chromosome 8 between 19.01–19.27 Mb (figures 3b and 4). This peak contains two known genes, COL11A and AMY2A (figure 4). COL11A—collagen type XII alpha—is associated with craniofacial development; mutations in this gene for humans result in Marshall's syndrome which is characterized by increased skull thickness and altered facial structure [47]. AMY2A—amylase alpha2 is part of the amylase gene family associated with adaptation to a higher starch diet in both humans and dogs [48,49].
4. Discussion
Our findings show that a high level of genomic divergence matches the phenotypic, behavioural and ecological differences between the European house and Bactrianus sparrows. Pairwise mean FST between the house and Bactrianus subspecies is half of that between the house and Spanish sparrow (electronic supplementary material, figure S4). High differentiation is unlikely to be a factor of distance between the house and Bactrianus; Spanish sparrow populations are similarly spatially isolated but show no evidence of population structuring. Instead, high divergence between the house lineages is likely due to the fact that they split 11 Kya ago. Divergence may have occurred prior to the widespread dissemination of agriculture and the evolution of commensalism in the house sparrow.
(a). Demography and gene flow in Eurasian Passer sparrows
Reanalysis of Eurasian sparrow genomic data alongside the Bactrianus has developed our understanding of population genomic structuring among these species. Admixture between the house and Spanish sparrow has played an important role in the hybrid origin of the Italian sparrow. We show that European populations of house and Spanish sparrows have experienced some level of admixture. Although our grouping of Spanish admixed populations is artificial, it still provides evidence of introgression. Our demographic inference suggests that gene flow has been ongoing since secondary contact, and as Spanish sparrows were already likely present in Southern Europe at the time the house sparrow was introduced alongside agriculture, this is a likely explanation. However, gene flow is still on-going and house × Spanish hybrids still occur in some regions—i.e. Spain and North Africa. The proportion of Spanish ancestry in the house appears to be tentatively lower in more northerly populations sampled in Norway, compared to those in France (K = 3, figure 1c). However, it is also possible that house/Spanish introgression is quite old; i.e. it could have occurred in the Middle and Near East where both species co-occur, prior to the expansion of the house into Europe. Concordance between signatures of divergent selection between the house and Bactrianus and measures of Spanish introgression into the house is consistent with this explanation. Most likely, Spanish ancestry in the house genome is due to both mechanisms, but more work is now necessary to distinguish them and quantify their relative importance.
The Bactrianus sparrow may have diverged from the main house lineage either prior to or as a result of the evolution of human-commensalism during the Neolithic revolution. The subspecies migrates, does not associate with human settlements and is less bold [13]—all traits that are common to non-human commensal sparrow species such as the Spanish. Bactrianus sparrows can therefore be considered a proxy for the ancestral, pre-commensal house sparrow. With the rapid expansion and spread of house sparrow populations following the invention of agriculture, it is likely the Bactrianus has remained a relict lineage confined to the Central Asian steppes. This is also supported by evidence of limited interbreeding between co-occurring house and Bactrianus sparrows in Kazakhstan [50,51]. It is possible that the Bactrianus sparrow is not truly wild, but feral, similar to Przewalski's horse [9]; we think this is unlikely given a divergence time consistent with the onset of the Neolithic revolution. Regardless, comparing the two lineages offers us a unique opportunity to look for signatures of strong positive selection that have occurred in the house lineage and that are absent from the Bactrianus—i.e. a comparative genome scan for adaptation to a human niche.
(b). Genomic signatures of selection in the house sparrow
Our comparative approach does indeed reveal multiple regions throughout the sparrow genome that show signatures of house-specific positive selective sweeps. We recognize that not all evidence of divergent selection between the house and the Bactrianus sparrow will be due to adaptation to an anthropogenic niche; nonetheless we argue that these are important candidate regions to follow up for further investigation. Genome scan approaches are prone to potential bias such as false positives due to recombination rate variation and demographic history [52]. However, our use of robust long-range haplotype tests that incorporate information on linkage disequilibrium and haplotype homozygosity means that we are able to identify strong patterns of selection which are not apparent using more standard measures such as FST [38]. Furthermore, our conservative outlier threshold means we have a very low FDR (0.001%) and higher confidence that the signatures of selective sweeps we have identified are likely to be genuine.
The sweep regions we detected harbour a number of candidate genes consistent with many of the phenotypic traits that are known to be divergent between house and non-commensal Bactrianus sparrows, such as plumage, migratory behaviour, dietary differences and skull morphology. An xpEHH peak on chromosome 12 is in close proximity to wnt7a, a gene that has previously shown to be involved in feather development and melanogenesis in birds [44,45]. This gene is also under apparent divergent selection between island populations of the Italian sparrow which differ in the darkness of their plumage [25]. Bactrianus sparrows too have a darker plumage than the house. A further peak on chromosome 9 harbours the PARL gene, which shows increased expression in the brains of white-crowned sparrows (Zonotrichia leucophyrs) during the migratory season [46]. This is also bolstered by our finding of regulation of circadian rhythm as an enriched GO pathway; importantly, Bactrianus sparrows migrate whereas house sparrows do not [13,19].
One of the strongest peaks of divergent selection sits on chromosome 8 and contains the COL11A and AMY2A genes (figure 4). COL11A is closely associated with craniofacial development in humans and is linked to Marshall's syndrome, a genetic disorder which leads to increased skull thickness and abnormal facial structure [47]. Skull morphology and craniofacial structure has be shown to differ between Bactrianus and house sparrows, with the latter exhibiting a more robust skull morphology and larger beak [27]. Craniofacial differences are further supported by an enrichment of cartilage condensation in our GO analysis. The shift in skull and beak morphology between sparrow lineages is commonly attributed to the dietary shift from natural seeds to agricultural food waste during the development of commensalism [27]. In contrast, introduction of human food resources appears to have weakened bite force and altered beak morphology in Darwin's finches [53]. Intriguingly, AMY2A, is part of a family of amylase genes that have been linked to the transition to starch-based diets in both humans and dogs during the Neolithic revolution [46,48]. In dogs, increased copy number of the closely related AMY2B gene is consistent with the spread of agriculture during this period [49,54,55]. Among human populations, AMY1 copy number is associated with dietary starch content and the frequency of AMY2A deletions is higher in groups with a low starch diet [48,56]. Our findings therefore add to the emerging picture that the Neolithic revolution introduced a common selective pressure that has resulted in parallel adaptations in similar genes for three very different taxa—humans, dogs and potentially, house sparrows.
At present, it is not clear whether AMY2A or COL11A or both genes are the target of selection at this region of the genome. However, since both genes occur just 154 Kb from one another, they may remain in linkage disequilibrium as a co-adapted gene complex. It is now necessary to investigate whether this is the case and to clearly test whether these genes are involved in adaptation to a human niche in house sparrows. Furthermore, determining the age of the selective sweep and testing whether selection is also apparent in the Italian sparrow is now necessary to conclusively link this adaptation to the onset of the Neolithic revolution. Nonetheless, our current findings place the origins of commensalism in house sparrows in an evolutionary context and show that understanding how this species came to be is informative for our understanding of our own recent evolutionary history.
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to Daniel Wegmann for his advice on designing demographic models and Joana Meier for her assistance with running SFS analysis. We thank Camilla Sætre, Juan Carlos Illera Cobo, Sonia Ramos, Caroline Øien Guldvog, Jo Skeie Hermansen, Stein Are Sæther, Richard Bailey, Fabrice Eroukhmanhoff, Anna Runemark, Sepand Riyahi, Almat Abayev and Syrymgul Zaripova for assistance with fieldwork. We are grateful to Fabrice Eroukhmanhoff for his comments on a draft version of this manuscript and to two anonymous reviewers and the editors for their constructive comments. Data analysis was conducted on the UiO Abel cluster [Norwegian Metacenter for High Performance Computing (NOTUR) and the University of Oslo] operated by the Research Computing Services group at The University Center for Information Technology (www.hpc.uio.no/) and the DDBJ Supercomputer at the National Institute of Genetics, Mishima, Japan.
Data accessibility
Scripts and data for analysis are available on Dryad: http://dx.doi.org/10.5061/dryad.mq25897 [34]. Whole genome resequencing data are available at the NCBI Sequence Read Archive, BioProject PRJNA255814 accession numbers SRR5369936-SRR5369966 and the European Nucleotide Archive with study accession number PRJEB27649 and sample accessions ERS2604996-ERS2605092.
Authors' contributions
M.R., T.O.E. and G.-P.S. designed the study. T.O.E., C.T. and M.R. conducted fieldwork and prepared samples for sequencing. G.-P.S., M.A. and A.G. organized sampling and conducted fieldwork. M.R. analysed the data. M.R. and T.O.E. wrote the manuscript. All authors commented on the manuscript and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by grants 240557 and 204523 to G.-P.S. from the Research Council of Norway.
References
- 1.Hulme-Beaman A, Dobney K, Cucchi T, Searle JB. 2016. An ecological and evolutionary framework for commensalism in anthropogenic environments. Trends Ecol. Evol. 31, 1–13. ( 10.1016/j.tree.2016.05.001) [DOI] [PubMed] [Google Scholar]
- 2.Vigne J-D. 2015. Early domestication and farming: what should we know or do for a better understanding? Anthropozoologica 50, 123–150. ( 10.5252/az2015n2a5) [DOI] [Google Scholar]
- 3.Zeder MA. 2015. Core questions in domestication research. Proc. Natl Acad. Sci. USA 112, 3191–3198. ( 10.1073/pnas.1501711112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Combs M, Puckett EE, Richardson J, Mims D, Munshi-South J. 2017. Spatial population genomics of the brown rat (Rattus norvegicus) in New York City. Mol. Ecol. 27, 83–98. ( 10.1111/mec.14437) [DOI] [PubMed] [Google Scholar]
- 5.Searle JB, et al. 2009. Of mice and (Viking?) men: phylogeography of British and Irish house mice. Proc. R. Soc. B 276, 201–207. ( 10.1098/rspb.2008.0958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray MM, Wegmann D, Haasl RJ, White MA, Gabriel SI, Searle JB, Cuthbert RJ, Ryan PG, Payseur BA. 2014. Demographic history of a recent invasion of house mice on the isolated Island of Gough. Mol. Ecol. 23, 1923–1939. ( 10.1111/mec.12715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson MTJ, Munshi-South J. 2017. Evolution of life in urban environments. Science 358, eaam8327 ( 10.1126/science.aam8327) [DOI] [PubMed] [Google Scholar]
- 8.Larson G, Fuller DQ. 2014. The evolution of animal domestication. Annu. Rev. Ecol. Syst. 45, 115–136. ( 10.1146/annurev-ecolsys-110512-135813) [DOI] [Google Scholar]
- 9.Gaunitz C, et al. 2018. Ancient genomes revisit the ancestry of domestic and Przewalski's horses. Science 360, 111–114. ( 10.1126/science.aao3297) [DOI] [PubMed] [Google Scholar]
- 10.Jones EP, Eager HM, Gabriel SI, Jóhannesdóttir F, Searle JB. 2013. Genetic tracking of mice and other bioproxies to infer human history. Trends Genet. 29, 298–308. ( 10.1016/j.tig.2012.11.011) [DOI] [PubMed] [Google Scholar]
- 11.Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH. 2004. Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2, e340-12 ( 10.1371/journal.pbio.0020340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falush D, et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585. ( 10.1126/science.1080857) [DOI] [PubMed] [Google Scholar]
- 13.Summers-Smith D. 1988. The sparrows. Calton, UK: T & AD Poyser. [Google Scholar]
- 14.Anderson TR. 2006. Biology of the ubiquitous house sparrow. Oxford, UK: Oxford University Press. [Google Scholar]
- 15.Jensen H, Jensen H, Sæther BE, Ringsby TH, Tufto J, Griffith SC, Ellegren H, Ellegren H. 2003. Sexual variation in heritability and genetic correlations of morphological traits in house sparrow (Passer domesticus). J. Evol. Biol. 16, 1296–1307. ( 10.1046/j.1420-9101.2003.00614.x) [DOI] [PubMed] [Google Scholar]
- 16.Ben Cohen S, Dor R. 2018. Phenotypic divergence despite low genetic differentiation in house sparrow populations. Sci. Rep. 8, 394 ( 10.1038/s41598-017-18718-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw LM, Chamberlain D, Evans M. 2008. The house sparrow Passer domesticus in urban areas: reviewing a possible link between post-decline distribution and human socioeconomic status. J. Ornithol. 149, 293–299. ( 10.1007/s10336-008-0285-y) [DOI] [Google Scholar]
- 18.Swaileh KM, Sansur R. 2006. Monitoring urban heavy metal pollution using the house sparrow (Passer domesticus). J. Environ. Monit. 8, 209–213. ( 10.1039/B510635D) [DOI] [PubMed] [Google Scholar]
- 19.Saetre GP, et al. 2012. Single origin of human commensalism in the house sparrow. J. Evol. Biol. 25, 788–796. ( 10.1111/j.1420-9101.2012.02470.x) [DOI] [PubMed] [Google Scholar]
- 20.Elgvin TO, Trier CN, Tørresen OK, Hagen IJ, Lien S, Nederbragt AJ, Ravinet M, Jensen H, Sætre G-P. 2017. The genomic mosaicism of hybrid speciation. Sci. Adv. 3, e1602996 ( 10.1126/sciadv.1602996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elgvin TO, Hermansen JS, Fijarczyk A, Bonnet T, Borge T, Saether SA, Voje KL, Sætre G-P. 2011. Hybrid speciation in sparrows II: a role for sex chromosomes? Mol. Ecol. 20, 3823–3837. ( 10.1111/j.1365-294X.2011.05182.x) [DOI] [PubMed] [Google Scholar]
- 22.Hermansen JS, Saether SA, Elgvin TO, Borge T, Hjelle E, Sætre G-P. 2011. Hybrid speciation in sparrows I: phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol. Ecol. 20, 3812–3822. ( 10.1111/j.1365-294X.2011.05183.x) [DOI] [PubMed] [Google Scholar]
- 23.Hermansen JS, Haas F, Trier CN, Bailey RI, Nederbragt AJ, Marzal A, Sætre G-P. 2014. Hybrid speciation through sorting of parental incompatibilities in Italian sparrows. Mol. Ecol. 23, 5831–5842. ( 10.1111/mec.12910) [DOI] [PubMed] [Google Scholar]
- 24.Trier CN, Hermansen JS, Sætre G-P, Bailey RI. 2014. Evidence for mito-nuclear and sex-linked reproductive barriers between the hybrid Italian sparrow and its parent species. PLoS Genet. 10, e1004075-10 ( 10.1371/journal.pgen.1004075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Runemark A, Trier CN, Eroukhmanoff F, Hermansen JS, Matschiner M, Ravinet M, Elgvin TO, Sætre G-P. 2018. Variation and constraints in hybrid genome formation. Nat. Ecol. Evol. 20, 229 ( 10.1038/s41559-017-0437-7) [DOI] [PubMed] [Google Scholar]
- 26.Paschou P, et al. 2014. Maritime route of colonization of Europe. Proc. Natl Acad. Sci. USA 111, 9211–9216. ( 10.1073/pnas.1320811111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riyahi S, Hammer Ø, Arbabi T, Sánchez A, Roselaar CS, Aliabadian M, Sætre G-P. 2013. Beak and skull shapes of human commensal and non-commensal house sparrows Passer domesticus. BMC Evol. Biol. 13, 200 ( 10.1186/1471-2148-13-200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutoit L, Vijay N, Mugal CF, Bossu CM, Burri R, Wolf J, Ellegren H. 2017. Covariation in levels of nucleotide diversity in homologous regions of the avian genome long after completion of lineage sorting. Proc. R. Soc. B 284, 20162756 ( 10.1098/rspb.2016.2756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. ( 10.1038/ng.806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27, 2156–2158. ( 10.1093/bioinformatics/btr330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danecek P, McCarthy SA. 2017. BCFtools/csq: haplotype-aware variant consequences. Bioinformatics 33, 2037–2039. ( 10.1093/bioinformatics/btx100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravinet M, Elgvin T, Trier C, Aliabadian M, Gavrilov A, Sætre G. 2018. Data from: Signatures of human-commensalism in the house sparrow genome Dryad Digital Repository. ( 10.5061/dryad.mq25897) [DOI] [PMC free article] [PubMed]
- 35.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 ( 10.1186/s13742-015-0047-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. ( 10.1101/gr.094052.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin SH, Davey JW, Jiggins CD. 2014. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257. ( 10.1093/molbev/msu269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitti JJ, Grossman SR, Sabeti PC. 2013. Detecting natural selection in genomic data. Annu. Rev. Genet. 47, 97–120. ( 10.1146/annurev-ge-47-120213-200001) [DOI] [PubMed] [Google Scholar]
- 39.O'Connell J, et al. 2014. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 10, e1004234-21 ( 10.1371/journal.pgen.1004234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautier M, Klassmann A, Vitalis R. 2016. REHH2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol. Ecol. Resour. 17, 78–90. ( 10.1111/1755-0998.12634) [DOI] [PubMed] [Google Scholar]
- 41.Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9, e1003905-17 ( 10.1371/journal.pgen.1003905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier JI, Sousa VC, Marques DA, Selz OM, Wagner CE, Excoffier L, Seehausen O. 2016. Demographic modelling with whole-genome data reveals parallel origin of similar Pundamilia cichlid species after hybridization. Mol. Ecol. 26, 123–141. ( 10.1111/mec.13838) [DOI] [PubMed] [Google Scholar]
- 43.Bindea G, et al. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. ( 10.1093/bioinformatics/btp101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo H, Xing Y, Liu Y, Luo Y, Deng F, Yang T, Yang K, Li Y. 2016. Wnt/β-catenin signaling pathway activates melanocyte stem cells in vitro and in vivo. J. Dermatol. Sci. 83, 45–51. ( 10.1016/j.jdermsci.2016.04.005) [DOI] [PubMed] [Google Scholar]
- 45.Noramly S, Freeman A, Morgan BA. 1999. β-catenin signaling can initiate feather bud development. Development 126, 3509–3521. [DOI] [PubMed] [Google Scholar]
- 46.Jones S, Pfister-Genskow M, Cirelli C, Benca RM. 2008. Changes in brain gene expression during migration in the white-crowned sparrow. Brain Res. Bull. 76, 536–544. ( 10.1016/j.brainresbull.2008.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffith AJ, Sprunger LK, Sirko-Osadsa DA, Tiller GE, Meisler MH, Warman ML. 1998. Marshall syndrome associated with a splicing defect at the COL11A1 locus. Am. J. Hum. Genet. 62, 816–823. ( 10.1086/301789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry GH, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260. ( 10.1038/ng2123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reiter T, Jagoda E, Capellini TD. 2016. Dietary variation and evolution of gene copy number among dog breeds. PLoS ONE 11, e0148899-19 ( 10.1371/journal.pone.0148899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gavrilov EI, Ispytateleley MKBO, Korelov MN. 1968. The Indian sparrow as a distinct good species. Byulleten Moskovskogo Obshchestva Ispyta- teleley Prirody Otdel Biologicheskiy 73, 115–122. [Google Scholar]
- 51.Yakobi VE. 1978. On the species independence of the Indian sparrow. Zoological Zhurnal 58, 136–137. [Google Scholar]
- 52.Hoban S, et al. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am. Nat. 188, 379–397. ( 10.1086/688018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De León LF, Raeymaekers J, Bermingham E. 2011. Exploring possible human influences on the evolution of Darwin's finches. Evolution 65, 2258–2272. ( 10.1111/j.1558-5646.2011.01297.x) [DOI] [PubMed] [Google Scholar]
- 54.Axelsson E, et al. 2013. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495, 360–364. ( 10.1038/nature11837) [DOI] [PubMed] [Google Scholar]
- 55.Arendt M, Cairns KM, Ballard JWO, Savolainen P, Axelsson E. 2016. Diet adaptation in dog reflects spread of prehistoric agriculture. Heredity 117, 301–306. ( 10.1038/hdy.2016.48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inchley CE, et al. 2016. Selective sweep on human amylase genes postdates the split with Neanderthals. Sci. Rep. 6, 1–10. ( 10.1038/srep37198) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ravinet M, Elgvin T, Trier C, Aliabadian M, Gavrilov A, Sætre G. 2018. Data from: Signatures of human-commensalism in the house sparrow genome Dryad Digital Repository. ( 10.5061/dryad.mq25897) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Scripts and data for analysis are available on Dryad: http://dx.doi.org/10.5061/dryad.mq25897 [34]. Whole genome resequencing data are available at the NCBI Sequence Read Archive, BioProject PRJNA255814 accession numbers SRR5369936-SRR5369966 and the European Nucleotide Archive with study accession number PRJEB27649 and sample accessions ERS2604996-ERS2605092.