Abstract
Introduction
Pediatric asthma has remained a health threat to children in recent years. The abnormal proliferation of airway smooth muscle (ASM) cells contributes to airway remodeling during development of asthma. MicroRNAs (miRNAs) are critical regulators of ASM cell proliferation during airway remodeling. miR-590-5p has been reported to regulate cell proliferation in various cell types. However, it remains unclear whether miR-590-5p regulates ASM cell proliferation. In this study, we aimed to investigate the potential role of miR-590-5p in regulating fetal ASM cell proliferation in vitro stimulated by platelet-derived growth factor (PDGF).
Material and methods
miRNA, mRNA, and protein expression was detected by real-time quantitative polymerase chain reaction and western blot. Cell proliferation was detected by CCK-8 and BrdU assays. The target of miR-590-5p was confirmed by dual-luciferase reporter assay.
Results
MiR-590-5p expression was significantly down-regulated in fetal ASM cells stimulated with PDGF (p < 0.05). Overexpression of miR-590-5p inhibited cell proliferation (p < 0.05), whereas the suppression of miR-590-5p promoted cell proliferation of fetal ASM cells stimulated with PDGF (p < 0.05). Signal transducer and activator of transcription 3 (STAT3) was identified as a target gene of miR-590-5p. In addition, miR-590-5p negatively regulated STAT3 expression (p < 0.05). Moreover, miR-590-5p also modulated downstream genes of STAT3 including cyclin D3 and p27 (p < 0.05). The restoration of STAT3 significantly reversed the inhibitory effect of miR-590-5p on fetal ASM cell proliferation.
Conclusions
MiR-590-5p inhibits proliferation of fetal ASM cells by down-regulating STAT3, thereby suggesting a novel therapeutic target for the treatment of pediatric asthma.
Keywords: airway smooth muscle cells, asthma, miR-590-5p, STAT3
Introduction
Asthma in the pediatric population has become a critical public health concern in recent years and has resulted in increasing morbidity and healthcare costs [1]. The increased airway hyper-responsiveness and remodeling are hallmarks of pediatric asthma [2]. Various inflammatory cytokines are highly upregulated in asthmatic children and airway inflammation is correlated with airway hyper-responsiveness [3, 4]. Numerous mitogens, including transforming growth factor-β, platelet-derived growth factor (PDGF), and connective tissue growth factor (CTGF), are elevated in asthmatic lungs, thereby inducing airway smooth muscle (ASM) cell proliferation and airway remodeling [5–7]. Airway smooth muscle cells are the main structural components of the airway, and hyperplasia of ASM cells is correlated with asthma severity [8]. Prevention of ASM proliferation may be a novel therapeutic approach in the treatment of asthma. However, molecular mechanisms underlying ASM hyper-proliferation remain poorly understood. Therefore, a better understanding of these mechanisms may shed light on the pathogenesis of asthma and help with the development of potential treatments for asthma.
MicroRNAs (miRNAs), a group of small noncoding RNAs, are emerging as critical regulators for gene expression [9]. MicroRNAs negatively modulate gene expression at the post-transcriptional level by binding the 3′-untranslated region (UTR) of target mRNA [9]. Various studies have shown that miRNAs are involved in various physiological and pathological processes; they regulate many biological processes, including cell proliferation, differentiation, and apoptosis [10]. A number of studies have shown the important role of miRNAs in the pathogenesis of asthma and the regulation of airway inflammation and hyper-responsiveness [11]. Therefore, miRNA-based therapy may have potential applications to the treatment of asthma.
Signal transducer and activator of transcription 3 (STAT3) is a cytokine and growth factor-inducible transcription factor that plays an important role in various biological processes, including cell proliferation, apoptosis, and differentiation [12]. Recent studies have suggested that STAT3 is an important regulator of asthma pathogenesis [13]. PDGF can induce STAT3 activation and promote proliferation of ASM cells [14]. Blocking STAT3 signaling impedes ASM cell proliferation induced by PDGF [14]. The inhibition of STAT3 suppresses lung inflammation and airway remodeling in a murine asthma model [15]. Therefore, STAT3 is a promising therapeutic target for preventing asthma.
Recent studies have revealed that miR-590-5p regulates cell proliferation of various cells such as cancer cells and vascular endothelial cells [16–18]. However, the role of miR-590-5p in ASM cell proliferation remains unclear. In this study, we aimed to investigate the potential role and underlying mechanism of miR-590-5p in regulating fetal ASM cell proliferation in vitro stimulated by PDGF. miR-590-5p expression was significantly down-regulated in fetal ASM cells stimulated with PDGF. Overexpression of miR-590-5p inhibited PDGF-induced fetal ASM cell proliferation. STAT3 was identified as a functional target gene of miR-590-5p in regulating fetal ASM cell proliferation. Our results demonstrate that miR-590-5p inhibits the proliferation of fetal ASM cells by down-regulating STAT3, thereby suggesting a potential therapeutic approach for the prevention of pediatric asthma.
Material and methods
Cell lines
Human fetal ASM cells were isolated from fetal tracheobronchial tissues (12–18 weeks gestation) via the enzymatic dissociation method, as previously described [19]. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Rockville, MD, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 2 mM glutamine, 1 mM sodium pyruvate, and 1% penicillin/streptomycin mix (Sigma, St. Louis, MO, USA). For tissue donation, written informed consent was obtained from each participant. The use of clinical tissue was approved by the Institutional Review Board of The First Hospital of Jilin University, and this study was performed in accordance with the Declaration of Helsinki. 293T cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM (Gibco) containing 10% FBS (Gibco) and 1% penicillin/streptomycin mix (Sigma). Cells were routinely maintained in a humidified atmosphere of 5% CO2 at 37°C.
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using Trizol reagent according to the manufacturer’s protocols. To detect miR-590-5p expression, cDNA was synthesized using the miScript Reverse Transcription Kit (Qiagen, Dusseldorf, Germany). To detect STAT3 mRNA expression, cDNA was synthesized using M-MLV Reverse Transcriptase (TaKaRa, Dalian, China). PCR amplification was done using a SYBR Green PCR kit (TaKaRa) in the ABI7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). Small nuclear RNA U6 served as an internal control to normalize miR-590-5p expression. GAPDH was used as an internal control to normalize expression of STAT3. Relative gene expression was analyzed via the 2– ΔΔCt method.
Cell transfection
Cells were transfected with miR-590-5p mimics, inhibitor, or negative control (GenePharma, Shanghai, China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cDNA fragment of the STAT3 open reading frame was inserted into the pcDNA3.1 vector (Sangon Biotech, Shanghai, China). This construct was transfected into cells using Lipofectamine 2000 (Invitrogen). After 48 h from transfection, cells were treated with 25 ng/ml PDGF (R&D Systems, Minneapolis, MN, USA) and incubated for 24 h.
Cell viability and growth assay
Cell viability and growth were detected by cell counting kit-8 (CCK-8) assay. Briefly, cells were seeded into 96-well plates and cultured overnight. After the indicated treatments, cells were treated with 10 µl of CCK-8 solution (Sigma) and cultured for 1 h. The optical density (OD) value at 490 nm was detected using a microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
BrdU (5-bromo-2′-deoxyuridine) assay
Cell proliferation was determined by BrdU assay using a BrdU kit (Sigma). Briefly, cells were seeded into 96-well plates. After the indicated treatments, cells were treated with BrdU labeling reagent for 2 h at 37°C. Then, cells were treated with FixDenat solution and incubated for 30 min at room temperature. Cells were treated with anti-BrdU-peroxidase and incubated for 90 min at room temperature. Afterwards, the substrate solution was added and incubated for 10 min. The OD value at 370 nm was measured by a microplate reader (BioTek Instruments).
Dual-luciferase reporter assay
Fragments of wild-type or mutant STAT3 3′-UTR containing the putative binding site of miR-590-5p was cloned into a pmirGLO vector (Promega, Madison, WI, USA). Then, 293T cells were co-transfected with the pmirGLO vector and miR-590-5p mimics using Lipofectamine 2000 (Invitrogen). After incubation for 48 h, cells were harvested and lysed. The relative activity was determined using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocols.
Western blot analysis
Equal amounts of protein were separated by 10% sodium dodecyl sulfate polyacrylamide gels, then transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Membranes were then blocked with 5% non-fat milk and incubated with anti-STAT3 and anti-GAPDH antibodies (Abcam, Cambridge, UK) at 4°C overnight. Membranes were probed with horseradish peroxidase-secondary antibody (1 : 5000; Sigma) for 1 h at room temperature. Protein bands were visualized by an enhanced chemiluminescence system (Millipore). The intensity of bands was quantified using the Image-Pro Plus 6.0 software.
Statistical analysis
Experimental data were presented as mean ± standard deviation. Statistical analyses were performed by Student’s t-test and one-way analysis of variance with the Bonferroni post hoc test using SPSS 18.0 statistical software (SPSS Inc., Chicago, IL, USA). Differences were considered statistically significant when p < 0.05.
Results
Expression of miR-590-5p is down-regulated in fetal ASM cells stimulated with PDGF
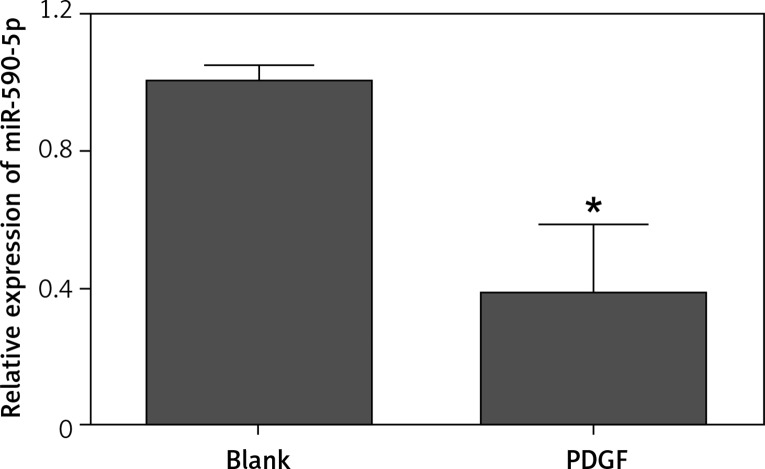
To investigate the potential role of miR-590-5p in regulating ASM cell proliferation, we examined the expression of miR-590-5p in fetal ASM cells stimulated with mitogen PDGF by RT-qPCR. The results showed that miR-590-5p expression was significantly decreased in fetal ASM cells stimulated with PDGF (Figure 1), thereby suggesting a critical role in the regulation of fetal ASM cell proliferation.
Figure 1.
Expression of miR-590-5p in fetal ASM cells. Human fetal ASM cells were treated with PDGF (25 ng/ml) for 24 h and then harvested for RT-qPCR analysis
*p < 0.05 vs. blank.
Overexpression of miR-590-5p inhibits proliferation of fetal ASM cells
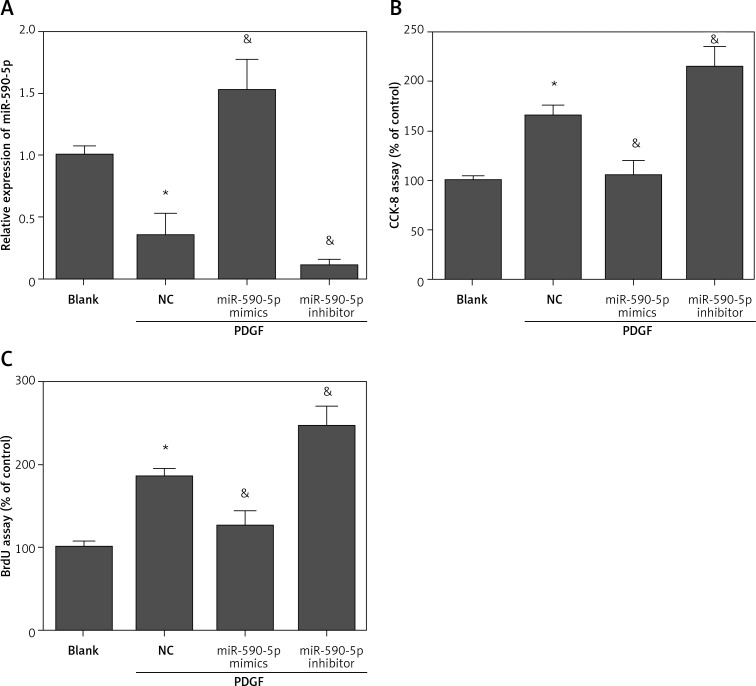
To investigate the biological function of miR-590-5p in fetal ASM cells, we performed gain- and loss-of-function experiments by transfecting miR-590-5p mimics or inhibitors. The results showed that miR-590-5p mimics-transfected cells showed higher expression levels of miR-590-5p, whereas miR-590-5p inhibitor-transfected cells showed lower expression levels of miR-590-5p compared with negative control (NC)-transfected cells (Figure 2 A). PDGF stimulation promoted ASM cell proliferation. However, overexpression of miR-590-5p significantly reduced PDGF-induced cell proliferation in fetal ASM cells (Figures 2 B and C). In contrast, suppression of miR-590-5p enhanced PDGF-induced cell proliferation (Figures 2 B and C). These results suggest that miR-590-5p can inhibit proliferation of fetal ASM cells.
Figure 2.
miR-590-5p inhibited the proliferation of fetal ASM cells. Human fetal ASM cells were transfected with miR-590-5p mimics or inhibitor for 48 h and then treated with PDGF (25 ng/ml) for 24 h. A – Expression level of miR-590-5p was detected by RT-qPCR. B – Cell viability and growth were detected by CCK-8 assay. C – Cell proliferation was detected by BrdU assay
*p < 0.05 vs. blank. &p < 0.05 vs. NC.
STAT3 is a potential target of miR-590-5p in fetal ASM cells
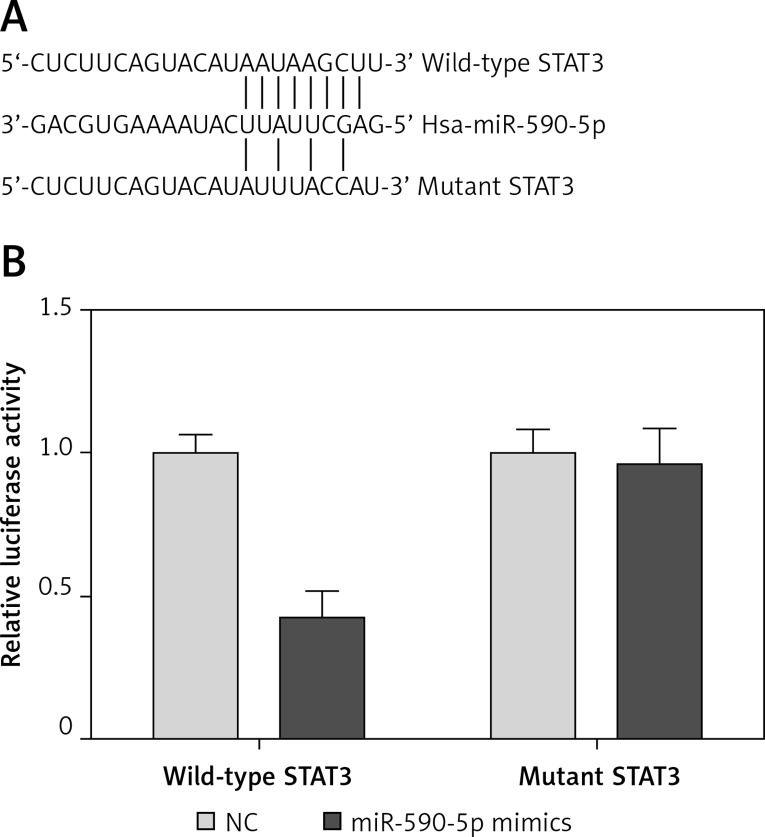
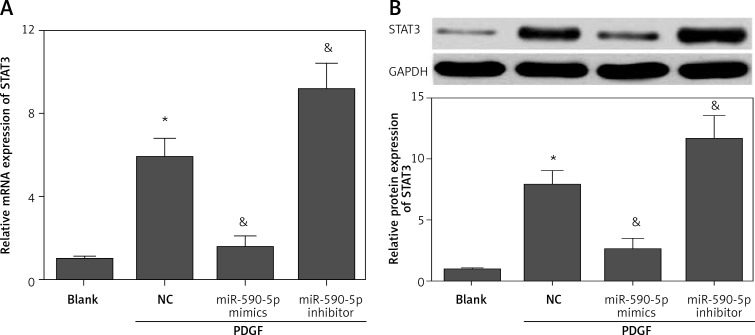
MicroRNAs exert functions by targeting downstream targets. To investigate the underlying mechanism by which miR-590-5p regulates ASM cell proliferation, we predicted the downstream target gene of miR-590-5p by bioinformatics analysis (TargetScan and microRNA.org). We found that STAT3, a critical regulator for ASM proliferation and asthma [14, 20], was a potential target gene of miR-590-5p. The 3′-UTR of STAT3 had a putative binding site for miR-590-5p (Figure 3 A). To confirm whether miR-590-5p directly targeted the 3′-UTR of STAT3, we performed a dual-luciferase reporter assay. The results showed that miR-590-5p significantly inhibited luciferase activity of the reporter vector containing the wild-type STAT3 3′-UTR (Figure 3 B). However, miR-590-5p showed no obvious effect on the luciferase activity of the reporter vector containing the mutant STAT3 3′-UTR (Figure 3 B). To further confirm that STAT3 is a target gene of miR-590-5p, we detected the effect of miR-590-5p on STAT3 expression in fetal ASM cells. Moreover, we found that both mRNA and protein expression of STAT3 were significantly inhibited by miR-590-5p overexpression, whereas the suppression of miR-590-5p promoted STAT3 expression (Figures 4 A and B). These results suggested that miR-590-5p directly targets the 3′-UTR of STAT3 and regulates its expression.
Figure 3.
miR-590-5p directly targeted 3′-UTR of STAT3. A – Diagram of the predicted miR-590-5p binding site in the 3′-UTR of STAT3. B – Dual-luciferase reporter assay was used to examine whether miR-590-5p directly binds to STAT3 3′-UTR
*p < 0.05 vs. NC.
Figure 4.
miR-590-5p inhibited the expression of STAT3. Fetal ASM cells were transfected with miR-590-5p mimics or inhibitor for 48 h and then treated with PDGF (25 ng/ml) for 24 h
*p < 0.05 vs. blank. &p < 0.05 vs. NC.
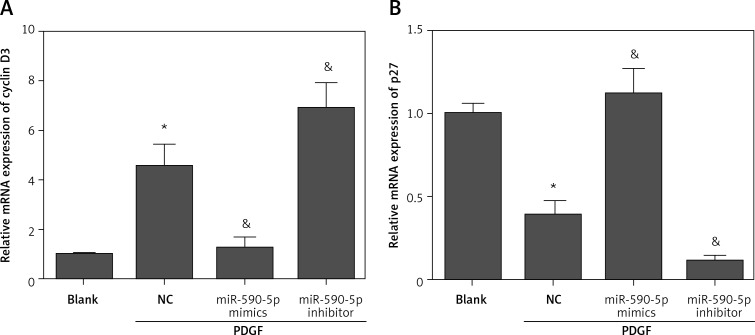
MiR-590-5p regulates expression of cyclin D3 and P27
A previous study reported that STAT3 regulates PDGF-induced ASM proliferation by regulating cyclin D3 and p27 [21]. Considering the regulatory effect of miR-590-5p on STAT3 expression, we speculated that miR-590-5p may regulate cyclin D3 and p27. To investigate whether miR-590-5p regulated cyclin D3 and p27, we detected the effect of miR-590-5p on the expression of cyclin D3 and p27 by RT-qPCR. The results showed that PDGF treatment significantly promoted cyclin D3 expression and suppressed p27 expression (Figures 5 A and B). As expected, miR-590-5p overexpression markedly inhibited cyclin D3 expression and promoted p27 expression (Figures 5 A and B). However, the suppression of miR-590-5p exhibited the opposite effect (Figures 5 A and B). Overall, these data suggest that miR-590-5p regulates the expression of cyclin D3 and p27 during PDGF-induced ASM proliferation.
Figure 5.
Fetal ASM cells were transfected with miR-590-5p mimics or inhibitor for 48 h and then treated with PDGF (25 ng/ml) for 24 h. The mRNA expression of cyclin D3 (A) and p27 (B) was detected by RT-qPCR
*p < 0.05 vs. blank. &p < 0.05 vs. NC.
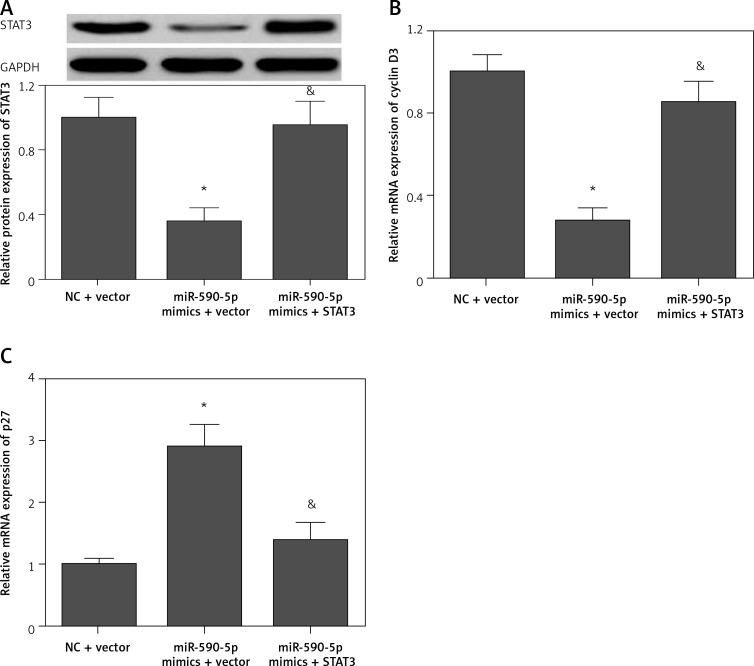
Restoration of STAT3 expression reverses the effect of miR-590-5p
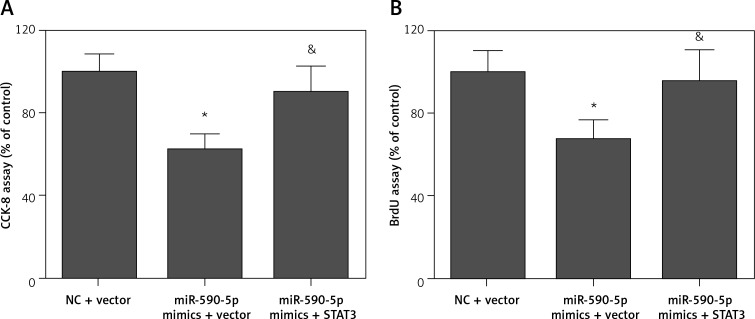
To confirm whether miR-590-5p exerts its function by targeting STAT3, we performed a rescue assay. Transfection of pcDNA3.1/STAT3 expression vector (without the 3′-UTR) significantly restored the expression of STAT3 in miR-590-5p mimics-transfected cells (Figure 6 A). The regulatory effect of miR-590-5p on cyclin D3 (Figure 6 B) and p27 (Figures 6 C) was markedly abrogated by STAT3 overexpression. Moreover, the inhibitory effect of miR-590-5p on fetal ASM cell proliferation was also significantly reversed by STAT3 overexpression (Figures 7 A and B). These data indicate that miR-590-5p inhibits ASM cell proliferation by downregulation of STAT3.
Figure 6.
Restoration of STAT3 expression in fetal ASM cells. Cells were co-transfected with pcDNA3.1/STAT3 vector and miR-590-5p mimics for 48 h and then treated with PDGF (25 ng/ml) for 24 h. A – Protein expression of STAT3 was detected by Western blot. mRNA expression of cyclin D3 (B) and p27 (C) was detected by RT-qPCR
*p < 0.05 vs. NC + vector. &p < 0.05 vs. miR-590-5p mimics + vector.
Figure 7.
Restoration of STAT3 reversed the inhibitory effect of miR-590-5p on ASM cell proliferation. Cells were co-transfected with pcDNA3.1/STAT3 vector and miR-590-5p mimics for 48 h and then treated with PDGF (25 ng/ml) for 24 h. A – Cell growth and viability were detected by CCK-8 assay. B – Cell proliferation was measured by BrdU assay
*p < 0.05 vs. NC + vector. &p < 0.05 vs. miR-590-5p mimics + vector.
Discussion
Airway smooth muscle cell proliferation is a critical process in the formation of airway remodeling in asthma. The mitogen PDGF promotes the proliferation of ASM cells in the bronchial tissues during the pathogenesis of asthma [22, 23]. Our study showed that miR-590-5p was an important regulator of PDGF-induced cell proliferation in fetal ASM cells, thereby suggesting a potential target for preventing ASM cell proliferation in pediatric asthma.
Recent studies suggested that miRNAs act as critical regulators in ASM cell proliferation. Perry et al. reported that miR-221 promoted proliferation of ASM cells in severe asthma [24]. The overexpression of miR-10a inhibited mitogen-induced ASM cell proliferation by down-regulation of the PI3K/Akt signaling pathway [25]. The expression of miR-138 is decreased in asthmatic ASM cells and overexpression of miR-138 suppresses proliferation of ASM cells by targeting 3-phosphoinositide-dependent protein kinase-1 [26]. Liu et al. revealed that miR-145 inhibited proliferation and migration of ASM cells induced by inflammatory cytokines through targeting Krüppel-like factor 4 [27]. miR-23b and miR-143-3p inhibited transforming growth factor-β1-induced cell proliferation of ASM cells [28–30]. All these findings suggest that miRNAs are critical regulators of ASM cell proliferation. In this study, we identified miR-590-5p as a novel miRNA involved in regulating fetal ASM cell proliferation, thereby suggesting a potential function of miR-590-5p in the pathogenesis of pediatric asthma.
miR-590-5p is emerging as an important regulator for cell proliferation. miR-590-5p reportedly inhibits proliferation of hepatocellular carcinoma cells by targeting S100A10 [16]. Zhou et al. reported that miR-590-5p suppressed the proliferation of colorectal cancer cells by targeting the nuclear factor 90/vascular endothelial growth factor A axis [17]. miR-590-5p inhibits the proliferation of breast cancer cells by targeting Sry-related high-mobility box 2 [31]. Moreover, miR-590-5p inhibits oxidized low-density lipoprotein induced proliferation of human umbilical vein endothelial cells by targeting lectin-like oxidized low-density lipoprotein receptor-1 [18]. These reports suggest that miR-590-5p functions as a negative regulator for cell proliferation. However, the positive effect of miR-590-5p has also been reported. Jiang et al. showed that miR-590-5p promoted the proliferation of hepatocellular carcinoma cells by targeting transforming growth factor-β RII [32]. Therefore, the role of miR-590-5p remains controversial. The precise role of miR-590-5p may be dependent on downstream target genes. miR-590-5p may bind to different target genes in different cell types after different stimulations. In this study, our results indicated that miR-590-5p was an inhibitor of ASM cell proliferation, thereby supporting the inhibitory role of miR-590-5p in regulating cell proliferation.
Accumulating studies have suggested that STAT3 is a pivotal regulator in the pathogenesis of asthma. Single nucleotide polymorphisms of the STAT3 gene were found in both adults and children with asthma, and are correlated with decreased lung function [33]. Knockdown of STAT3 impeded allergic inflammation and airway hyper-responsiveness in a murine model of asthma [13]. STAT3 inhibitor administration prevents lung inflammation and airway remodeling in a murine asthma model [15]. STAT3 is essential for the expression of pro-inflammatory factors in ASM cells, which is involved in airway inflammation and hyper-responsiveness in asthma [34–36]. Knockdown of STAT3 abolishes IgE-induced ASM cell proliferation [20]. PDGF induces the proliferation of ASM cells by activating STAT3 [14]. Furthermore, knockdown of STAT3 inhibits PDGF-induced ASM cell proliferation through the down-regulation of cyclin D3 and up-regulation of p27 expression [21]. A recent study reported that STAT3 promotes the pro-angiogenesis ability of ASM cells through up-regulation of vascular endothelial growth factor [37]. These studies suggest that STAT3 is a promising molecular target in preventing ASM cell proliferation. Various studies have shown that STAT3 is epigenetically regulated by miRNAs [38, 39]. In this study, we identified STAT3 as a target gene of miR-590-5p in fetal ASM cells, in which miR-590-5p negatively regulated STAT3 expression. The decreased expression of miR-590-5p may contribute to increased STAT3 expression, thus leading to increased ASM cell proliferation. Therefore, miR-590-5p may serve as a novel inhibitor for STAT3.
STAT3 regulates cell proliferation by promoting expression of cyclin D3, which is critical for cell cycle progression [40, 41]. In addition, STAT3 promotes cell proliferation by inhibiting expression of p27, a cell cycle inhibitor [42, 43]. Previous research has shown that the knockdown of STAT3 inhibits proliferation of ASM cells associated with the down-regulation of cyclin D3 and up-regulation of p27. In accordance with these findings, our results demonstrated that the inhibition of STAT3 by miR-590-5p also inhibited the expression of cyclin D3 and promoted the expression of p27 in fetal ASM cells.
In conclusion, our study suggests that miR-590-5p is a novel endogenic inhibitor for STAT3 in fetal ASM cells. The inhibition of STAT3 by miR-590-5p may be a potential and promising therapeutic approach in the treatment of pediatric asthma. However, the precise role of miR-590-5p/STAT3 in regulating asthma requires further investigation in animal models in vivo.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Asher MI, Montefort S, Bjorksten B, et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet. 2006;368:733–43. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 2.Palmer LJ, Rye PJ, Gibson NA, et al. Airway responsiveness in early infancy predicts asthma, lung function, and respiratory symptoms by school age. Am J Respir Crit Care Med. 2001;163:37–42. doi: 10.1164/ajrccm.163.1.2005013. [DOI] [PubMed] [Google Scholar]
- 3.Majak P, Jerzynska J, Bojo M, et al. Cytokine profiling in exhaled breath condensate after exercise challenge in asthmatic children with post-exercise symptoms. Arch Med Sci. 2016;12:778–84. doi: 10.5114/aoms.2015.48547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attanasi M, Consilvio NP, Rapino D, et al. Bronchial hyperresponsiveness to mannitol, airway inflammation and Asthma Control Test in atopic asthmatic children. Arch Med Sci. 2016;12:137–44. doi: 10.5114/aoms.2016.57589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbato A, Turato G, Baraldo S, et al. Airway inflammation in childhood asthma. Am J Respir Crit Care Med. 2003;168:798–803. doi: 10.1164/rccm.200305-650OC. [DOI] [PubMed] [Google Scholar]
- 6.Brown SD, Baxter KM, Stephenson ST, et al. Airway TGF-beta1 and oxidant stress in children with severe asthma: association with airflow limitation. J Allergy Clin Immunol. 2012;129:388–396. doi: 10.1016/j.jaci.2011.11.037. 396 e381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin SC, Chou HC, Chiang BL, et al. CTGF upregulation correlates with MMP-9 level in airway remodeling in a murine model of asthma. Arch Med Sci. 2017;13:670–6. doi: 10.5114/aoms.2016.60371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin JG, Ramos-Barbon D. Airway smooth muscle growth from the perspective of animal models. Respir Physiol Neurobiol. 2003;137:251–61. doi: 10.1016/s1569-9048(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 9.Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 10.Adams BD, Parsons C, Walker L, et al. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–71. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshpande DA, Dileepan M, Walseth TF, et al. MicroRNA regulation of airway inflammation and airway smooth muscle function: relevance to asthma. Drug Dev Res. 2015;76:286–95. doi: 10.1002/ddr.21267. [DOI] [PubMed] [Google Scholar]
- 12.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 13.Simeone-Penney MC, Severgnini M, Tu P, et al. Airway epithelial STAT3 is required for allergic inflammation in a murine model of asthma. J Immunol. 2007;178:6191–9. doi: 10.4049/jimmunol.178.10.6191. [DOI] [PubMed] [Google Scholar]
- 14.Simon AR, Takahashi S, Severgnini M, et al. Role of the JAK-STAT pathway in PDGF-stimulated proliferation of human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1296–304. doi: 10.1152/ajplung.00315.2001. [DOI] [PubMed] [Google Scholar]
- 15.Gavino AC, Nahmod K, Bharadwaj U, et al. STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy. 2016;71:1684–92. doi: 10.1111/all.12937. [DOI] [PubMed] [Google Scholar]
- 16.Shan X, Miao Y, Fan R, et al. MiR-590-5P inhibits growth of HepG2 cells via decrease of S100A10 expression and inhibition of the Wnt pathway. Int J Mol Sci. 2013;14:8556–69. doi: 10.3390/ijms14048556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Q, Zhu Y, Wei X, et al. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axis. Cell Death Dis. 2016;7:e2413. doi: 10.1038/cddis.2016.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai Y, Zhang Z, Cao Y, et al. MiR-590-5p inhibits oxidized-LDL induced angiogenesis by targeting LOX-1. Sci Rep. 2016;6:22607. doi: 10.1038/srep22607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandya HC, Snetkov VA, Twort CH, et al. Oxygen regulates mitogen-stimulated proliferation of fetal human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1220–30. doi: 10.1152/ajplung.00268.2001. [DOI] [PubMed] [Google Scholar]
- 20.Redhu NS, Shan L, Al-Subait D, et al. IgE induces proliferation in human airway smooth muscle cells: role of MAPK and STAT3 pathways. Allergy Asthma Clin Immunol. 2013;9:41. doi: 10.1186/1710-1492-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simeone-Penney MC, Severgnini M, Rozo L, et al. PDGF-induced human airway smooth muscle cell proliferation requires STAT3 and the small GTPase Rac1. Am J Physiol Lung Cell Mol Physiol. 2008;294:L698–704. doi: 10.1152/ajplung.00529.2007. [DOI] [PubMed] [Google Scholar]
- 22.Ohno I, Nitta Y, Yamauchi K, et al. Eosinophils as a potential source of platelet-derived growth factor B-chain (PDGF-B) in nasal polyposis and bronchial asthma. Am J Respir Cell Mol Biol. 1995;13:639–47. doi: 10.1165/ajrcmb.13.6.7576701. [DOI] [PubMed] [Google Scholar]
- 23.Aubert JD, Hayashi S, Hards J, et al. Platelet-derived growth factor and its receptor in lungs from patients with asthma and chronic airflow obstruction. Am J Physiol. 1994;266:L655–63. doi: 10.1152/ajplung.1994.266.6.L655. [DOI] [PubMed] [Google Scholar]
- 24.Perry MM, Baker JE, Gibeon DS, et al. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am J Respir Cell Mol Biol. 2014;50:7–17. doi: 10.1165/rcmb.2013-0067OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu R, Pan W, Fedulov AV, et al. MicroRNA-10a controls airway smooth muscle cell proliferation via direct targeting of the PI3 kinase pathway. FASEB J. 2014;28:2347–57. doi: 10.1096/fj.13-247247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Yang K, Sun X, et al. MiR-138 suppresses airway smooth muscle cell proliferation through the PI3K/AKT signaling pathway by targeting PDK1. Exp Lung Res. 2015;41:363–9. doi: 10.3109/01902148.2015.1041581. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Sun X, Wu Y, et al. Effects of miRNA-145 on airway smooth muscle cells function. Mol Cell Biochem. 2015;409:135–43. doi: 10.1007/s11010-015-2519-7. [DOI] [PubMed] [Google Scholar]
- 28.Chen M, Huang L, Zhang W, et al. MiR-23b controls TGF-beta1 induced airway smooth muscle cell proliferation via TGFbetaR2/p-Smad3 signals. Mol Immunol. 2016;70:84–93. doi: 10.1016/j.molimm.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Chen M, Shi J, Zhang W, et al. MiR-23b controls TGF-beta1 induced airway smooth muscle cell proliferation via direct targeting of Smad3. Pulm Pharmacol Ther. 2017;42:33–42. doi: 10.1016/j.pupt.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Cheng W, Yan K, Xie LY, et al. MiR-143-3p controls TGF-beta1-induced cell proliferation and extracellular matrix production in airway smooth muscle via negative regulation of the nuclear factor of activated T cells 1. Mol Immunol. 2016;78:133–9. doi: 10.1016/j.molimm.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Zhou L, Zhao LC, Jiang N, et al. MicroRNA miR-590-5p inhibits breast cancer cell stemness and metastasis by targeting SOX2. Eur Rev Med Pharmacol Sci. 2017;21:87–94. [PubMed] [Google Scholar]
- 32.Jiang X, Xiang G, Wang Y, et al. MicroRNA-590-5p regulates proliferation and invasion in human hepatocellular carcinoma cells by targeting TGF-beta RII. Mol Cells. 2012;33:545–51. doi: 10.1007/s10059-012-2267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litonjua AA, Tantisira KG, Lake S, et al. Polymorphisms in signal transducer and activator of transcription 3 and lung function in asthma. Respir Res. 2005;6:52–61. doi: 10.1186/1465-9921-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamasaki A, Saleh A, Koussih L, et al. IL-9 induces CCL11 expression via STAT3 signalling in human airway smooth muscle cells. PLoS One. 2010;5:e9178. doi: 10.1371/journal.pone.0009178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saleh A, Shan L, Halayko AJ, et al. Critical role for STAT3 in IL-17A-mediated CCL11 expression in human airway smooth muscle cells. J Immunol. 2009;182:3357–65. doi: 10.4049/jimmunol.0801882. [DOI] [PubMed] [Google Scholar]
- 36.Shan L, Redhu NS, Saleh A, et al. Thymic stromal lymphopoietin receptor-mediated IL-6 and CC/CXC chemokines expression in human airway smooth muscle cells: role of MAPKs (ERK1/2, p38, and JNK) and STAT3 pathways. J Immunol. 2010;184:7134–43. doi: 10.4049/jimmunol.0902515. [DOI] [PubMed] [Google Scholar]
- 37.Lv J, Sun B, Mai Z, et al. STAT3 potentiates the ability of airway smooth muscle cells to promote angiogenesis by regulating VEGF signalling. Exp Physiol. 2017;102:598–606. doi: 10.1113/EP086136. [DOI] [PubMed] [Google Scholar]
- 38.Yan XT, Ji LJ, Wang Z, et al. MicroRNA-93 alleviates neuropathic pain through targeting signal transducer and activator of transcription 3. Int Immunopharmacol. 2017;46:156–62. doi: 10.1016/j.intimp.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 39.Liu S, Hu C, Wang Y, et al. miR-124 inhibits proliferation and invasion of human retinoblastoma cells by targeting STAT3. Oncol Rep. 2016;36:2398–404. doi: 10.3892/or.2016.4999. [DOI] [PubMed] [Google Scholar]
- 40.Stout BA, Bates ME, Liu LY, et al. IL-5 and granulocyte-macrophage colony-stimulating factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3 protein expression in human eosinophils. J Immunol. 2004;173:6409–17. doi: 10.4049/jimmunol.173.10.6409. [DOI] [PubMed] [Google Scholar]
- 41.Amin HM, McDonnell TJ, Ma Y, et al. Selective inhibition of STAT3 induces apoptosis and G(1) cell cycle arrest in ALK-positive anaplastic large cell lymphoma. Oncogene. 2004;23:5426–34. doi: 10.1038/sj.onc.1207703. [DOI] [PubMed] [Google Scholar]
- 42.Wang ST, Ho HJ, Lin JT, et al. Simvastatin-induced cell cycle arrest through inhibition of STAT3/SKP2 axis and activation of AMPK to promote p27 and p21 accumulation in hepatocellular carcinoma cells. Cell Death Dis. 2017;8:e2626. doi: 10.1038/cddis.2016.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu TY, Pang JH, Wu KP, et al. Platelet-rich plasma increases proliferation of tendon cells by modulating Stat3 and p27 to up-regulate expression of cyclins and cyclin-dependent kinases. Cell Prolif. 2015;48:413–20. doi: 10.1111/cpr.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]