Abstract
Although the inhibitor of apoptosis protein-like protein-2 (ILP-2) has been shown as a serological biomarker for breast cancer, its effect on breast cancer cell growth remains elusive. The present study aimed to determine the role of ILP-2 in breast cancer cell growth. We used immunohistochemistry to analyze ILP-2 expression in 59 tissue paraffin-embedded blocks, which included 35 breast cancer tissues and 24 galactophore hyperplasia tissues. Western blot analysis was used to detect protein expression levels of ILP-2 in breast cancer cell lines such as HCC-1937, MX-1 and MCF-7 as well as breast gland cell line MCF 10A. ILP-2 was silenced by siRNA in HCC-1937, MX-1 and MCF-7 cell lines. MTT assays, scratch assays and AO-EB double staining analysis were conducted to evidence the role of ILP-2 on breast cancer cell growth. Results from this study showed increased ILP-2 expression in breast cancer tissues and breast cancer cell lines such as HCC-1937, MX-1 and MCF-7. Cell viability or rate of cell migration of HCC-1937, MX-1 and MCF-7 cell lines was significantly inhibited when ILP-2 was knocked down by siRNA. The apoptosis rate of HCC-1937, MX-1 and MCF-7 cell lines was increased when compared with that of the control group. Thus, ILP-2 plays an active role in the growth of breast cancer cells.
Keywords: ILP-2, breast cancer cells, HCC-1937, MX-1, MCF-7
Introduction
Inhibitors of apoptosis proteins (IAPs) are involved in cell death, migration and cell cycle (1). They exhibit an important role in maintaining the balance between cell death and cell growth (2,3). IAPs may effectively suppress cell apoptosis and promote cell growth (4). Additionally, IAP deregulation may accelerate cell canceration and provide a new target for anticancer therapy (5).
Following identification of the first human IAP protein, neuronal apoptosis inhibitory protein (NAIP) (6), eight proteins of IAP family have been identified: protein (NAIP/BIRC1/NLRB), cellular IAP1/human IAP2/BIRC2, cellular IAP2/human IAP1/BIRC3, X-linked IAP (XIAP)/BIRC4, survivin/BIRC5, baculoviral IAP repeat-containing ubiquitin-conjugating enzyme/apollon/BIRC6, Livin/melanoma-IAP/BIRC7/KIAP and inhibitory of apoptosis proteins-like protein-2 (ILP-2)/testis-specific IAP (Ts-IAP)/hILP-2/BIRC8 (5,7,8).
ILP-2 was initially detected in the human testis (9,10) and in our previous study, we showed that ILP-2 is a novel serological biomarker for breast cancer (8). In this study, we identified whether ILP-2 led to breast cancer progression. We analyzed the expression levels of ILP-2 in breast cancer tissue samples by immunohistochemistry, followed by detection of ILP-2 expression in breast cancer cell lines through western blot analysis. We further investigated the influence of ILP-2 in breast cancer growth via MTT assay, scratch assay and acridine orange/ethidium bromide (AO/EB)double-staining analysis in ascertaining the specific role of ILP-2 in breast cancer. Furthermore, we knocked down ILP-2 by siRNA to elucidate its role in breast cancer.
Materials and methods
Statement of Ethics
The present study was approved by Jishou University Ethics Committee (Jishou, China). All participated patients received and approved the written informed consent before joining the study.
Patient samples
Fifty-nine pathological samples were provided by the First Affiliated Hospital, College of Medical Science, Jishou University (Jishou, China). These samples included 35 breast cancer and 24 galactophore hyperplasia tissues. The average age of breast cancer patients and galactophore hyperplasia patients was 47 and 32 years, respectively. These samples were collected from July 2013 to October 2013. Breast cancer samples were collected from stage II patients diagnosed according to cancer pathology standards, which are scored on the formation of the gland tumor, the polymorphism of nucleus and the fission count, and designated the infiltrating ductal carcinoma for level I on the total score 3–5, level II on 6–7 and level III on 8–9. In addition, galactophore hyperplasia samples were from breast fibroadenoma patients.
Immunohistochemistry
Tissue paraffin-embedded blocks were cut into 4 µm-thick slices and pasted on the glass slide. These blocks were dewaxed by dimethylbenzene and dehydrated in graded series of alcohol. Peroxidase was inactivated by incubation with 3% hydrogen peroxide at 25°C for 10 min. Antigens were retrieved by heating a citrate-buffered solution in the microwave for 10 min. Tissue sections were incubated with 5% bovine serum albumin (BSA) for 13 min at room temperature and then incubated with ILP-2 (rabbit IgG; dilution 1:500; cat. no. sc-130107; Santa Cruz Biotechnology, Inc., CA, USA) overnight at 4°C. Slices were visualized with 3,3′-diaminobenzidine kit (Beyotime Institute of Biotechnology, Shanghai, China) for 5 min, and then stained by hematoxylin and examined under light microscopy.
Western blot analysis
HCC-1937, MX-1, MCF-7 and MCF-10A cell lines (5×106 cells/cell line) were separately collected and western blot analysis was performed. Briefly, total proteins were extracted from cells with RIPA buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS and 1% deoxycholate) containing protease inhibitors. The total protein concentration was determined using a BCA Protein Assay kit (Applygen Technologies Inc., Beijing, China). Thirty micrograms of total protein was separated on a 10% SDS-polyacrylamide gel and then transferred onto polyvinylidene fluoride (PVDF) membranes (Merck KGaA, Darmstadt, Germany). The membranes were blocked with TBST (Tris-buffered saline and Tween-20) and 5% skim milk powder for 2 h, then incubated with primary antibodies against ILP-2 (rabbit IgG, 1:1,000; cat. no. Ab9664; Abcam, Cambrige, UK) and tubulin (rabbit IgG, 1:1,000; cat. no. 11224-1-AP; Proteintech Group, Wuhan, China) overnight at 4°C. On the second day, the membranes were washed by TBST and incubated with a goat anti-rabbit immunoglobulin G-horseradish peroxidase (IgG-HRP) (anti-rabbit IgG, 1:5,000; cat. no. ZB-2301; ZSGB-BIO, Beijing, China) for 1 h at room temperature. An enhanced chemiluminescence detection system (Super ECL Plus; Applygen Technologies) was used to visualize immunoreactive proteins. The protein band was visualized by chemical chemiluminescence imaging system (Beijing Sage Creation Science Co., Ltd., Beijing, China). The images were analyzed with ImageJ (National Institutes of Health, Bethesda, MD, USA). Tubulin was used as an internal control.
RNA interference (RNAi)
According to the manufacturer's instructions, Invitrogen™ Lipofectamine 2000 (Thermo Fisher Scientific, Inc., MA, USA) and small interfering RNA [siRNA-5, siRNA-3, siRNA-1519, siRNA-1643 and negative control (NC)] (Shanghai GenePharma Co., Ltd., Shanghai, China) were separately gently mixed at a volume ratio of 1:1 and stored for 20 min at room temperature. Mixed Lipofectamine 2000 and small interfering RNA were added to MCF-7 cells in the logarithmic growth phase and cultivated in 5% CO2 for 24 h at 37°C, wherein the small interfering RNA had a final concentration of 50 nM. Interference effects on ILP-2 expression were analyzed via western blot analysis.
Mixed Lipofectamine 2000 and small interfering RNA (siRNA-5, siRNA-3 and NC) were correspondingly added to the HCC-1937, MX-1 and MCF-7 cells in the logarithmic growth phase and cultivated in 5% CO2 for 24, 48 and 72 h at 37°C. Total proteins were separately extracted. After determining protein concentration using the BCA kit (Beyotime Institute of Biotechnology, Shanghai, China), proteins were analyzed by western blot analysis.
MTT assays
HCC-1937, MX-1 and MCF-7 cells in the logarithmic phase were simultaneously digested with trypsin, re-suspended in complete culture medium and arranged in four groups: The siRNA-5 group, the siRNA-3 group, the NC group and HCC-1937 or MX-1 or MCF-7 cell group. Four groups of cells were separately seeded on 2×103 cells/well in three 96-well plates, wherein every cell group was arranged in 6-wells, incubated in 5% CO2 for 24 h at 37°C and mixed with the small interfering RNA. Following cultivation of the four cell groups for 24, 48 and 72 h, 10 µl of MTT (5 mg/ml) (Beijing Dingguo Biotechnology, Beijing, China) was separately added to each well. The medium was discarded 4 h later and 100 µl dimethyl sulfoxide (DMSO; Shanghai Pharmaceutical Group Co., Ltd., Shanghai, China) was added to each well to stop the reaction. After vortexing for 10 min at room temperature, optical density was measured at 490 nm using a microplate reader (Biotek ELx800; BioTek Instruments, Winooski, VT, USA).
Scratch assays
Five parallel lines were scratched on the back of a 6-well plate per well. Four groups of HCC-1937, MX-1 and MCF-7 cells in the logarithmic phase, analogous to MTT assay procedure, were simultaneously seeded on 1×104 cells/well in a 6-well plate, cultivated in 5% CO2 for 24 h at 37°C, mixed with small interfering RNA and incubated for 24 h. Three vertical scratch wounds were separately made on parallel lines by a sterile pipette tip in the four cell groups. Cells were cultivated in a 5% CO2 at 37°C. Three scratch wound areas were measured by an Olympus CKX41 inverted microscope (Olympus Corp., Tokyo, Japan) for 0, 24, 48 and 72 h. Experiments were triplicated.
AO/EB double staining analysis
HCC-1937, MX-1 and MCF-7 cells in the logarithmic growth phase were adhered to coverslips in the well of a 6-well plate (1×104 cells/well) and cultivated in 5% CO2 for 24 h at 37°C. Mixed small interfering RNA (siRNA-5, siRNA-3 and NC) and Lipofectamine 2000 were simultaneously added to the HCC-1937, MX-1 and MCF-7 cells and subsequently cultivated in 5% CO2 for 24, 48 and 72 h at 37°C. Culture medium was removed and cells were washed with phosphate-buffered saline (PBS) twice. Dye Reagents 1 and 2 were mixed at a volume ratio of 1:1 and diluted by PBS at a ratio of 1:25. Diluted staining agent was dropped into breast cancer cells and cultivated in the darkroom for 5 min at room temperature. Coverslips were examined under an Olympus BX51 inverted fluorescence microscope (Olympus Corp.) at an excitation/emission wavelength of 498/516 nm.
Statistical analysis
Histograms were constructed by GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical analysis was performed by conducting χ2 test, t-test, one-way ANOVA, or post hoc Bonferroni tests by SPSS statistics 17.0 (SPSS, Inc., Chicago, IL, USA). A level of P<0.05 was considered statistically significant.
Results
Breast cancer tissues have high ILP-2 expression
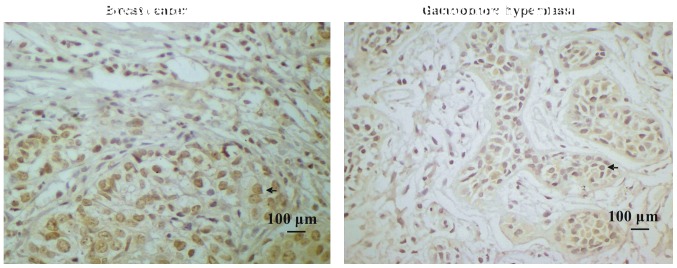
Paraffin-embedded blocks were analyzed using immunohistochemical analysis to investigate ILP-2 protein expression in the tissues. As shown in Fig. 1A and B, expression levels of ILP-2 in breast cancer tissues were significantly increased in comparison to levels noted in the galactophore hyperplasia tissues (P<0.05). In addition, positive rates of ILP-2 expression in breast cancers were higher than that in the galactophore hyperplasias (Table I) (χ2=8.183, P<0.01). Our results indicate high ILP-2 expression levels in breast cancer tissues.
Figure 1.
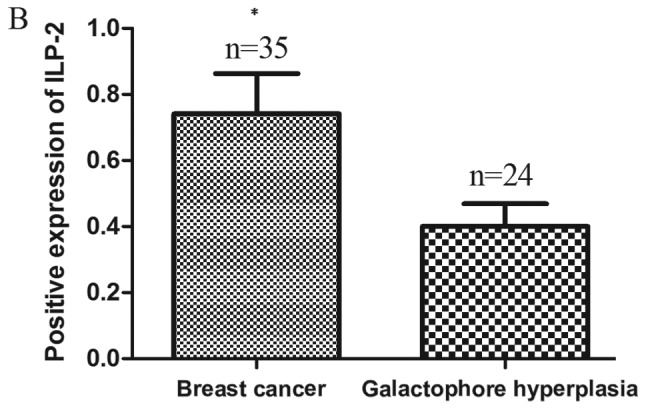
(A) Immunohistochemistry of ILP-2 expression in tissues. Samples of 35 breast cancers and 24 galactophore hyperplasias were analyzed. Scale bar, 100 µm. (B) The positive rate of breast cancers and galactophore hyperplasias was 62.9 and 25%, respectively. Groups were compared by conducting χ2 test and t-test. *P<0.05 vs. the galactophore hyperplasia samples; n=59.
Table I.
Immunohistochemical analysis of ILP-2 in breast tissues.
| Protein expression of ILP-2 | ||||
|---|---|---|---|---|
| Group | n | Positive | Negative | Positive rate (%) |
| Breast cancer | 35 | 22 | 13 | 62.9a |
| Galactophore hyperplasia | 24 | 6 | 18 | 25 |
χ2=8.183
P<0.01. ILP-2, inhibitor of apoptosis protein-like protein-2.
ILP-2 is highly expressed in breast cancer cells
Western blot analysis was employed to analyze ILP-2 expression in the 4 breast cancer cell lines. As shown in Fig. 2, protein expression levels of ILP-2 in the breast cancer cell lines such as HCC-1937 (P<0.05), MX-1 (P<0.01) and MCF-7 (P<0.01) were significantly increased when compared with that of the breast epithelial cell line MCF-10A. These results corroborate the immunohistochemical results, evidencing the highly expressed levels of ILP-2 breast cancer cells.
Figure 2.
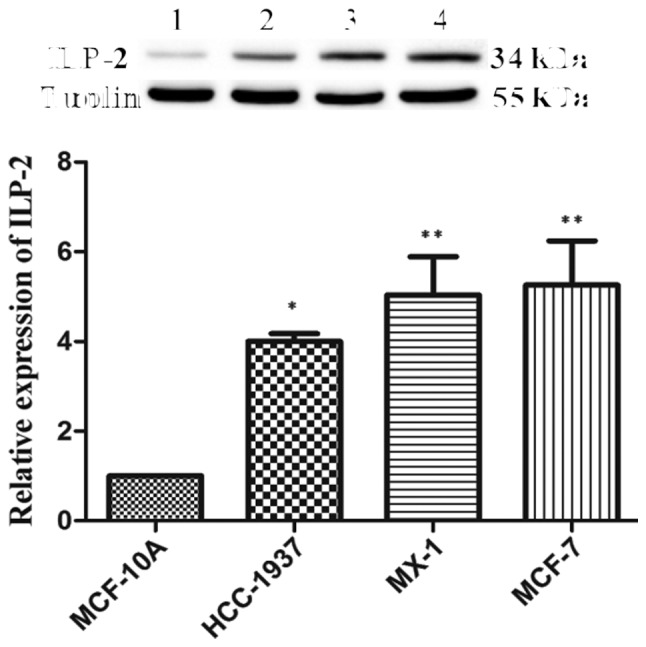
Western blot analysis indicating ILP-2 protein expression in MCF-10A, HCC-1937, MX-1 and MCF-7 cell lines. The results showed that ILP-2 expression in the HCC-1937, MX-1 and MCF-7 cell lines was significantly increased when compared to that of the MCF-10A cells. Lanes 1, 2, 3 and 4 separately indicate the protein expression level in the MCF-10A, HCC-1937, MX-1 and MCF-7 cell lines. *P<0.05, **P<0.01 vs. the MCF-10A cells. Data are expressed as mean ± SEM by ANOVA. Groups were compared by conducting Bonferroni test; n=3.
siRNA-5 sequence inhibits ILP-2 expression in breast cancer cell lines
We employed western blot analysis to analyze the interference effect of five siRNAs on ILP-2 expression in the MCF-7 cell line. As shown in Fig. 3, when ILP-2 was specifically knocked down by five siRNAs (siRNA-5, siRNA-3, NC, siRNA-1519 and siRNA-1643), which follow the sequences as depicted in Table II, ILP-2 expression was notably decreased in the siRNA-5 group (P<0.001) in comparison to that of the MCF-7 group. These results indicate that the siRNA-5 sequence show the highest interference efficiency on ILP-2 expression.
Figure 3.
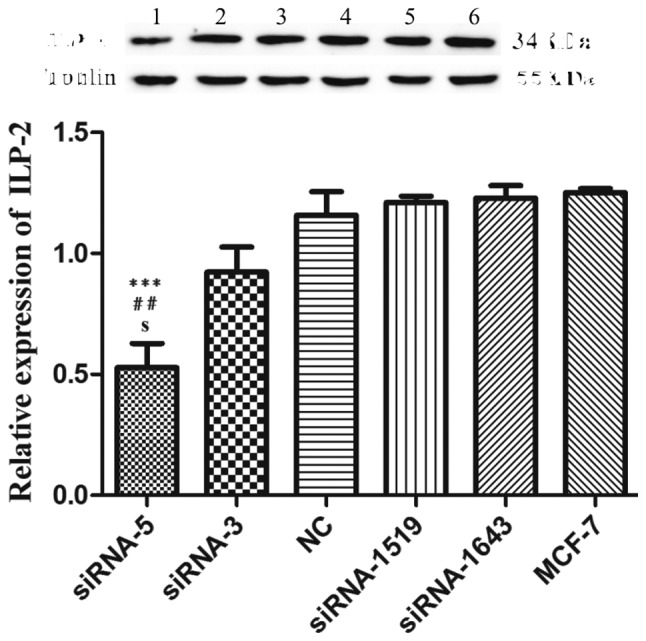
Western blot analysis showing ILP-2 protein expression in MCF-7 cells following knockdown of ILP-2. The data indicate that knockdown by the siRNA-5 sequence resulted in decreased ILP-2 expression in MCF-7 cells in comparison to the MCF-7 group. Lanes 1, 2, 3, 4, 5 and 6 separately indicate the expression of ILP-2 in the siRNA-5, siRNA-3, NC, siRNA-1519, siRNA-1643 and MCF-7 group. ***P<0.001 vs. the MCF-7 group; ##P<0.01 vs. the NC group; sP<0.05 vs. the siRNA-3 group. Data are expressed as mean ± SEM by ANOVA. Groups were compared by conducting Bonferroni test; n=3.
Table II.
siRNA sequences.
| Name | Sense (5′-3′) | Antisense (5′-3′) |
|---|---|---|
| siRNA-5 | CUAUACGAAUGGGAUUUGATT | UCAAAUCCCAUUCGUAUAGTT |
| siRNA-3 | UGGUACAAACUACCAAGAATT | UUCUUGGUAGUUUGUACCATT |
| siRNA-1519 | GGUACAAACUACCAAGAAATT | UUUCUUGGUAGUUUGUACCTT |
| siRNA-1643 | GAGGAAAGAAUUCAAACAUTT | AUGUUUGAAUUCUUUCCUCTT |
| Negative control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
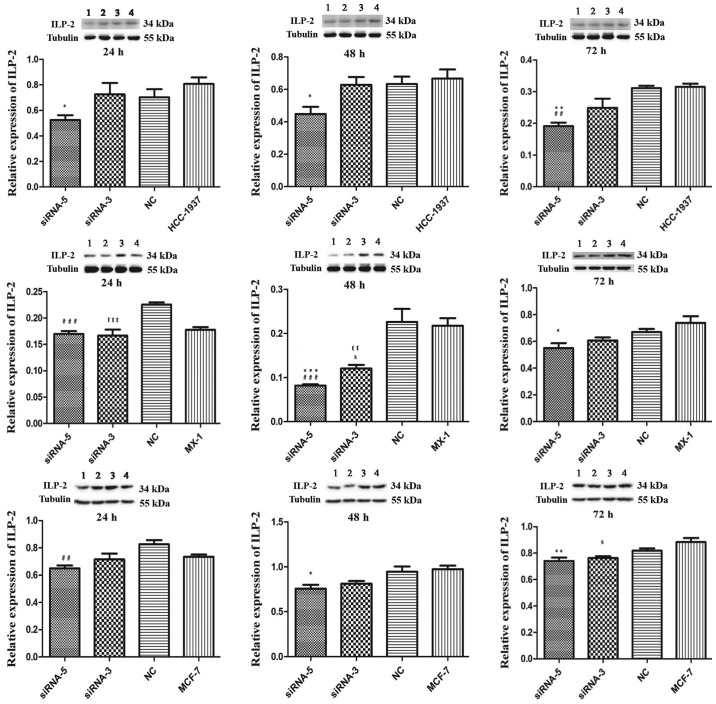
As illustrated in Fig. 4, in the HCC-1937 cell line, the siRNA-5 group exhibited significantly decreased ILP-2 expression similar to that of the control HCC-1937 group (24 and 48 h, P<0.05; 72 h, P<0.01). Likewise in the MX-1 cell line, the siRNA-5 group exhibited significantly decreased ILP-2 expression at 48 h (P<0.001) and 72 h (P<0.05) when compared with that of the control MX-1 group. In the MCF-7 cell line, ILP-2 expression in the siRNA-5 group was significantly decreased at 48 h (P<0.05) and 72 h (P<0.01) when compared with that of the MCF-7 group.
Figure 4.
Western blot analysis showing the expression of ILP-2 in the HCC-1937, MX-1 and MCF-7 cell lines following knockdown of ILP-2 for 24, 48 and 72 h. Lanes 1, 2, 3 and 4 separately indicate ILP-2 expressions in siRNA-5, siRNA-3, NC and HCC-1937 or MX-1 or MCF-7 group. In regards to the HCC-1937 cell line, expression of ILP-2 was significantly decreased in the siRNA-5 group compared to the HCC-1937 group. *P<0.05, **P<0.01 vs. the HCC-1937 group; ##P<0.01 vs. the NC group. In regards to the MX-1 cell line, ILP–2 expression was significantly decreased when compared to the MX-1 group following the knockdown by the siRNA-5 sequence for 48 and 72 h. *P<0.05, ***P<0.001 vs. the MX-1 group; ###P<0.001 vs. the NC group; ttP< 0.01, tttP< 0.001 vs. the NC group; sP<0.05 vs. the MX-1group. In regards to the MCF-7 cell line, ILP-2 expression was significantly decreased when compared to MCF-7 group after knockdown by the siRNA-5 sequence for 48 and 72 h. *P<0.05, **P<0.01 vs. the MCF-7 group; ##P<0.01 vs. the NC group; sP<0.05 vs. the MCF-7 group. Data are expressed as mean ± SEM by ANOVA. Groups were compared by conducting Bonferroni test; n=3.
ILP-2 positively influences the cell viability of breast cancer cell lines
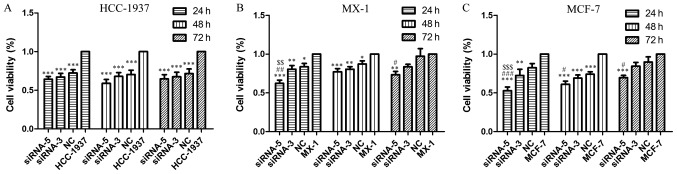
MTT assays were performed to examine cell viability when ILP-2 expression was knocked down by siRNA sequences for 24, 48 and 72 h in the HCC-1937, MX-1 and MCF-7 cell lines respectively so as to determine the role of ILP-2 in breast cancer cell growth. As shown in Fig. 5, cell viability of the siRNA-5 groups was significantly reduced when compared to that of the control group (Fig. 5A and C, P<0.001; Fig. 5B, 24 and 48 h, P<0.001, 72 h, P<0.01), thus, indicating that ILP-2 positively promotes the growth of breast cancer cells.
Figure 5.
MTT assay demonstrating the cell viability of HCC-1937, MX-1 and MCF-7 cell lines when ILP-2 was separately knocked down for 24, 48 and 72 h. In regards to the HCC-1937 cell line, the cell viability of the siRNA-5 group was notably decreased when compared to the HCC-1937 group. ***P<0.001 vs. the HCC-1937 group. In regards to the MX-1 cell line, cell viability of the siRNA-5 group was significantly decreased when compared to the MX-1 group. *P<0.05, **P<0.01, ***P<0.001 vs. the MX-1 group; #P<0.05, ##P<0.01 vs. the NC group; $$P<0.01 vs. the siRNA-3 group. In regards to the MCF-7 cell line, cell viability of the siRNA-5 group was significantly decreased when compared to the MCF-7 group. **P<0.01, ***P<0.001 vs. the MCF-7 group; #P<0.05, ###P<0.001 vs. the NC group; $$$P<0.001 vs. the siRNA-3 group. Data are expressed as mean ± SEM by ANOVA. Groups were compared by conducting Bonferroni test; n=6.
ILP-2 is actively involved in the cell migration of breast cancer cell lines
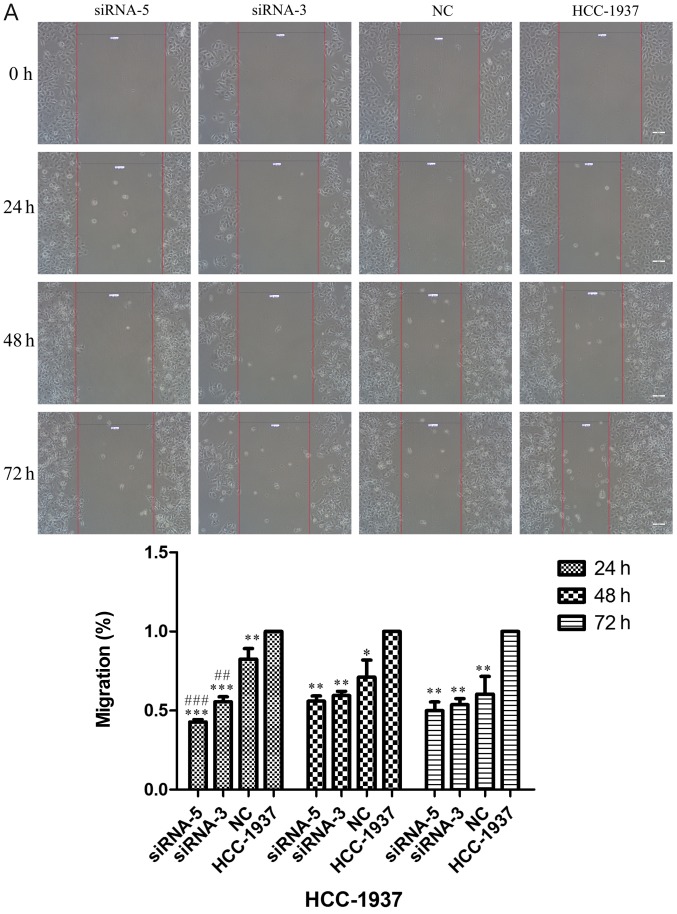
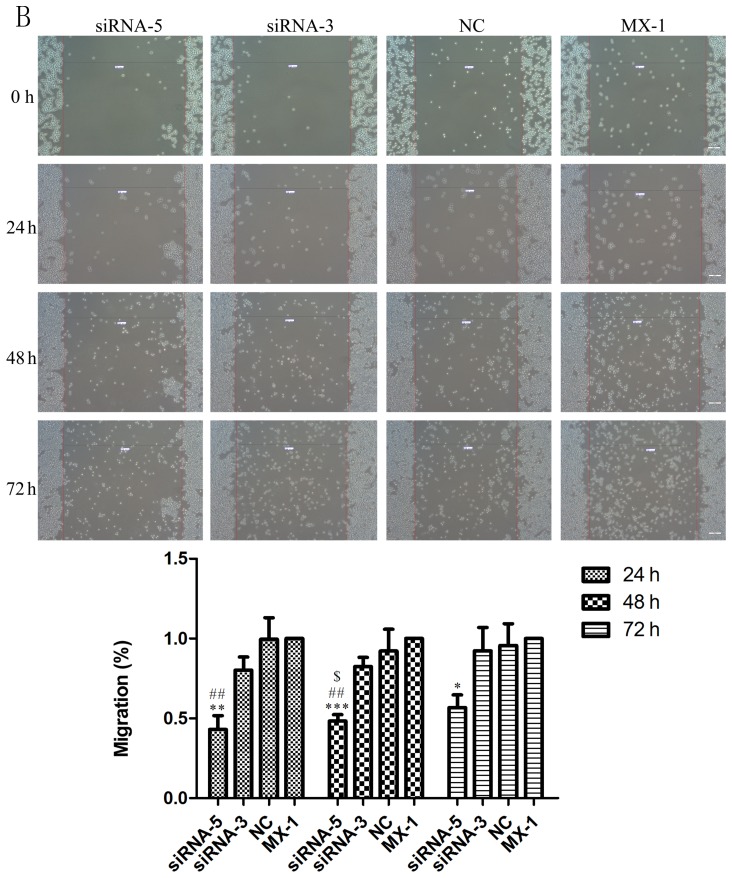
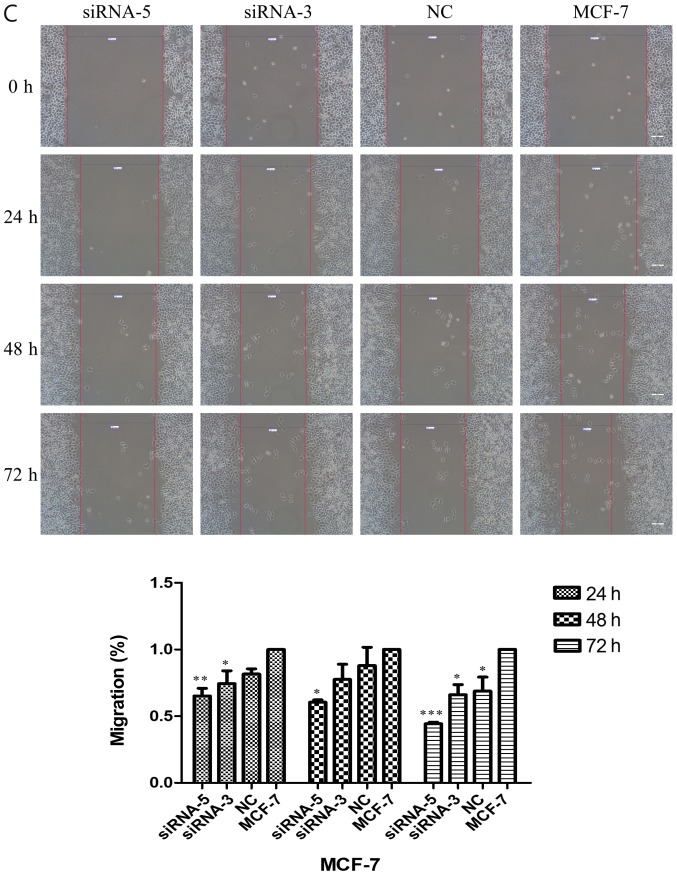
Scratch assays were performed to study the rate of cell migration in ILP-2 siRNA knockdown cell lines such as HCC-1937, MX-1 and MCF-7 so as to ascertain whether ILP-2 is involved in breast cancer cell migration. As shown in Fig. 6A-C, the rate of cell migration of the siRNA-5 group was significantly decreased in comparison to that of the control group (Fig. 6A, 24 h, P<0.001, 48 and 72 h, P<0.01; Fig. 6B, 24 h, P<0.01, 48 h, P<0.001, 72 h, P<0.05; Fig. 6C, 24 h, P<0.01, 48 h, P<0.05, 72 h, P<0.001), with results implying that ILP-2 positively accelerates the migration of breast cancer cells.
Figure 6.
Scratch analysis showing the rate of cell migration of the HCC-1937, MX-1 and MCF-7 cell lines when ILP-2 was separately knocked down for 24, 48 and 72 h. (A) In regards to the HCC-1937 cell line, the rate of cell migration of the siRNA-5 group was notably decreased compared to the HCC-1937 group. *P<0.05, **P<0.01, ***P<0.001 vs. the HCC-1937 group, ##P<0.01, ###P<0.001 vs. the NC group. (B) In regards to the MX-1 cell line, the rate of cell migration of the siRNA-5 group was notably decreased compared to the MX-1 group. *P<0.05, **P<0.01, ***P<0.001 vs. the MX-1 group; ##P<0.01 vs. the NC group; $P<0.05 vs. the siRNA-3 group. (C) In regards to the MCF-7 cell line, the rate of cell migration of the siRNA-5 group was notably decreased compared to the MCF-7 group. *P<0.05, **P<0.01, ***P<0.001 vs. the MCF-7 group. Data are expressed as mean ± SEM by ANOVA. Groups were compared by conducting Bonferroni test. Scale bar, 200 µm; n=3.
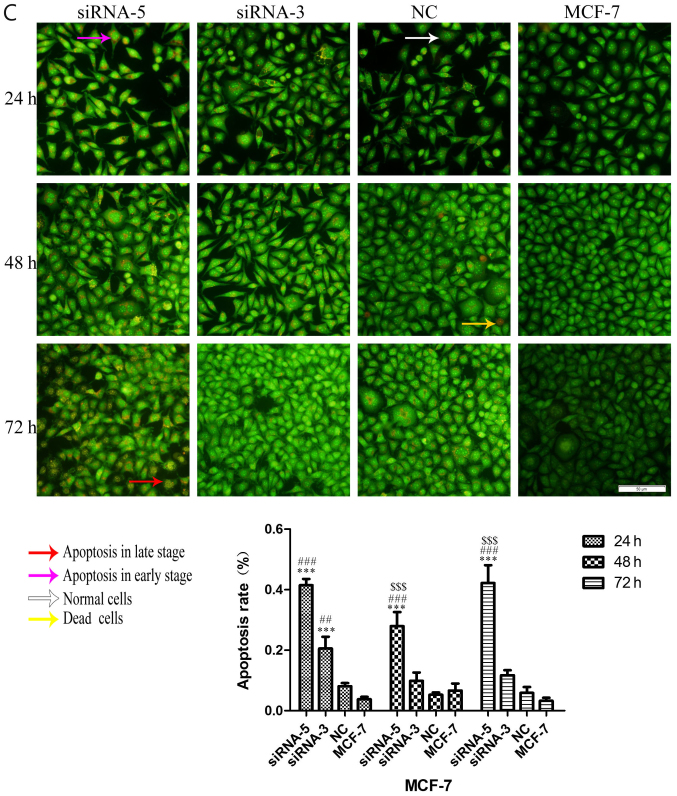
ILP-2 inhibits the apoptosis of breast cancer cells
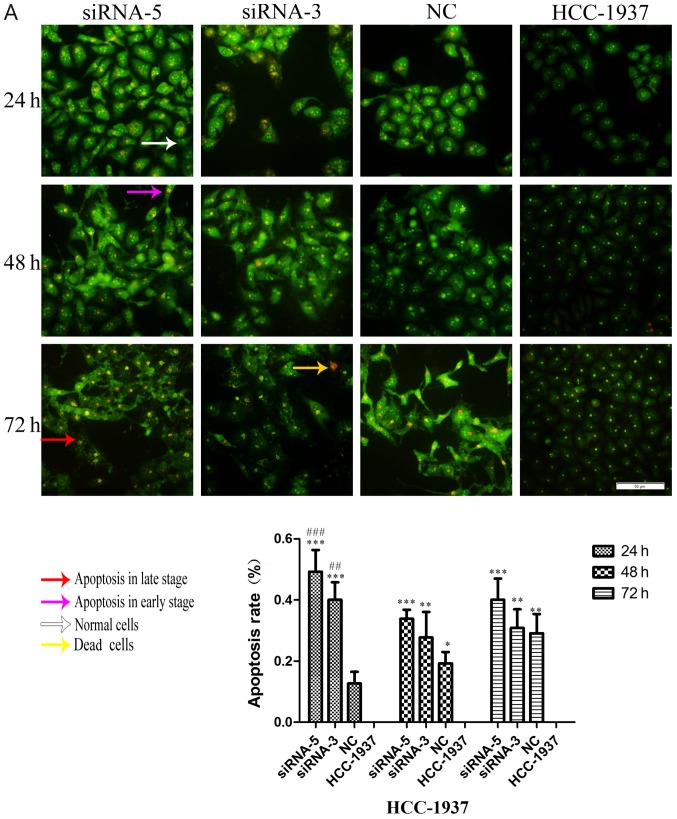
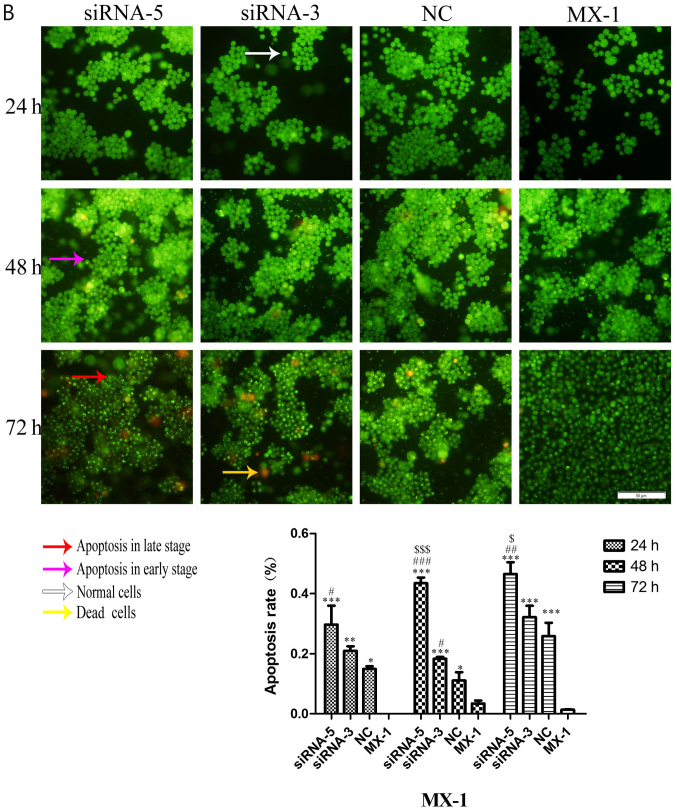
Considering the effect of ILP-2 on the apoptosis of breast cancer cells, dual AO/EB staining analysis was separately used to detect HCC-1937, MX-1 and MCF-7 cell apoptosis when ILP-2 was knocked down for 24, 48 and 72 h to further determine the role of ILP-2 on breast cancer cell growth. As shown in Fig. 7A-C, the apoptosis rate of the siRNA-5 group was significantly increased when compared with that of the control group (P<0.001 at all times for all cell lines), with results indicating that ILP-2 positively inhibits the apoptosis of breast cancer cells and promotes the growth of breast cancer cells.
Figure 7.
AO-EB double staining analysis showing the apoptotic rate of HCC-1937, MX-1 and MCF-7 cell lines when ILP-2 was separately knocked down for 24, 48 and 72 h. (A) In regards to the HCC-1937 cell line, the apoptosis rate of the siRNA-5 group was notably increased when compared to the HCC-1937 group. *P<0.05, **P<0.01, ***P<0.001 vs. the HCC-1937 group; ##P<0.01, ###P<0.001 vs. the NC group. (B) In regards to the MX-1 cell line, the apoptotic rate of the siRNA-5group was notably increased when compared to the MX-1 group. *P<0.05, **P<0.01, ***P<0.001 vs. the MX-1 group; #P<0.05, ##P<0.01, ###P<0.001 vs. the NC group; $P<0.05, $$$P<0.001 vs. the siRNA-3 group. (C) In regards to the MCF-7 cell line, the apoptotic rate of the siRNA-5 group was notably increased when compared to the MCF-7 group. ***P<0.001 vs. the MCF-7 group; ##P<0.01, ###P<0.001 vs. the NC group; $$$P<0.001 vs. the siRNA-3 group. Data are expressed as mean ± SEM by ANOVA. Groups were compared by conducting Bonferroni test. Scale bar, 100 µm; n=5.
Discussion
ILP-2 is a novel apoptotic inhibitory protein closely correlated to caspase-9 (9). ILP-2/BIRC-8 was found to inhibit HepG2 cell apoptosis and promote cell growth (11). Based on the results from the present study, we hypothesize that ILP-2 accelerates the growth of breast cancer cells.
Initially, ILP-2 was found to be solely expressed in the testis of normal tissues (9,10). It was later detected in lymphoblastoid cells (9). In our prior published study, we demonstrated high ILP-2 expression in breast cancer patient sera (8). In the present study, we found high ILP-2 expression in the breast cancer tissues and breast cancer cells. Livin protein expression is elevated in breast cancer tissues and cell lines, which in turn promotes the progression and metastasis of breast cancer (12). Our present results indicate that ILP-2 plays an active role in breast cancer cell growth.
XIAP (ILP-1) and Livin are members of the IAP family, with a very similar domain to ILP-2 (4). RNAi technology can distinctively inhibit ILP-1 and Livin expression (7,13–19). We applied RNAi technology to knock down ILP-2 expression and analyzed its role in breast cancer cell growth. Our results demonstrated that siRNA-5 can inhibit the protein expression of ILP-2 in breast cancer cells.
Livin participates in proliferation, migration and invasion of breast cancer (20). Overexpression of Livin promotes the migratory and invasive abilities of MCF-7, with knockdown of Livin exhibiting contrasting effect (12). After inhibiting ILP-2 expression using siRNA-5 in breast cancer cells such as HCC-1937, MX-1 and MCF-7 cell lines, cell viability and rate of cell migration were respectively significantly decreased, the cell apoptosis rate was separately significantly increased. These results suggest that ILP-2 vigorously promotes breast cancer cell growth.
In conclusion, our experiment confirms the hypothesis that ILP-2 plays a significant role in the growth of breast cancer cells and is a novel growth enhancer for breast cancer. However, further mechanistic studies need to be carried out to examine how ILP-2 promotes breast cancer cell growth. Accordingly, we plan to further explore the underlying mechanism of the growth-promoting activity of ILP-2 in breast cancer cells. First, we will apply the co-immunoprecipitation technology to search the protein which can interact with ILP-2 and verify it. In addition, we will construct Lenti-ILP-2-shRNA which knocks down the expression of ILP-2, transfect it into MCF-7 cells and filtrate the stably transfected MCF-7 cells by addition of the puromycin to MCF-7 cells. The stably transfected MCF-7 cells can be applied to establish human breast cancer xenografts in nude mice. When the solid tumors are successfully grown, we will excise them, draw growth cures after measuring the solid tumor size and analyze the expression of the ILP-2 downstream gene in its pathway by using the solid tumors.
Acknowledgements
We thank Dr Benson O.A. Botchway and Dr Akhileshwar Namani (Zhejiang University School of Medicine) for having critically revised the manuscripts.
Funding
The present study was supported by the National Natural Science Foundation of China (no. 81360397), the Scientific Research Project in Jishou University (no. Jdy16024) and the Scientific Research Project for Graduates in JiShou University (no. JGY201772).
Availability of data and materials
All data generated or analyzed in this study are included in this published article.
Authors' contributions
MX designed the manuscript and was also involved in the conception of the study; LZ, WZ, XZ and SX wrote the manuscript, collected clinical information and performed the statistical analyses; MW, XP, YP, BL, PT, QC and DY assisted with the immunohistochemistry and western blotting; ZC, ZS, SW and KY assisted with MTT assays, scratch assays and AO/EB double staining analysis. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work is appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Jishou University (Jishou, China). All participated patients received and approved the written informed consent before joining the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lopez J, Meier P. To fight or die-inhibitor of apoptosis proteins at the crossroad of innate immunity and death. Curr Opin Cell Biol. 2010;22:872–881. doi: 10.1016/j.ceb.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.de Almagro MC, Vucic D. The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp Oncol. 2012;34:200–211. [PubMed] [Google Scholar]
- 4.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 5.Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T, Ahmad B. Inhibitors of apoptotic proteins: New targets for anticancer therapy. Chem Biol Drug Des. 2013;82:243–251. doi: 10.1111/cbdd.12176. [DOI] [PubMed] [Google Scholar]
- 6.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda JE, MacKenzie A, et al. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 7.Hiscutt EL, Hill DS, Martin S, Kerr R, Harbottle A, Birch-Machin M, Redfern CP, Fulda S, Armstrong JL, Lovat PE. Targeting X-linked inhibitor of apoptosis protein to increase the efficacy of endoplasmic reticulum stress-induced apoptosis for melanoma therapy. J Invest Dermatol. 2010;130:2250–2258. doi: 10.1038/jid.2010.146. [DOI] [PubMed] [Google Scholar]
- 8.Xiang M, Zhou W, Gao D, Fang X, Liu Q. Inhibitor of apoptosis protein-like protein-2 as a novel serological biomarker for breast cancer. Int J Mol Sci. 2012;13:16737–16750. doi: 10.3390/ijms131216737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter BW, Mir SS, Eiben LJ, Lewis J, Reffey SB, Frattini A, Tian L, Frank S, Youle RJ, Nelson DL, et al. Molecular cloning of ILP-2, a novel member of the inhibitor of apoptosis protein family. Mol Cell Biol. 2001;21:4292–4301. doi: 10.1128/MCB.21.13.4292-4301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Zhang Q, Liu B, Han M, Shan B. Challenge and promise: roles for Livin in progression and therapy of cancer. Mol Cancer Ther. 2008;7:3661–3669. doi: 10.1158/1535-7163.MCT-08-0480. [DOI] [PubMed] [Google Scholar]
- 11.Chuturgoon AA, Phulukdaree A, Moodley D. Fumonisin B1 inhibits apoptosis in HepG2 cells by inducing Birc-8/ILP-2. Toxicol Lett. 2015;235:67–74. doi: 10.1016/j.toxlet.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Han Y, Zhang L, Wang W, Li J, Song M. Livin promotes the progression and metastasis of breast cancer through the regulation of epithelial-mesenchymal transition via the p38/GSK3β pathway. Oncol Rep. 2017;38:3574–3582. doi: 10.3892/or.2017.6017. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Xiao XQ, Deng CM, Su XS, Li GY. Downregulation of xIAP expression by small interfering RNA inhibits cellular viability and increases chemosensitivity to methotrexate in human hepatoma cell line HepG2. J Chemother. 2006;18:525–531. doi: 10.1179/joc.2006.18.5.525. [DOI] [PubMed] [Google Scholar]
- 14.Buneker CK, Yu R, Deedigan L, Mohr A, Zwacka RM. IFN-γ combined with targeting of XIAP leads to increased apoptosis-sensitisation of TRAIL resistant pancreatic carcinoma cells. Cancer Lett. 2012;316:168–177. doi: 10.1016/j.canlet.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Bai L, Lu J, Liu L, Yang CY, Sun H. Targeting inhibitors of apoptosis proteins (IAPs) for new breast cancer therapeutics. J Mammary Gland Biol Neoplasia. 2012;17:217–228. doi: 10.1007/s10911-012-9265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Wang S, Sun H, Pan Z, Zhou W, Wu M. Inhibition of tumorigenesis and invasion of hepatocellular carcinoma by siRNA-mediated silencing of the livin gene. Mol Med Rep. 2010;3:903–907. doi: 10.3892/mmr.2010.355. [DOI] [PubMed] [Google Scholar]
- 17.Wang TS, Ding QQ, Guo RH, Shen H, Sun J, Lu KH, You SH, Ge HM, Shu YQ, Liu P. Expression of livin in gastric cancer and induction of apoptosis in SGC-7901 cells by shRNA-mediated silencing of livin gene. Biomed Pharmacother. 2010;64:333–338. doi: 10.1016/j.biopha.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Dasgupta A, Alvarado CS, Xu Z, Findley HW. Expression and functional role of inhibitor-of-apoptosis protein livin (BIRC7) in neuroblastoma. Biochem Biophys Res Commun. 2010;400:53–59. doi: 10.1016/j.bbrc.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Yang D, Song X, Zhang J, Ye L, Wang S, Che X, Wang J, Zhang Z, Wang L. Suppression of livin gene expression by siRNA leads to growth inhibition and apoptosis induction in human bladder cancer T24 cells. Biosci Biotechnol Biochem. 2010;74:1039–1044. doi: 10.1271/bbb.90934. [DOI] [PubMed] [Google Scholar]
- 20.Li CJ, Cong Y, Liu XZ, Zhou X, Shi X, Wu SJ, Zhou GX, Lu M. Research progress on the livin gene and osteosarcomas. Asian Pac J Cancer Prev. 2014;15:8577–8579. doi: 10.7314/APJCP.2014.15.20.8577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed in this study are included in this published article.