Abstract
Gefitinib, an epidermal growth factor receptor (EGFR)-specific drug, is effective for ~1 year, after which resistance is inevitable. Calpain 2 (CAPN2) is known to serve a role in the drug response and resistance in certain cancer therapies. However, the full function of CAPN2, particularly in non-small cell lung cancer, has not yet been elucidated. In the present study, CAPN2 expression in gefitinib-resistant lung adenocarcinoma cells was investigated. CAPN2 function in these cells was further evaluated using gene knockdown both in vitro and in vivo. The results demonstrated that CAPN2 was strongly associated with gefitinib-resistance, and CAPN2 mRNA and protein expression levels were significantly increased in gefitinib-resistant cell lines. Furthermore, CAPN2 knockdown inhibited gefitinib-resistant cell proliferation in vitro and in vivo. CAPN2 conferred gefitinib-resistance by inhibiting cell apoptosis and arresting the cell cycle. CAPN2 knockdown also induced caspase activation and mitochondrial dysfunction, and its function in gefitinib resistance appeared to be largely mediated by EGFR/protein kinase B/survivin signaling pathway activation. These results suggest that CAPN2 is responsible for EGFR-tyrosine kinase inhibitor resistance, and CAPN2 inhibition may be used to provide therapeutic benefits in the treatment of gefitinib resistance.
Keywords: non-small cell lung cancer, calpain 2, gefitinib, apoptosis, tyrosine kinase inhibitor resistance
Introduction
Targeted therapies for lung cancer harboring the epidermal growth factor receptor (EGFR) fusion gene generally involve the use of EGFR-tyrosine kinase inhibitors (TKIs) and have a large influence on patient outcome. The EGFR-TKI gefitinib is the first-line treatment for EGFR-positive non-small cell lung cancer (NSCLC) patients, however, almost all patients invariably acquire resistance to the drug (1,2). The mechanisms underlying this acquired resistance to gefitinib have been extensively explored and mainly stem from secondary mutations, such as Thr790Met, Asp761Tyr and Thr854Ala, as well as the activation of bypass or alternative pathways, including MET gene amplification and histological/phenotypic transformation (3,4). Although the T790M mutation and MET gene amplification have been identified as the major mechanisms of resistance, the full signaling pathway involved in the process is largely unknown.
The calpain (CAPN) protein family is an important group of proteases that are activated by calcium, and serves an important role in cell migration, survival and apoptosis (5–7). CAPN2, also known as m-CAPN, is one of the most widely investigated members of this family. Experimental and clinical evidence indicates that CAPN2 is aberrantly expressed in various tumors, and has a pivotal function in tumorigenesis, disease progression and therapy resistance (8–10). It has also been demonstrated that CAPNs are responsible for activating various downstream proteins that further facilitate cancer progression. In breast cancer, for instance, CAPN has been associated with the cleavage of human EGFR2, which is responsible for cancer pathogenesis (11). Furthermore, the expression of CAPN was associated with relapse-free survival in HER2-positive breast cancer patients treated with trastuzumab following adjuvant chemotherapy (12). A more recent study supported these functions (13), indicating that high CAPN2 expression had a clear adverse effect in ovarian cancer patients and was also important in platinum-based therapy resistance. These results highlight the predictive significance of CAPN2 in metastasis and progression in multiple types of cancer. However, the function of CAPN2 in NSCLC, particularly in EGFR-TKI resistance, has not yet been elucidated.
In our previous studies, we identified that PLIN2 and Annexin A5 confer gefitinib resistance (12,14). In the present study, CAPN2 expression was investigated in gefitinib-resistant lung adenocarcinoma cells. CAPN2 function in these cells was further evaluated using gene knockdown in vitro and in vivo. To the best of our knowledge, this is the first investigation of CAPN2 expression and function during EGFR-TKI resistance.
Materials and methods
Cell lines and culture
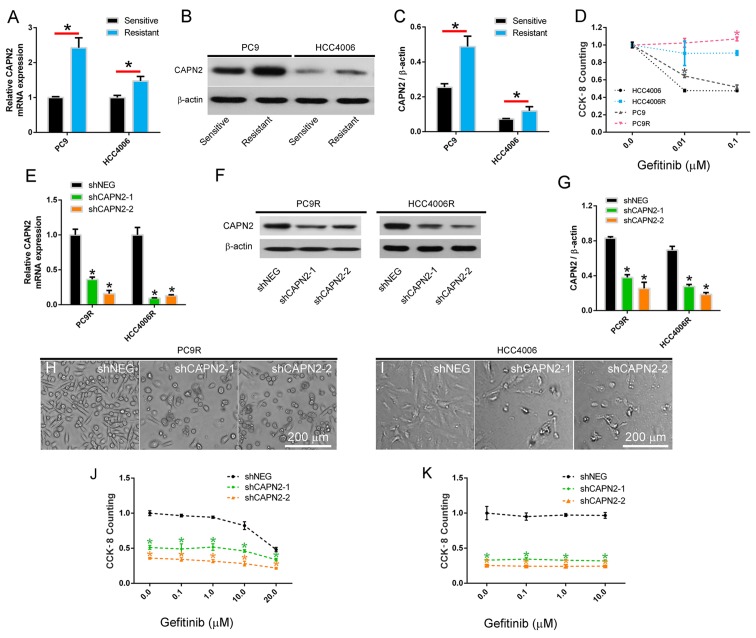
Gefitinib-sensitive PC9 and HCC4006 human lung adenocarcinoma cells were purchased from the American Type Culture Collection (Manassas, VA, USA). To induce gefitinib resistance, the cells were exposed to increasing concentrations of gefitinib (ranging from 0.01 to 10.0 mM; Selleck Chemicals, Houston, TX, USA). The viability of gefitinib-resistant PC9R and HCC4006R cells was unaffected up to 10 µM gefitinib (Fig. 1J and K), whereas the viability of gefitinib-sensitive PC9 and HCC4006 cells was significantly reduced in the presence of 0.1 µM gefitinib (Fig. 1D). Gefitinib-sensitive and -resistant cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum and 100 U/ml penicillin/streptomycin. In order to maintain gefitinib resistance, 1 µM gefitinib was added to the cells every 2 weeks. The cells were grown as monolayers in a humidified atmosphere of 5% CO2 at 37°C.
Figure 1.
CAPN2 is upregulated in gefitinib-resistant cells and mediates gefitinib resistance. (A) CAPN2 mRNA expression was measured by reverse transcription-quantitative polymerase chain reaction in gefitinib-sensitive and gefitinib-resistant NSCLC cell lines. (B) Representative western blots highlighting CAPN2 protein expression and (C) changes in CAPN2 protein levels quantified by scanning densitometry in gefitinib-sensitive and gefitinib-resistant NSCLC cell lines. (D) Cytotoxic effects of gefitinib on PC9, PC9R, HCC4006 and HCC4006R cells measured by cell counting kit-8 (n=4). *P<0.05 vs. the corresponding gefitinib-sensitive cells. (E) CAPN2 mRNA expression following transfection of PC9R and HCC4006R cells with shNEG, or with shCAPN2-1 and shCAPN2-2 to silence CAPN2 expression. (F) Representative immunoblots and (G) scanning densitometry-quantified levels of CAPN2 protein expression following shRNA transfection. Morphological examination of shRNA-transfected (H) PC9R and (I) HCC4006R cells, indicating that CAPN2 knockdown reduces the number of resistant cells in gefitinib-resistant cultures. Cytotoxic effects of gefitinib on shRNA-transfected (J) PC9R and (K) HCC4006R cells (n=4). *P<0.05 vs. shNEG group. CAPN2, calpain 2; NSCLC, non-small cell lung cancer; shRNA, short hairpin RNA; shNEG, negative control shRNA; shCAPN2, CAPN2 shRNA.
Cell proliferation and viability assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assess cell proliferation. Briefly, cells were plated in 96-well plates at ~1,000 cells/well with 200 µl culture medium. After 24 h, 10 µl of the CCK-8 solution was added to each well, and the plates were incubated for 1 h at 37°C. Finally, the absorbance values at 450 nm were determined using a microplate reader (Multiskan; Thermo Fisher Scientific, Inc.), with a reference wavelength of 650 nm. All the experiments were conducted at least in triplicate.
Lentiviral construction and infection
For gene expression silencing, two short hairpin RNA (shRNA) vectors against the CAPN2 genes, namely shCAPN2-1 (TRCN0000272831) and shCAPN2-2 (TRCN0000003543), were obtained from the RNAi Consortium (Broad Institute, Cambridge, MA, USA). Lentiviral plasmids, containing GV112-shCAPN2-1 and shCAPN2-2, and negative control (shNEG) plasmids were obtained from GeneChem Co., Ltd. (Shanghai, China). For overexpression of EGFR, lentiviruses carrying human EGFR (GenBank accession no. NM_005228) and empty vector were also purchased from GeneChem Co., Ltd. The lentiviral particles were produced by transfecting 293T cells with the lentiviral plasmids. For viral infection, gefitinib-resistant PC9R or HCC4006R cells (5×105 cells/well) were plated in 6-well plates, grown to 50–70% confluence in a humidified atmosphere of 5% CO2 at 37°C, and subsequently incubated with RPMI-1640 medium containing the virus and 4 µg/ml polybrene at a multiplicity of infection of 10. After incubation for 6 h, the transfection medium was replaced with normal RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum and 100 U/ml penicillin/streptomycin. After incubation for another 18 h, the infected cells were then incubated with gefitinib (ranging from 0 to 20.0 µM) for 96 h. The transfection protocol was used in all the following experiments. For EGFR overexpression rescue experiments, EGFR overexpressed (PC9R-EGFR) and empty vector overexpressed (PC9R-vector) PC9R cells were transfected with shNEG and shCAPN2-1. For survivin or PI3K/AKT inhibition experiments, PC9R cells following transfection with shNEG, shCAPN2-1 or shCAPN2-2, were followed by incubation with 10 nM YM155 (a survivin inhibitor) or 20 µM LY294002 (a phosphoinositide 3-kinase/AKT inhibitor) for 24 h before CCK8 measurement.
5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay
Following transfection for 96 h, gefitinib-resistant PC9R and HCC4006R cells were incubated with 10 µM EdU (Thermo Fisher Scientific, Inc.) for 4 h before being fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature. EdU incorporation was detected according to manufacturer's protocol. After EdU staining, Hoechst 33342 (Thermo Fisher Scientific, Inc.) staining was detected according to manufacturer's protocol. The cells were then imaged using a Nikon A1R confocal laser scanning microscope system (Nikon Corporation, Tokyo, Japan). Gefitinib-resistant PC9R and HCC4006R cells that were positive for EdU incorporation and Hoechst 33342 staining were counted using ImageJ software (version 1.42; National Institutes of Health, Bethesda, MD, USA), and then the percentage of EdU-positive cells was calculated.
Detection of apoptosis by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining
A TUNEL assay was performed to measure cell apoptosis using the In Situ Cell Death Detection Kit, Fluorescein (Roche Molecular Diagnostics, Pleasanton, CA, USA). Briefly, transfected PC9R cells (5×105 cells/well) were seeded on glass coverslips coated with poly-L-lysine (ScienCell Research Laboratories, Inc., San Diego, CA, USA) and treated with 1 µM gefitinib. Following transfection for 96 h, cells were fixed in 3.7% formaldehyde in PBS for 15 min at room temperature. TUNEL staining was performed according to manufacturer's protocol. After TUNEL staining, DAPI staining (Thermo Fisher Scientific, Inc.) was performed. Slides were scanned (Pannoramic P250; 3DHistech Ltd., Budapest, Hungary) and viewed using the Pannoramic Viewer software (3DHistech Ltd.). PC9R cells positive for TUNEL and DAPI staining were counted using ImageJ software (version 1.42), and the percentage of TUNEL-positive cells was calculated.
Detection of apoptosis by flow cytometry
An Annexin V-APC and DAPI double staining kit (Thermo Fisher Scientific, Inc.) was used to analyze cellular apoptosis. Transfected PC9R cells were seeded in 6-well plates (5×105 cells/well) and treated with 1 µM gefitinib. Cells were then digested with trypsin (Gibco® trypsin-EDTA; Thermo Fisher Scientific, Inc.), washed with PBS three times, suspended in 500 µl binding buffer and then incubated with 5 µl APC-conjugated Annexin V and 3 µl DAPI for 15 min at room temperature in the dark. The stained cells were detected using a BD FACSAria II flow cytometer (BD Biosciences, San Jose, CA, USA).
Cell cycle analysis
Transfected PC9R cells were seeded in 6-well plates (5×105 cells/well) and treated with 1 µM gefitinib. Subsequently, cells were collected, washed with PBS and fixed in 70% ethanol for 24 h at 4°C. The fixed cells were then stained with propidium iodide and RNase (FS9527-100; Cell Cycle Fast detecting kit; Fusion Biotech, Shanghai, China) in the dark for 30 min at room temperature. Finally, the cell cycle distribution was analyzed by flow cytometry using a BD FACSAria II device (BD Biosciences).
Measurement of mitochondrial membrane potential
In order to examine changes in the mitochondrial membrane potential, a MitoProbe™ JC-1 assay kit (Thermo Fisher Scientific, Inc.) was used, according to the manufacturer's protocol. A BD FACSAria II flow cytometer was used to obtain the results. In healthy mitochondria, JC-1 forms J-aggregates emitting red fluorescence at 590 nm, while J-monomers emit green fluorescence at 490 nm in depolarized mitochondria; thus, mitochondria damage was indicated by an increase in the ratio of J-monomers.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.) and quantified using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). An amount of 1 µg RNA was used for reverse transcription by PrimeScript™ RT Reagent Kit (RR037A; Takara, Osaka, Japan). The cDNA (20 ng) was subsequently used as the template for qPCR. The amplification cycling parameters (40 cycles) were as follows: 15 sec at 95°C, 15 sec at 60°C and 45 sec at 72°C. The following primer sequences were used in the present study: CAPN2 sense, 5′-CGAGAGGGCCATCAAGTACC-3′ and antisense, 5′-TAGGGCCCCAACTCCTTGAA-3′; cyclin-dependent kinase inhibitor 1A (CDKN1A) sense, 5′-CTGGGGATGTCCGTCAGAAC-3′ and antisense, 5′-CATTAGCGCATCACAGTCGC-3′; growth arrest and DNA damage inducible α (GADD45A) sense, 5′-CCATGCAGGAAGGAAAACTATG-3′ and antisense, 5′-CCCAAACTATGGCTGCACACT-3′; cyclin-dependent kinase 1 (CDK1) sense, 5′-TAGCGCGGATCTACCATACC-3′ and antisense, 5′-CATGGCTACCACTTGACCTG-3′; CDK2 sense, 5′-GCCCTATTCCCTGGAGATTC-3′ and antisense, 5′-CAAGCTCCGTCCATCTTCAT-3′; and β-actin sense, 5′-CTGGCACCCAGCACAATG-3′ and antisense, 5′-CCGATCCACACGGAGTACTTG-3′. Gene expression was normalized to that of β-actin and calculated with the 2−ΔΔCq method (15). The RT-qPCR assay was performed at least three separate times in triplicate.
Western blot assay
Total protein from PC9, PC9R, HCC4006 and HCC4006R cells was extracted using RIPA lysis buffer and the protein concentration was determined using BCA assay (Shanghai Zhuoli Biotechnology Co., Ltd., Shanghai, China). Next, total protein was separated on polyacrylamide gels (5% stacking gel and 12% separating gel), and transferred to polyvinylidene difluoride membranes (PVDF). The membranes were then incubated at 4°C overnight with anti-CAPN2 (1:2,000) and keratin-5 (KRT5, 1:1,000) antibodies purchased from Abcam (Cambridge, MA, USA), as well as EGFR (1:1,000), protein kinase B (AKT, 1:1,000), phosphorylated AKT (pAKT; Ser473, 1:1,000), surviving (1:1,000), cleaved caspase-3 (Asp175, 1:1,000), cleaved poly-ADP-ribose polymerase (PARP; Asp214, 1:1,000) and β-actin (1:2,000) antibodies obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). Subsequently, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit (1:2,000) or anti-mouse immunoglobulin G secondary antibodies (1:2,000) obtained from Jackson ImmunoResearch, Inc. (West Grove, PA, USA) at room temperature for 1 h. Proteins were visualized using Pierce ECL western blotting substrate (Thermo Fisher Scientific, Inc.) and autoradiography. The blots were analyzed using Quantity One, version 4.6 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
cDNA array screening
Total RNA was extracted using RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA) from PC9R cells expressing shCAPN2-1 or negative control shRNA, reverse transcribed, and amplified using OneArray Plus RNA Amplification Kit (Phalanx Biotech Group, Taiwan). Cy5-labeled amplicons were hybridized to Human Whole Genome OneArray (Phalanx Biotech Group), imaged on a G2505C Agilent Microarray Scanner (Agilent Technologies, Santa Clara, CA, USA), and analyzed in Resolver (Rosetta Biosoftware, Seattle, WA, USA).
Evaluation of tumorigenicity
The animal experiments performed in the present study were approved by the Institutional Animal Care and Use Committee at Zhongshan Hospital, Fudan University (Shanghai, China). Eight male BALB/c nude mice (age, 4–6 weeks; weight, 18–20 g) were obtained from the Shanghai Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China), and were housed in cages with access to food and water in a temperature-controlled room with a 12 h dark/light cycle. PC9R cells transfected with shNEG or shCAPN2-1 were subcutaneously injected into the right flanks of nude mice without any treatment. Tumor volume was monitored and calculated according to the following formula: Volume=length × width2/2. After ~1 month, nude mice were sacrificed by cervical dislocation after anesthesia, and the tumors were isolated, weighed and embedded in paraffin. Ki67 immunohistochemical staining was performed on the sections of the paraffin-embedded tissue to identify the proliferating cells in the xenograft tumors. Ki67-positive cells were quantified in randomly selected fields from each tissue section using ImageJ software.
Statistical analysis
Data are expressed as the mean ± standard deviation of at least three independent experiments. One-way analysis of variance followed by Bonferroni's multiple comparison test was used to compare different groups, while Student's t-test was used for comparisons between two groups. A P-value of <0.05 was considered to denote a statistically significant difference.
Results
CAPN2 is upregulated in gefitinib-resistant cells and mediates gefitinib resistance
Two gefitinib resistant NSCLC cell lines, designated PC9R and HCC4006R, were established by continuously exposing PC9 and HCC4006 cells to increasing concentrations of gefitinib. Next, CAPN2 mRNA and protein expression was investigated, and the results revealed that CAPN2 was significantly upregulated in the PC9R and HCC4006R cell lines compared with the normal PC9 and HCC4006 cell lines, respectively (Fig. 1A-C), indicating that CAPN2 may serve an important role in gefitinib resistance. To confirm this, gefitinib resistance was examined in these four cell lines by CCK-8 assay, and it was observed that PC9 cells, which expressed relatively higher CAPN2 protein in comparison with HCC4006 cells, had a higher proliferation rate compared with that of HCC4006 cells in the presence of 0.01 µM gefitinib. By contrast, PC9R cells, which expressed relatively higher CAPN2 protein in comparison with HCC4006R cells, had a higher proliferation rate than that of HCC4006R cells in the presence of 0.1 µM gefitinib (Fig. 1D).
To further evaluate the role of CAPN2 in gefitinib resistance, PC9R and HCC4006R cells were transfected with shCAPN2-1 or shCAPN2-2 to silence CAPN2 expression. shNEG was transfected into cells as a control. The RT-qPCR and western blot analyses demonstrated that shCAPN2-1 and shCAPN2-2 transfection significantly inhibited CAPN2 expression compared with that in shNEG transfected cells (Fig. 1E-G). Furthermore, a morphological examination indicated that CAPN2 knockdown reduced the number of gefitinib-resistant cells, as observed by the increase in gefitinib effectiveness (1 µM, for 96 h) in PC9R and HCC4006R cells infected (24 h) with shCAPN2-1 or shCAPN2-2 compared with that in the shNEG (Fig. 1H and I). It was also confirmed that CAPN2 knockdown reduced the viability of gefitinib-resistant cells by treating silenced and control cells with different gefitinib concentrations (ranging between 0.1 and 20.0 µM; Fig. 1J and K).
CAPN2 knockdown inhibits gefitinib-resistant cell proliferation in vitro and in vivo
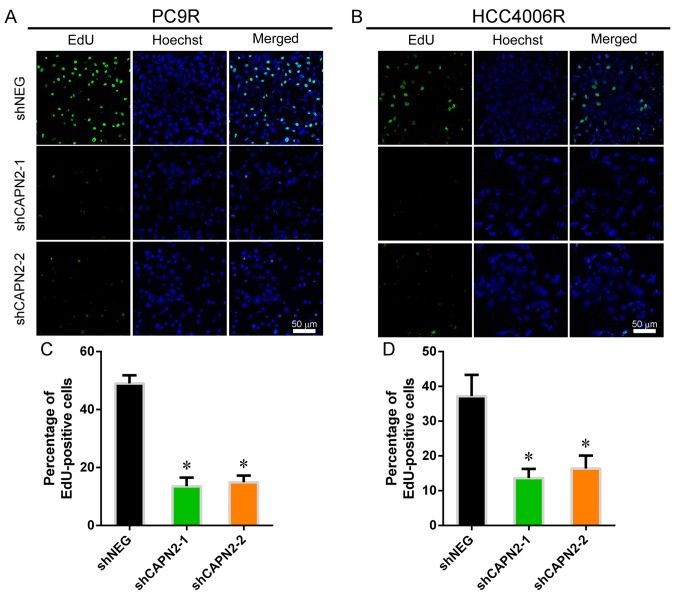
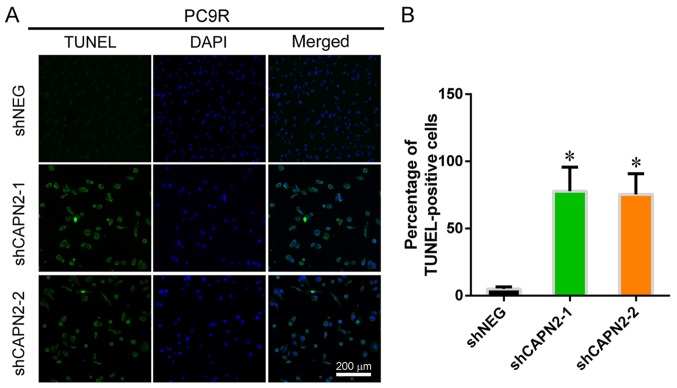
To investigate the effect of CAPN2 on gefitinib-resistant cell proliferation, EdU incorporation assay was performed (Fig. 2A and B). CAPN2 knockdown appeared to suppress the proliferation of gefitinib-resistant cells in the presence of gefitinib (Fig. 2C and D), implying that CAPN2 promotes proliferation in gefitinib-resistant cells. Furthermore, TUNEL staining demonstrated that CAPN2 knockdown significantly increased the apoptosis of gefitinib-resistant PC9R cells in the presence of gefitinib (Fig. 3A and B).
Figure 2.
CAPN2 knockdown inhibits gefitinib-resistant cell proliferation in vitro. Representative images showing EdU incorporation in (A) PC9R and (B) HCC4006R cells. Statistical analysis of EdU incorporation in (C) PC9R and (D) HCC4006R cells. CAPN2, calpain 2; EdU, 5-ethynyl-2′-deoxyuridine; sh, short hairpin RNA; NEG, negative control. *P<0.05 vs. negative control.
Figure 3.
CAPN2 knockdown induces gefitinib-resistant cell apoptosis in vitro. (A) Representative images demonstrating TUNEL staining in PC9R cells. (B) Statistical analysis of TUNEL-positive PC9R cells. CAPN2, calpain 2; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; sh, short hairpin RNA; NEG, negative control. *P<0.05 vs. negative control.
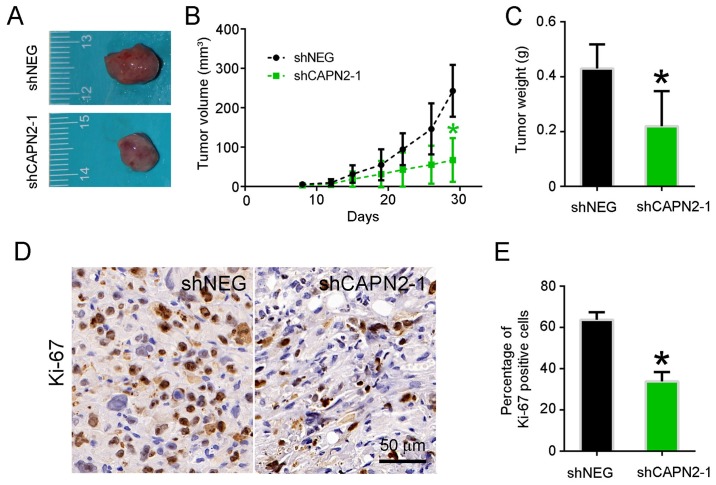
To determine whether CAPN2 knockdown suppresses lung tumor growth in vivo, PC9R cells transfected with shCAPN2-1 and shNEG were injected into nude mice. Visible tumors were observed in animals after 1 week; however, the volume of tumors derived from CAPN2-silenced cells was evidently smaller in comparison with that of tumors derived from control cells (Fig. 4A). A similar trend was observed at 1 month after transfection in terms of the tumor volume and weight (Fig. 4B and C). Accordingly, the abundance of proliferating Ki67-positive cells was significantly lower in CAPN2-deficient tumors as compared with that in control xenografts (Fig. 4D and E).
Figure 4.
CAPN2 knockdown inhibits gefitinib-resistant cell proliferation in vivo. (A) Representative images of tumors, (B) quantified tumor volume and (C) quantified tumor weight at 1 month after subcutaneous implantation of PC9R cells transfected with shCAPN2-1 or shNEG. (D) Ki-67 immunohistochemical staining of xenograft tumors, and (E) statistical analysis of Ki-67 staining. CAPN2, calpain 2; sh, short hairpin RNA; NEG, negative control. *P<0.05 vs. negative control.
CAPN2 silencing promotes apoptosis and cell cycle arrest in gefitinib-resistant cells via caspase activation and mitochondrial dysfunction
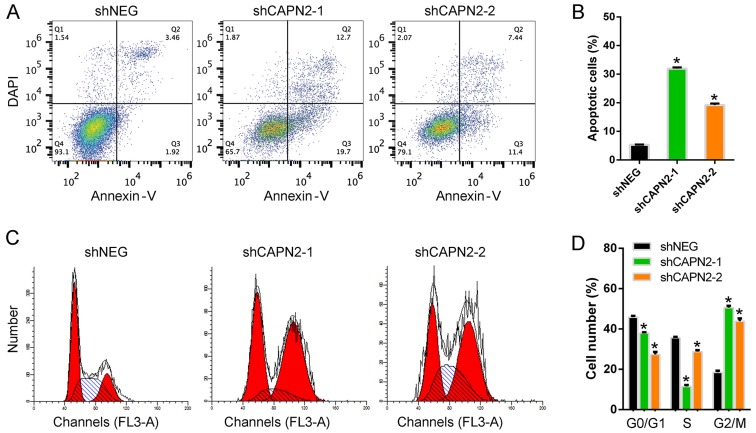
To further confirm the function of CAPN2 during gefitinib resistance, the cell apoptosis levels were analyzed. Upon treatment with gefitinib, the apoptotic rate in CAPN2-silenced PC9R cells was significantly higher than that of control cells (Fig. 5A and B). Furthermore, the number of cells in the G0/G1 and S phases was markedly decreased upon CAPN2 silencing, while the number of cells in the G2/M phase was increased, indicating cell cycle arrest (Fig. 5C and D).
Figure 5.
CAPN2 silencing promotes apoptosis and cell cycle arrest in gefitinib-resistant cells. (A) Scatterplots of cellular apoptosis detected by flow cytometry in PC9R cells transfected with shNEG, shCAPN2-1 or shCAPN2-2, and treated with 1 µM gefitinib. (B) Percentage of apoptotic cells. (C) Representative histograms highlighting the distribution of cells in each phase of the cell cycle for PC9R cells transfected with shNEG, shCAPN2-1 or shCAPN2-2, and treated with 1 µM gefitinib. (D) Statistical analysis of cell cycle distribution for each treatment. CAPN2, calpain 2; sh, short hairpin RNA; NEG, negative control. *P<0.05 vs. negative control.
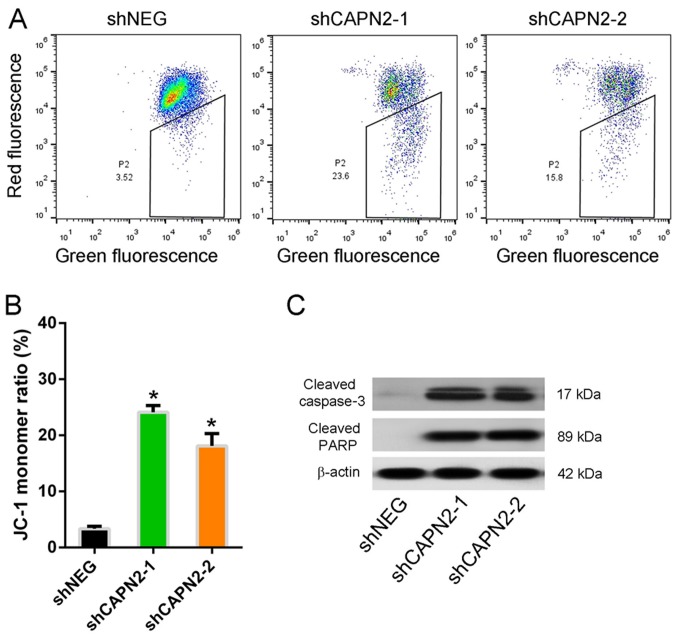
The mitochondrial integrity was also assessed by staining cells with JC-1, a dye that emits red fluorescence as it aggregates in intact mitochondria, but emits green fluorescence as a monomer in damaged mitochondria. The JC-1 monomer ratio in CAPN2-silenced PC9R cells was higher compared with that of control cells in the presence of gefitinib (Fig. 6A and B), indicating mitochondrial dysfunction. Furthermore, cleaved caspase-3 and cleaved PARP, two apoptotic proteins, were upregulated in CAPN2-depleted cells compared with the levels in control cells (Fig. 6C), indicating that CAPN2-depleted cells undergo apoptosis (16).
Figure 6.
CAPN2 regulates mitochondrial dynamics and apoptotic protein expression. (A) Scatterplots of JC-1 expression detected by flow cytometry following shNEG, shCAPN2-1 or shCAPN2-2 transfection. (B) Statistical analysis of JC-1 distribution using the J-monomer ratio. (C) Representative western blots demonstrating cleaved caspase-3 and cleaved PARP expression in transfected cells. CAPN2, calpain 2; sh, short hairpin RNA; NEG, negative control; PARP, poly-ADP-ribose polymerase. *P<0.05 vs. negative control.
CAPN2 regulates gefitinib-resistance via the EGFR/AKT/survivin signaling pathway
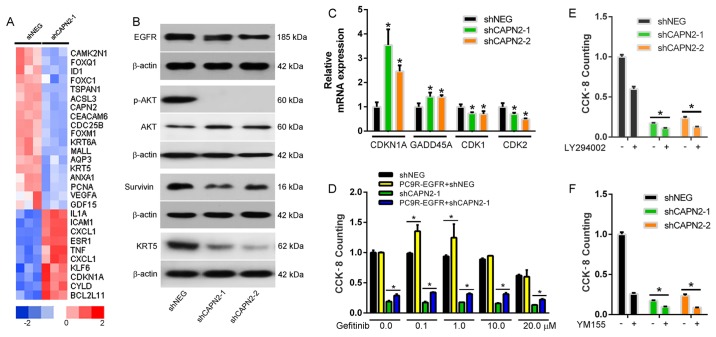
To evaluate the molecular mechanism underlying CAPN2-mediated gefitinib resistance, a whole human cDNA array was used, and changes in CAPN2-associated downstream signaling pathways upon CAPN2 silencing were investigated (Fig. 7A). Western blotting confirmed that EGFR, pAKT, survivin and KRT5 levels were evidently decreased in cells transfected with shCAPN2-1 or shCAPN2-2 compared with those in cells transfected with shNEG (Fig. 7B). Furthermore, RT-qPCR analysis confirmed that the expression levels of the cell cycle genes CDKN1A and GADD45A were significantly increased, whereas CDK1 and CDK2 levels were decreased in PC9R cells transfected with shCAPN2-1 or shCAPN2-2 compared with those in cells transfected with shNEG (Fig. 7C). Given that the EGFR/AKT/survivin pathway serves a crucial role in NSCLC progression (17,18), it is likely that CAPN2 confers gefitinib resistant through EGFR signaling.
Figure 7.
CAPN2 regulates gefitinib-resistance via the EGFR/AKT/survivin signal pathway. (A) Heatmap of downstream CAPN2 target proteins determined by a whole human cDNA array. (B) EGFR, pAKT, AKT, survivin and KRT5 protein expression was measured by western blotting. (C) CDKN1A, GADD45A, CDK1 and CDK2 gene expression was measured by reverse transcription-quantitative polymerase chain reaction. *P<0.05 vs. negative control. (D) The viability of PC9R cells transfected with EGFR overexpressing vector and/or shCAPN-1 in the presence of various concentrations of gefitinib was determined by cell counting kit-8 assay. Viability of PC9R cells following transfection with shNEG, shCAPN2-1 or shCAPN2-2, followed by incubation with (E) 20 µM LY294002 (a phosphoinositide 3-kinase/AKT inhibitor) or (F) 10 nM YM155 (a survivin inhibitor). *P<0.05 between two groups. CAPN2, calpain 2; sh, short hairpin RNA; NEG, negative control; EGFR, epidermal growth factor receptor; AKT, protein kinase B; KRT5, keratin-5; CDKN1A, cyclin-dependent kinase inhibitor 1A; GADD45A, growth arrest and DNA damage inducible α; CDK, cyclin-dependent kinase.
To evaluate this possibility, a rescue assay was performed in the PC9R cells. It was observed that overexpression of EGFR in PC9R cells increased cell viability in the presence of gefitinib (Fig. 7D). However, overexpression of EGFR in the PC9R-shCAPN2 cells also rescued the cell viability in the presence of gefitinib. These results suggest that EGFR overexpression partly reverses CAPN2 depletion-induced apoptosis. Furthermore, inhibition of phosphoinositide 3-kinase (PI3K)/AKT (Fig. 7E) or survivin (Fig. 7F) in the PC9R-shCAPN2 cells was able to further induce cell apoptosis. Taken together, all these findings indicated that CAPN2 stimulates gefitinib resistance mainly via activation of the EGFR/AKT/survivin signaling pathway.
Discussion
Drug resistance is a major cause of cancer treatment failure. Targeted EGFR drugs, such as gefitinib, appear to be effective for approximately 1 year, but resistance is inevitably developed shortly after this period. CAPN2 has been demonstrated to serve a role in altering the cellular response to certain cancer therapies (9,10,19). However, the events and mechanisms involved in these processes remain incompletely understood. In the present study, it was observed that CAPN2 is strongly associated with gefitinib-resistance, with CAPN2 mRNA and protein expression levels being significantly increased in gefitinib-resistant cell lines. In addition, CAPN2 was found to regulate gefitinib-resistant cell proliferation, mitochondrial function and apoptosis, largely involving dysregulation of the PI3K/AKT/survivin pathway.
Previously, high CAPN2 expression was reported to be significantly associated with platinum resistant tumors in ovarian cancer (13), while CAPN2 degradation mediated irinotecan secondary resistance in colorectal cancer xenografts (20). Consistent with these previous studies, CAPN2 was observed to be upregulated in gefitinib-resistant cells in the present study. Furthermore, CAPN2 knockdown inhibited gefitinib-resistant cell proliferation in vitro and in vivo.
In addition to its effect on proliferation in drug resistant cells, CAPN2 knockdown also appears to affect the mitochondrial function. The mitochondria are essential during various cellular processes and function to protect cells from apoptosis, genomic instability, inflammation, abnormal bioenergetics, oxidative stress and metastasis (21). Cleaved caspase-3 and cleaved PARP are two critical targets of the mitochondria-mediated apoptosis pathway (22). Indeed, a previous study has demonstrated that mitochondria-mediated caspase-dependent apoptosis is involved in the anticancer activity against human lung cancer cells (23). In the present study, the results revealed that CAPN2 knockdown induced gefitinib-resistant cell apoptosis and cell cycle arrest in conjunction with caspase activation and increased mitochondrial dysfunction. This is not altogether surprising as CAPNs are known to be responsible for mitochondrial membrane permeabilization (24), which subsequently alters various mitochondrial processes.
Notably, using a whole human cDNA array in the current study, EGFR was identified to be a downstream signaling molecule of CAPN2. The EGFR downstream signaling route includes the MAPK, JAK/STAT and PI3K/AKT/mTOR pathways among others (6). Previous studies have demonstrated that AKT activation contributes to EGFR-TKI resistance in EGFR mutation-positive lung cancer (3,20,25), indicating that the PI3K/AKT pathway is an important target for overcoming EGFR-TKI resistance. It has also been reported that CAPN2 activates AKT signaling in pulmonary artery smooth muscle cells (9), which is consistent with the changes in AKT phosphorylation observed in the present study. Survivin is an apoptosis inhibitor involved in tumorigenesis, and inhibition of the EGFR/PI3K/AKT pathway appears to downregulate its expression in EGFR mutation-positive NSCLC cells (17). This is consistent with the findings of the present study, which demonstrated that CAPN2 knockdown in gefitinib-resistant cells repressed survivin expression. Furthermore, KRT5 is a well-recognized marker for basal cells (26,27) and is overexpressed in various cancer types (7). Similar to surviving expression, the current study observed that CAPN2 inhibition reduced KRT5 expression, suggesting that CAPN2 may regulate cell differentiation via KRT5 inhibition.
In conclusion, the present study investigated the role of CAPN2 in gefitinib-resistant lung adenocarcinoma cells and revealed that CAPN2 expression is upregulated during drug resistance. CAPN2 appeared to regulate PC9R cell proliferation and apoptosis by triggering mitochondrial caspase-dependent cell death pathways and mainly targeting EGFR/AKT/survivin signaling. Furthermore, downregulation of CAPN2 was observed to inhibit PC9R cell proliferation and induce cell apoptosis both in vitro and in vivo. These data demonstrate that CAPN2 functions as a facilitator of gefitinib resistance, largely via the activation of EGFR/AKT/survivin signaling pathway. Thus, the present study supports the development of drugs targeting CAPN2 as a means to enhance the effectiveness of cancer therapy and reduce drug resistance.
Acknowledgements
The authors would like to thank Mr. Lei Gao, a member of Bai Lab, for suggestions and discussions.
Funding
This study was supported by grants from the National Key R&D Plan (no. 2016YFC1304104), the National Major Scientific and Technological Special Project for ‘Significant New Drugs Development’ (no. 2018ZX09201002-006), the Natural Science Foundation of China (nos. 81400018, 81570028 and 81770039), the Shandong Province Natural Science Foundation (no. ZR2017PH066), the Shanghai Science and Technology Committee Grant (15DZ1941100), the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (no. 2013BAI09B00), the Zhongshan Hospital Clinical Research Foundation (no. 2016ZSLC05), and the Shanghai Three-Year Plan of the Key Subjects Construction in Public Health-Infectious Diseases and Pathogenic Microorganism (no. 15GWZK0102).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JZ, CB and QH were involved in the conception of the study and supervised the conduction of the entire project. GZ, TF, MC and JL performed the research, analyzed the data and prepared the manuscript. All authors have read the final version of the manuscript and are in agreement for publication upon acceptance.
Ethics approval and consent to participate
Experiments were approved by the Research Ethics Committee of Zhongshan Hospital, Fudan University (Shanghai, China) and performed according to relevant guidelines and regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16:e447–e459. doi: 10.1016/S1470-2045(15)00246-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhong WZ, Zhou Q, Wu YL. The resistance mechanisms and treatment strategies for EGFR-mutant advanced non-small-cell lung cancer. Oncotarget. 2017;8:71358–71370. doi: 10.18632/oncotarget.20311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvayrac O, Mazieres J, Figarol S, Marty-Detraves C, Raymond-Letron I, Bousquet E, Farella M, Clermont-Taranchon E, Milia J, Rouquette I, et al. The RAS-related GTPase RHOB confers resistance to EGFR-tyrosine kinase inhibitors in non-small-cell lung cancer via an AKT-dependent mechanism. EMBO Mol Med. 2017;9:238–250. doi: 10.15252/emmm.201606646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B. 2015;5:390–401. doi: 10.1016/j.apsb.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreira JB, Wohlwend M, Alves MN, Wisløff U, Bye A. A small molecule activator of AKT does not reduce ischemic injury of the rat heart. J Transl Med. 2015;13:76. doi: 10.1186/s12967-015-0444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 7.Camilo R, Capelozzi VL, Siqueira SA, Del Carlo Bernardi F. Expression of p63, keratin 5/6, keratin 7, and surfactant-A in non-small cell lung carcinomas. Hum Pathol. 2006;37:542–546. doi: 10.1016/j.humpath.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Ho WC, Pikor L, Gao Y, Elliott BE, Greer PA. Calpain 2 regulates Akt-FoxO-p27(Kip1) protein signaling pathway in mammary carcinoma. J Biol Chem. 2012;287:15458–15465. doi: 10.1074/jbc.M112.349308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P, Miao C, Liang C, Shao P, Wang Z, Li J. Silencing CAPN2 expression inhibited castration-resistant prostate cancer cells proliferation and invasion via AKT/mTOR signal pathway. Biomed Res Int. 2017;2017:2593674. doi: 10.1155/2017/2593674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B, Zhou Y, Lu D, Liu Y, Zhang SQ, Xu Y, Li W, Gu X. Comparison of the protein expression of calpain-1, calpain-2, calpastatin and calmodulin between gastric cancer and normal gastric mucosa. Oncol Lett. 2017;14:3705–3710. doi: 10.3892/ol.2017.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li ST, Chen NN, Qiao YB, Zhu WL, Ruan JW, Zhou XZ. SC79 rescues osteoblasts from dexamethasone though activating Akt-Nrf2 signaling. Biochem Biophys Res Commun. 2016;479:54–60. doi: 10.1016/j.bbrc.2016.09.027. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Chang M, Li J, Fang T, Hu J, Bai C. Knockdown of annexin A5 restores gefitinib sensitivity by promoting G2/M cell cycle arrest. Respir Res. 2018;19:96. doi: 10.1186/s12931-018-0804-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storr SJ, Safuan S, Woolston CM, Abdel-Fatah T, Deen S, Chan SY, Martin SG. Calpain-2 expression is associated with response to platinum based chemotherapy, progression-free and overall survival in ovarian cancer. J Cell Mol Med. 2012;16:2422–2428. doi: 10.1111/j.1582-4934.2012.01559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang M, Hu J, Fang T, Li J, Song Y, Bai C, Zhou J. PLIN2 confers gefitinib resistance by inhibiting cell apoptosis via activation of EGFR/AKT/survivin in PC9R cells. Oncotarget. 2018 Jan 10; (Epub ahead of print). doi: 10.18632/oncotarget.24153. [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis: Lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto K, Okamoto I, Hatashita E, Kuwata K, Yamaguchi H, Kita A, Yamanaka K, Ono M, Nakagawa K. Overcoming erlotinib resistance in EGFR mutation-positive non-small cell lung cancer cells by targeting survivin. Mol Cancer Ther. 2012;11:204–213. doi: 10.1158/1535-7163.MCT-11-0638. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto K, Okamoto I, Okamoto W, Tanaka K, Takezawa K, Kuwata K, Yamaguchi H, Nishio K, Nakagawa K. Role of survivin in EGFR inhibitor-induced apoptosis in non-small cell lung cancers positive for EGFR mutations. Cancer Res. 2010;70:10402–10410. doi: 10.1158/0008-5472.CAN-10-2438. [DOI] [PubMed] [Google Scholar]
- 19.Miao C, Liang C, Tian Y, Xu A, Zhu J, Zhao K, Zhang J, Hua Y, Liu S, Dong H, et al. Overexpression of CAPN2 promotes cell metastasis and proliferation via AKT/mTOR signaling in renal cell carcinoma. Oncotarget. 2017;8:97811–97821. doi: 10.18632/oncotarget.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobsen K, Bertran-Alamillo J, Molina MA, Teixidó C, Karachaliou N, Pedersen MH, Castellví J, Garzón M, Codony-Servat C, Codony-Servat J, et al. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat Commun. 2017;8:410. doi: 10.1038/s41467-017-00450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim W, Yang C, Jeong M, Bazer FW, Song G. Coumestrol induces mitochondrial dysfunction by stimulating ROS production and calcium ion influx into mitochondria in human placental choriocarcinoma cells. Mol Hum Reprod. 2017;23:786–802. doi: 10.1093/molehr/gax052. [DOI] [PubMed] [Google Scholar]
- 22.Sivalingam KS, Paramasivan P, Weng CF, Viswanadha VP. Neferine potentiates the antitumor effect of cisplatin in human lung adenocarcinoma cells Via a mitochondria-mediated apoptosis pathway. J Cell Biochem. 2017;118:2865–2876. doi: 10.1002/jcb.25937. [DOI] [PubMed] [Google Scholar]
- 23.Losuwannarak N, Sritularak B, Chanvorachote P. Cycloartobiloxanthone induces human lung cancer cell apoptosis via mitochondria-dependent apoptotic pathway. In Vivo. 2018;32:71–78. doi: 10.21873/invivo.11206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clinic Proc. 2008;83:584–594. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Jiang H, Wang L, Chen X, Wu K, Zhang S, Ma S, Xia B. Increased MIR31HG lncRNA expression increases gefitinib resistance in non-small cell lung cancer cell lines through the EGFR/PI3K/AKT signaling pathway. Oncol Lett. 2017;13:3494–3500. doi: 10.3892/ol.2017.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.