Abstract
Orthodenticle homolog 1 (OTX1) has previously been revealed to be tightly associated with the development and progression of several human tumors. However, the functional roles and underlying molecular mechanisms of OTX1 in gastric cancer (GC) remain poorly understood. In the present study, we observed that OTX1 was highly expressed in GC tissues compared with adjacent non-tumor tissues based on a large cohort of samples from The Cancer Genome Atlas (TCGA) database. An immunohistochemical analysis indicated that OTX1 levels were increased in tumors that became metastatic compared with those in tumors that did not. This finding was significantly associated with patients who had shorter overall survival times. The knockdown of OTX1 significantly inhibited the proliferation, migration and invasion of SGC-7901 and MGC-803 cells. Furthermore, the knockdown of OTX1 induced cell cycle arrest in the G0/G1 phase and reduced the expression of cyclin D1. In addition, the inhibition of OTX1 led to increased GC cell apoptosis by upregulating cleaved PARP, cleaved caspase-3 and Bax. In conclusion, our data indicated that OTX1 functions as a key regulator in tumor growth and metastasis of GC cells. Thus, OTX1 may be a promising novel target for molecular therapy directed toward GC.
Keywords: OTX1, gastric cancer, proliferation, metastasis, apoptosis
Introduction
Gastric cancer (GC) is the fourth most commonly diagnosed malignancy worldwide and is characterized by an adverse clinical outcome (1–3). Due to the lack of effective biomarkers for an early diagnosis, the 5-year survival rate of patients with advanced GC is only 5–15% (4,5). Surgical resection may be the only hope for advanced-stage GC patients, but has a high rate of recurrence (6,7). Therefore, it is imperative to explore novel biomarkers and therapeutic targets that may be helpful in developing targeted therapies for GC.
Orthodenticle homolog 1 (OTX1), an OTX family (OTX1, OTX2, OTX3 and CRX) protein, is a transcription factor that specifically binds to TAATCC/T elements on target genes (8). OTX1 is comprised of a bicoid-like homeodomain and belongs to the homeobox (HB) family of genes. Previous studies have revealed that OTX1 plays a crucial role in the development of the brain, sensory organs, early human fetal retina and mammary gland (9–11). OTX1 has been recently reported to be frequently overexpressed in various cancers, including medulloblastomas, breast and colorectal cancer and hepatocellular carcinoma (12,13), indicating that OTX1 may be a key regulator in the development and progression of human carcinogenesis. Further studies have revealed that OTX1 promotes tumor proliferation and migration in colorectal cancer and hepatocellular carcinoma (14,15). However, the role and underlying mechanism of OTX1 in the development and progression of GC remains to be elucidated.
In the present study, we observed that OTX1 was overexpressed in GC samples and that there was a significant correlation between a high expression level of OTX1 and poor prognosis in GC patients. We investigated the effects of OTX1 silencing on GC cell proliferation, migration and invasion. Furthermore, cell cycle distribution and cell apoptosis were examined following OTX1 knockdown. This study indicated that OTX1 could be a therapeutic target for the treatment of GC.
Materials and methods
Differential expression analysis of OTX1 using UALCAN
The expression data of OTX1 in GC and normal samples were retrieved and analyzed using the online web portal UALCAN (http://ualcan.path.uab.edu) (16) based on The Cancer Genome Atlas (TCGA) level 3 RNA-seq and clinical data from stomach adenocarcinomas.
Patients and specimens
The present study was approved by the Ethics Committee of Haimen People's Hospital, and all the patients provided written informed consent. Cancer tissue specimens were obtained from 50 GC patients who underwent radical gastrectomy without prior radiotherapy or chemotherapy between June 2010 and July 2013 at the Department of General Surgery, Haimen People's Hospital (Jiangsu, China). In addition, 50 patients with benign gastric diseases who underwent simple polypectomy via endoscopy were included as controls. All diagnoses of GC, gastric polyps and lymph node metastasis were confirmed by histopathological examination. The tissue specimens were fixed in 4% formalin immediately after removal and were embedded in paraffin for immunohistochemical staining. Each sample was frozen and stored at −80°C. Among the 50 GC cases, there were 34 males and 16 females with ages ranging from 44 to 90 years (mean age, 68 years). All the specimens were confirmed by pathological diagnoses and were staged according to the 7th AJCC-TNM Classification of Malignant Tumors. The median follow-up period was 32.90 months (range, 2.93–43.83 months).
Immunohistochemical analysis and evaluation of OTX1 expression
Immunohistochemical staining was performed using a standard immunoperoxidase staining procedure. OTX1 expression in benign and malignant specimens was evaluated according to methods described by Terrinoni et al (13). Sections were semi-quantitatively scored for the extent of immunoreactivity as follows: 0, 0% immunoreactive cells; 1, <5% immunoreactive cells; 2, 5–50% immunoreactive cells; and 3, >50% immunoreactive cells. Additionally, the staining intensity was semi-quantitatively scored as 0 (negative), 1 (weak), 2 (intermediate), or 3 (strong). The final immunoreactivity score was defined as the sum of both parameters, and the samples were grouped as having negative (0), weak (1–2), moderate (3), or strong (4–6) staining. For statistical purposes, only the final immunoreactivity scores of the moderate and strong groups were considered positive, and the other final scores were considered negative.
Cell culture
The SGC-7901 and MGC-803 cell lines were purchased from the Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China). Cells were cultured with Dulbecco's modified Eagle's medium (DMEM; HyClone Laboratories; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Life Technologies, Paisley, UK) and antibiotics (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in a humidified incubator containing 5% CO2 at 37°C.
Lentivirus-mediated RNA interference
The shRNA targeted to OTX1 and a negative control shRNA (shNC) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Recombinant lentiviruses expressing OTX1-shRNA (shOTX1) or shNC were provided by Shanghai Genechem Co., Ltd. (Shanghai, China). For the construction of stable cell lines, the cells were selected with puromycin for 7 days after infection with a lentivirus for 72 h. Quantitative real-time polymerase chain reaction (RT-qPCR) and western blot analyses were performed to determine the expression level of OTX1.
RT-qPCR
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. Subsequently, 1 µg of total RNA was used to synthesize first-strand cDNA using a reverse transcription reagent kit (Takara Biotechnology, Co., Ltd., Dalian, China). The RT-qPCR assay was performed with SYBR Green Premix Ex Taq (Takara Biotechnology) using the following thermocycling conditions: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, and the data were analyzed using the ∆∆Cq method with GAPDH as an internal control. The primer sequences were as follows: OTX1 forward, 5′-CTGCTCTTCCTCAATCAATGG-3′ and reverse, 5′-ACCCTGACTTGTCTGTTTCC-3′; GAPDH forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-GGCATGGACTGTGGTCATGAG-3′. The experiment was repeated three times.
Cell proliferation assay
SGC-7901 and MGC-803 cells were infected with shNC or shOTX1 for 72 h. Cell proliferation was determined using Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Tokyo, Japan). Briefly, 2×103 SGC-7901 or MGC-803 cells/well were seeded into 96-well plates. At each of the indicated time-points (0, 1, 2, 3, 4 and 5 days), 10 µl of CCK-8 reagent was added to each well, followed by incubation for 2 h at 37°C. The optical density (OD) values at 450 nm for each well were determined using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Colony formation assay
SGC-7901 and MGC-803 cells were seeded into 6-well plates at a density of 400 cells/well following infection with an shNC- or shOTX1-expressing virus. The cells were then cultured for 14 days, and the media were replaced every three days. The cells were stained with 0.1% crystal violet (Beyotime Institute of Biotechnology, Shanghai, China) for 25 min after fixation with 4% paraformaldehyde for 20 min. The plates were washed with ddH2O three times and air-dried. Finally, colonies with >50 cells were captured with an ×200 magnification using an inverted microscope (Leica Microsystems, Wetzlar, Germany) and counted manually.
Migration/invasion assay
Matrigel-coated Transwell chambers were used for the invasion assay, and uncoated Transwell chambers (8-µm pore; Corning Inc., Lowell, MA, USA) were used for the migration assay. SGC-7901 and MGC-803 cells were infected with the desired virus and were trypsinized after 72 h of infection. Subsequenlty, 1.2×105 SGC-7901 cells or 3×104 MGC-803 cells were seeded into the upper chamber of an 8-µm pore-size insert with or without Matrigel, while the lower chamber was filled with DMEM containing 10% FBS. After a 48-h culture period, the cells were fixed and stained with 0.1% crystal violet for 20 min. Cells that had passed through the 8-µm pore were counted in five random fields using a light microscope (Leica Microsystems) at an ×100 magnification. Three independent experiments were performed.
Wound healing assay
The wound healing assay was performed as described by Wang et al (17). Briefly, GC cells infected with the corresponding virus were seeded into 6-well plates to form a single confluent cell layer. The plates were scratched with 200-µl pipettes to form a wound within the confluent cell layer. The cells were then incubated in serum-free medium for 0, 24 or 72 h. The movement of cells into the scratched area was photographed using the inverted Leica DMI3000B microscope (Leica Microscopes).
Cell cycle analysis
For the cell cycle assays, SGC-7901 or MGC-803 cells were collected via trypsinization and fixed in ice-cold 70% ethanol overnight. The cells were then washed three times with PBS and incubated with 10 mg/ml RNase and 1 mg/ml propidium iodide (PI) (Sigma-Aldrich; Merck) at 37°C for 30 min in the dark. All the samples were assessed using flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) and were analyzed using CellQuest acquisition software version 3.3 (BD Biosciences). Each experiment was performed in triplicate.
Cell apoptosis assay
Cell apoptosis was detected using the Annexin V/PI apoptosis kit following the manufacturer's instructions (Invitrogen; Thermo Fisher Scientific). Briefly, GC cells were harvested by EDTA-free trypsinization and were washed twice with ice-cold PBS. Subsequently, the cells were resuspended in 100 µl of binding buffer at a density of 1×106 cells/ml, followed by the addition of 5 µl of Annexin V-FITC and 5 µl of PI for 15 min at room temperature in the dark. Finally, 400 µl of binding buffer was added to each sample, and the cells were immediately analyzed by flow cytometry. The experiment was repeated three times. The experiment was repeated three times. The perentages of Q3 quadrant (Annexin V−/PI−), Q4 quadrant (Annexin V+/PI−) and Q2 quadrant (Annexin V+/PI+) were as following: 96.93±0.75, 1.3±0.06 and 0.8±0.05% in the shNC group and 85.73±0.31, 3.7±0.16 and 4±0.21% in the shOTX1 group in SGC-7901 cells; 96.7±0.85, 1.5±0.08 and 0.9±0.04% in the shNC group and 88.63±2.71, 4.8±0.32 and 4.2±0.13% in the shOTX1 group in MGC-803 cells.
Western blot analysis
The total protein content of the GC cells was extracted using ice-cold radio-immunoprecipitation assay buffer (RIPA; Beyotime Institute of Biotechnology) supplemented with PMSF and was incubated on ice for 30 min. Following centrifugation at 10,000 × g, the supernatants were harvested, and the protein concentration was determined using the BCA protein assay kit (Beyotime Institute of Biotechnology). Equal amounts of protein (30 µg) were separated via 10% SDS-PAGE and were transferred onto polyvinylidene difluoride membranes (PVDF) (EMD Millipore, Billerica, MA, USA). Then, the PVDF membranes were blocked with 5% fat-free milk (GuangMing, Shanghai, China) and incubated at 4°C overnight with the following primary antibodies: Anti-OTX1 (1:500; cat. no. sc-517000; Santa Cruz Biotechnology), anti-GAPDH (1:1,000; cat. no. ab181602; Abcam, Cambridge, UK), anti-cleaved caspase-3 (1:1,000; cat. no. 9661), anti-cleaved PARP (1:1,000; cat. no. 5625), anti-Bax (1:1,000; cat. no. 14796), anti-N-cadherin (1:1,000; cat. no. 13116), anti-Bcl-2 (1:1,000; cat. no. 3498; all from Cell Signaling Technology), anti-proliferating cell nuclear antigen (PCNA) (1:1,000; cat. no. ab92552; Abcam), anti-Slug (1:1,000; cat. no. ab27568; Abcam), anti-vimentin (1:2,000; cat. no. ab92547; Abcam) and anti-Snail (1:1,000; cat. no. 3879; Cell Signaling Technology). The membranes were then incubated with goat anti-rabbit IgG H&L (HRP) (1:5,000; cat. no. ab6721; Abcam) after washing three times with TBST buffer. The protein bands were visualized using ECL detection reagents (EMD Millipore). GAPDH served as the loading control.
Statistical analysis
All statistical analyses were calculated using the SPSS 18.0 software for Windows (SPSS, Inc., Chicago, IL, USA). An independent Student's t-test was used to compare the means of two groups. Pearson's χ2 test was used to analyze the association of OTX1 expression with the clinicopathological parameters. Kaplan-Meier plots and log-rank tests were used for the survival analysis. Univariate and multivariate Cox proportional hazards regression models were used to analyze the independent prognostic factors. The data were presented as the mean ± SD. Student's t-test (two-tailed) was used to analyze continuous variables. The results were considered to indicate a statistically significant difference when P<0.05.
Results
Clinical significance of OTX1 in GC tissues
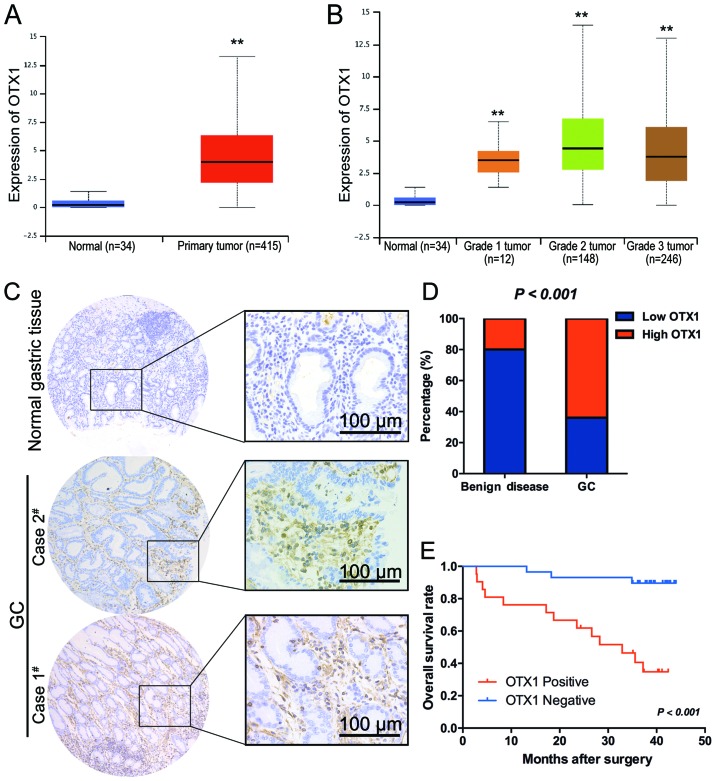
Using microarray analysis, a previous study revealed that OTX1 was upregulated in GC tissues (18), indicating an oncogenic role for OTX1 in GC. To further confirm this result in a large cohort of patient samples, we retrieved data from TCGA database and analyzed the expression of OTX1 in stomach adenocarcinomas, comprised of 415 GC tissues and 34 non-cancerous gastric tissues, using the online web portal UALCAN (http://ualcan.path.uab.edu) (16). As clearly indicated in Fig. 1A, OTX1 was significantly upregulated in GC tissues compared with normal tissues. In addition, OTX1 had the highest expression level in grade II tumors (Fig. 1B).
Figure 1.
OTX1 is highly expressed in gastric tumor samples. (A) Boxplot showing the relative expression of OTX1 in normal and GC samples; **P<0.01. (B) Boxplot showing the relative expression of OTX1 in normal, grade I, grade II and grade III GC patients; **P<0.01. (C) Representative image of GC staining using an anti-OTX1 antibody. (Case #2) Moderate expression of OTX1 in GC tissues; (Case #1) strong expression of OTX1 in poorly differentiated GC tissues (scale bar, 100 µm). (D) The bar graph revealing the statistical results for the staining intensity in GC tissues and benign-diseased tissues. (E) Kaplan-Meier curve for the OS of GC patients was correlated with OTX1 expression. OS, overall survival.
Furthermore, the protein expression levels of OTX1 were determined by immunohistochemistry in 50 samples of archived paraffin-embedded GC tissues and 50 gastric polyp epithelial tissues (Fig 1C). The expression of OTX1 was significantly higher in the tumor tissues than in the benign disease tissues (P<0.001) (Fig. 1D). A clinicopathological association study of 50 GCs revealed that OTX1 was significantly associated with the Borrmann type (P=0.012) and lymph node metastasis (P<0.001) (Table I), indicating that OTX1 may play a role in metastasis. Notably, positivity for OTX1 was significantly correlated with a shorter overall survival (OS) time (log rank, 16.61; P<0.001) (Fig. 1E). A multivariate Cox regression analysis further revealed that OTX1 was an independent prognostic marker for the OS time of GC patients (hazard ratio, 0.126; 95% confidence interval, 0.035–0.448; P=0.001) (Table II).
Table I.
Association of the expression of OTX1 with the clinicopathological characteristics of GC patients.
| Relative OTX1 expression | |||||
|---|---|---|---|---|---|
| Variable | Category | Negative (n=29) | Positive (n=21) | χ2 | P-value |
| Age | <60 years | 13 | 6 | 1.366 | 0.242 |
| ≥60 years | 16 | 15 | |||
| Sex | Male | 18 | 16 | 1.116 | 0.291 |
| Female | 11 | 5 | |||
| Location of tumor | Upper stomach | 2 | 0 | 2.236 | 0.525 |
| Middle stomach | 8 | 4 | |||
| Lower stomach | 17 | 15 | |||
| Mixed | 2 | 2 | |||
| Borrmann type | Early stage | 8 | 0 | 8.812 | 0.012a |
| I+II type | 8 | 4 | |||
| III+IV type | 13 | 17 | |||
| Histological differentiation | Well | 4 | 0 | 5.991 | 0.05 |
| Moderate | 9 | 3 | |||
| Poor | 16 | 18 | |||
| Tumor invasion (AJCC) | Tis-T2 | 17 | 0 | 18.652 | <0.001a |
| T3-T4 | 12 | 21 | |||
| Lymph node metastasis | Yes | 13 | 20 | 13.973 | <0.001a |
| No | 16 | 1 | |||
| TNM stage (AJCC) | I–II | 25 | 4 | 22.552 | <0.001a |
| III–IV | 4 | 17 | |||
Bold values indicate statistical significance
P<0.05. OTX1, orthodenticle homolog 1; GC, gastric cancer.
Table II.
Univariate and multivariate analyses of clinical variables contributing to OS.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (<60 vs. ≥60 years) | 3.004 (0.847–10.657) | 0.074 | – | – |
| Sex (male vs. female) | 0.786 (0.250–2.473) | 0.680 | – | – |
| Location of tumor (upper or middle stomach vs. lower stomach) | 0.373 (0.134–1.039) | 0.049 | – | – |
| Histological differentiation (well or moderate vs. poor) | 2.209 (0.732–6.665) | 0.334 | – | – |
| Tumor invasion (AJCC) (Tis-T2 vs. T3-T4) | 9.625 (1.263–73.330) | 0.007a | 2.884 (0.255–32.578) | 0.392 |
| Lymph node metastasis (yes vs. no) | 4.122 (0.929–18.295) | 0.043a | 1.085 (0.144–8.154) | 0.937 |
| TNM stage (AJCC) (I–II vs. III–IV) | 5.113 (1.619–16.145) | 0.002 | 1.413 (0.283–7.059) | 0.673 |
| Type of surgery (curative resection vs. palliative) | 0.431 (0.097–1.921) | 0.255 | – | – |
| OTX1 expression in tumor (negative vs. positive) | 0.126 (0.035–0.448) | <0.001a | 0.126 (0.035–0.448) | 0.001 |
Bold values indicate statistical significance
P<0.05. CI, confidence interval; HR, hazard ratio; OTX1, orthodenticle homolog 1; OS, overall survival.
Lentivirus-mediated knockdown of OTX1 in GC cell lines
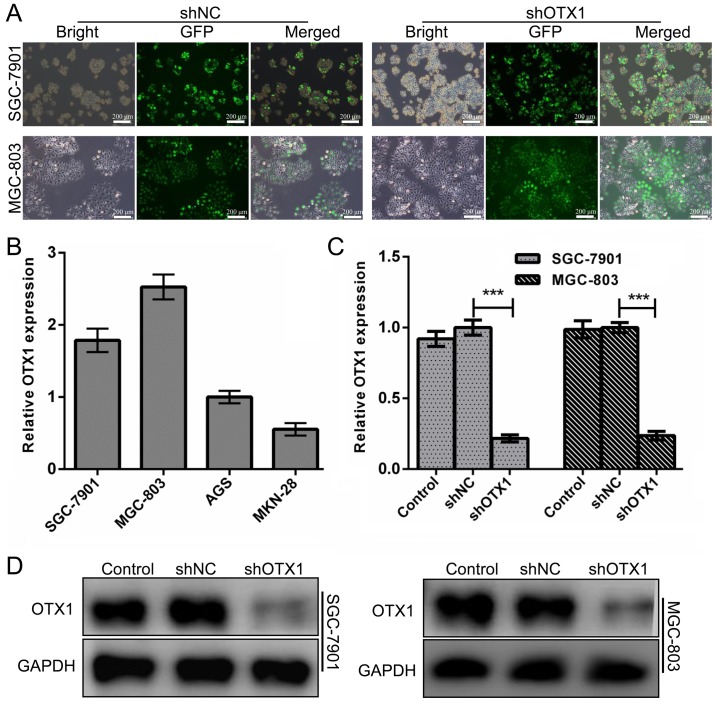
To explore the biological role of OTX1 in the tumorigenesis of GC, we determined the mRNA expression of OTX1 in four GC cell lines and found that SGC-7901 and MGC-803 cells harbor relatively higher expression of OTX1 (Fig. 2B). Therefore, we suppressed OTX1 with a specific shRNA lentiviral vector in SGC-7901 and MGC-803 cells. As displayed in Fig. 2A, the infection efficiency was greater than 90%, as determined by the EGFP signal. Real-time PCR and western blot assays at 72 h following infection revealed that the expression of OTX1 in both cell lines was significantly downregulated at the mRNA and protein levels (Fig. 2C and D, P<0.01).
Figure 2.
Lentivirus-mediated depletion of OTX1 in GC cells. (A) Microscopy images of SGC-7901 and MGC-803 cells infected with control lentivirus (shNC) and shOTX1 lentivirus (scale bar, 200 µm). (B) The level of OTX1 mRNA in four GC cell lines was determined using an RT-qPCR assay. GAPDH was used as an internal control. (C) The level of OTX1 mRNA in the indicated cells was determined using an RT-qPCR assay. GAPDH was used as an internal control. ***P<0.001 compared with the negative control group (shNC). (D) Western blot analysis of OTX1 in SGC-7901 and MGC-803 cells infected with an shNC or shOTX1 lentiviral vector. GAPDH served as the loading control.
Depletion of OTX1 significantly suppresses the proliferation of GC cells
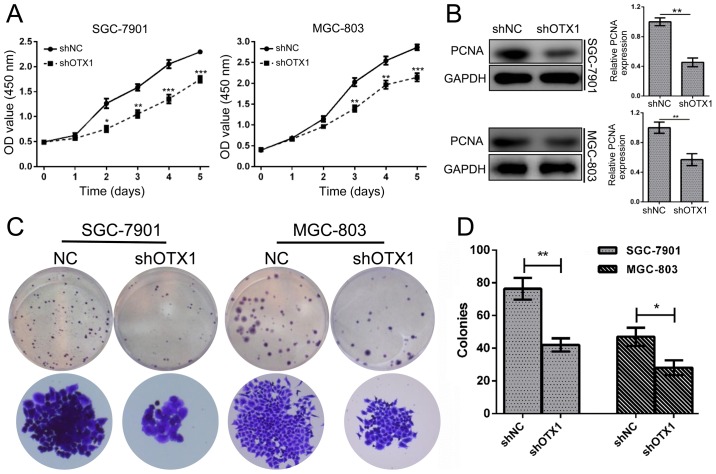
To determine the effect of OTX1 on GC cell proliferation, a CCK-8 assay was conducted in SGC-7901 and MGC-803 cells following infection with shOTX1 or shNC. The results revealed that OTX1 knockdown significantly inhibited the proliferation of GC cells (P<0.001, P<0.01, P<0.05; Fig. 3A). Consistently, the knockdown of OTX1 markedly reduced the expression of PCNA (Fig. 3B). In addition, the colony formation assay revealed that shOTX1-infected cells formed smaller and fewer colonies than the shNC-infected cells (Fig. 3C and D; P<0.01). Collectively, these data indicated that OTX1 promoted the proliferation of GC cells.
Figure 3.
OTX1 knockdown inhibits the growth of GC cells. (A) The proliferation rates of SGC-7901 and MGC-803 cells were determined using CCK-8 assays following transfection with shOTX1 and shNC lentiviral vectors. (B) Western blot analysis of PCNA expression in SGC-7901 and MGC-803 cells following OTX1 knockdown (left). Ratio of PCNA to GAPDH determined by band density, presented as the mean ± SD, compared with shNC (designated as 1.00) (right). (C) Colony-forming ability was examined following the knockdown of OTX1 in GC cells. Representative images of the crystal violet staining of colonies are displayed. (D) The number of colonies was counted and statistically analyzed. The data are expressed as the mean ± SD for three independent experiments. *P<0.05; **P<0.01; ***P<0.001 compared with the negative control group (shNC).
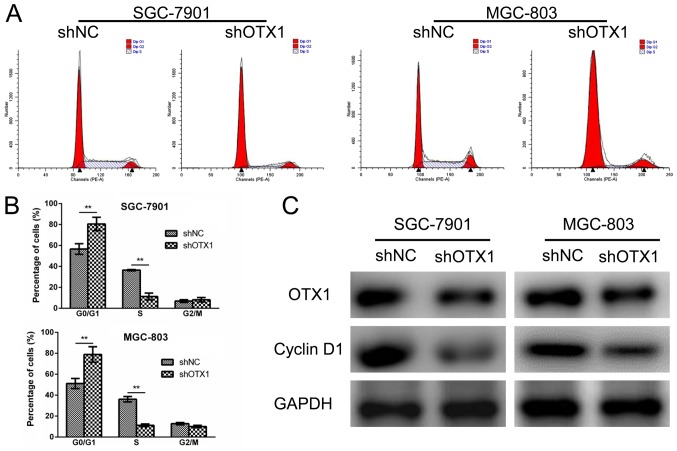
Effect of OTX1 knockdown on the cell cycle of GC cells
To further investigate the role for OTX1 in tumor growth, a cell cycle analysis was performed using flow cytometry. SGC-7901 and MGC-803 cells infected with shOTX1 exhibited a notable increase in the percentage of cells in the G0/G1 phase compared with the shNC groups (Fig. 4A and B), indicating that OTX1 knockdown led to a G0/G1-phase arrest. Consistently, cyclin D1 protein expression was markedly downregulated in the shOTX1-infected GC cells (Fig. 4C). Collectively, these results indicated that the knockdown of OTX1 inhibited cell proliferation by inducing a G0/G1-phase arrest.
Figure 4.
Effect of OTX1 knockdown on the cell cycle in GC cells. (A) The cell cycle distributions of SGC-7901 and MGC-803 cells were determined via flow cytometry following OTX1 depletion. (B) The percentage of cells in the G0/G1, S and G2/M phases was calculated. The data are presented as the mean ± SD, **P<0.01. (C) The expression levels of OTX1 and cyclin D1 in SGC-7901 cells (left panel) and MGC-803 cells (right panel) were examined by western blot analysis with GAPDH as the internal control.
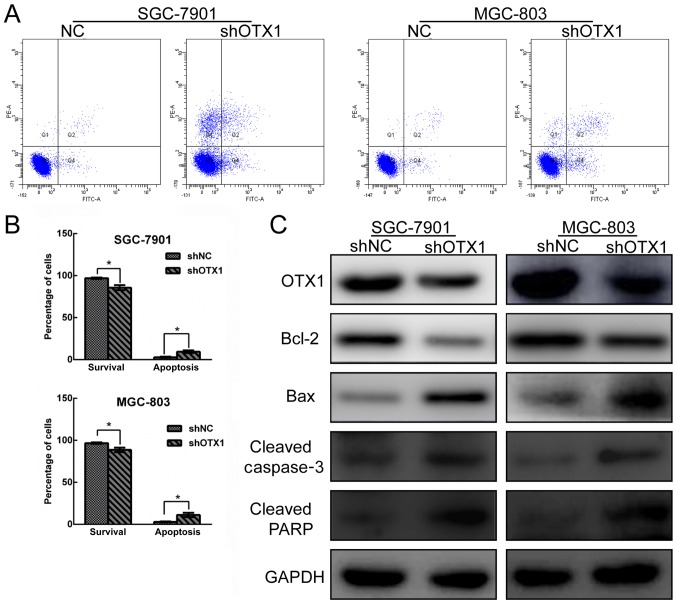
Effect of OTX1 on GC cell apoptosis
The effect of OTX1 knockdown on cell apoptosis was determined by Annexin V-FITC/PI staining using flow cytometry. As revealed in Fig. 5A and B, OTX1-silenced GC cells exhibited an increased rate of apoptosis compared with the shNC groups. Subsequently, a western blot assay was used to further examine the effect of OTX1 on the expression of apoptosis-related proteins. As displayed in Fig. 5C, the depletion of OTX1 resulted in the upregulation of cleaved caspase-3, cleaved PARP and Bax expression and the reduction of Bcl-2 expression, further supporting the observation that OTX1 knockdown induced GC cell apoptosis. Collectively, these data indicated that OTX1 knockdown induced cell apoptosis by modulating apoptosis-related proteins.
Figure 5.
Depletion of OTX1 induces cell apoptosis in GC cells. SGC-7901 and MGC-803 cells were infected with shNC- or shOTX1-expressing virus. (A) Flow cytometric quantification of apoptosis using dual Annexin V-FITC and PI staining in the indicated cells. (B) Percentages of surviving cells and apoptotic cells are presented as the mean ± SD (n=3). *P<0.05 compared with the negative control group (shNC). (C) The expression levels of OTX1, Bcl-2, Bax, cleaved caspase-3 and cleaved PARP in SGC-7901 (left panel) and MGC-803 cells (right panel) were determined by western blot analysis. GAPDH served as a loading control.
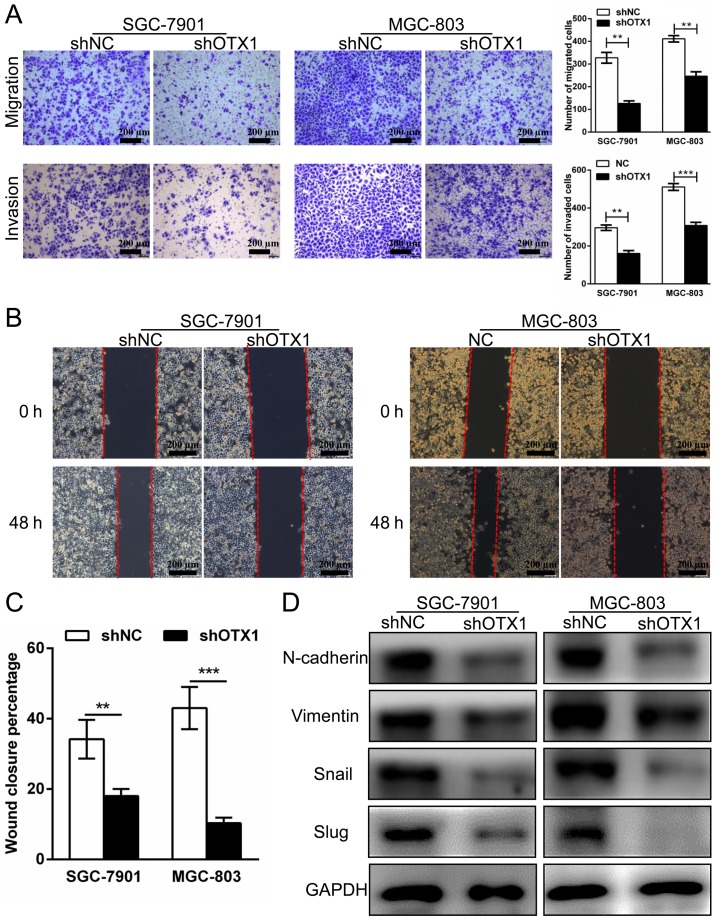
Effect of OTX1 on GC cell migration and invasion
To investigate whether OTX1 contributed to the migration and invasion of GC cells, wound healing, Transwell migration and Matrigel invasion assays were performed. As displayed in Fig. 6A, the knockdown of OTX1 significantly attenuated cell migration and invasive abilities compared with those in the control group. In addition, we observed that the percentage of wound closure in the OTX1-silenced cells was markedly decreased compared with the shNC-infected cells, indicating that the migration of the SGC-7901 and MGC-803 cells was inhibited by OTX1 silencing (Fig. 6B and C). Subsequently, we used western blot analysis to examine whether the expression of epithelial-mesenchymal transition (EMT)-related proteins was altered following OTX1 knockdown. Consistent with our previous observations, we found that the OTX1-silenced cells exhibited a reduction in the expression of N-cadherin, vimentin, Snail and Slug (Fig. 6D). In summary, these results indicated that OTX1 promoted GC cell migration and invasion by altering the expression of EMT-related proteins.
Figure 6.
Knockdown of OTX1 attenuates GC cell migration and invasion. (A) Migration (upper panel) and invasion (bottom panel) assays for shNC-transfected and shOTX1-transfected GC cells. Scale bars, 100 µm. Quantitative results for the migrated and invaded GC cells (right panel) are shown. Three independent experiments were conducted, and the results are presented as the mean ± SD. **P<0.01, ***P<0.001. (B) A wound healing assay was performed to determine the migration of SGC-7901 and MGC-803 cells following infection with an shNC- or shOTX1-expressing virus. (C) Quantitative analysis of the percentage of wound closure. **P<0.01, ***P<0.001. (D) Western blot analysis of the levels of EMT marker proteins in SGC-7901 and MGC-803 cells following infection with shNC- and shOTX1-expressing viral vectors. GAPDH served as an internal control.
Discussion
The overexpression of OTX1 is a common event in various types of cancer, including medulloblastomas, breast and colorectal cancer and hepatocellular carcinoma (12–15). The overexpression of OTX1 in GC tissues was reported in a genome-wide expression profiling analysis (18). Our clinical association study revealed that the overexpression of OTX1 was significantly associated with the Borrmann type, lymph node metastasis, and a shorter OS time in GC patients. Cox proportional hazard regression analysis further identified OTX1 as an independent factor for a poor prognosis, demonstrating that OTX1 overexpression in GC may serve as a biomarker for early detection and precise prognoses. However, to our knowledge, the biological role of OTX1 in GC cells and the underlying mechanisms of this factor remain largely unknown. In the present study, we provided evidence that OTX1 plays an important role in GC carcinogenesis.
OTX1 has been previously only implicated in embryonic development. Recently, increasing evidence has demonstrated that OTX1 participates in the progression of numerous malignancies (9,12–15). In breast cancer cells, it was revealed that p53 directly induced the expression of OTX1 by acting on the OTX1 promoter (13). The present study demonstrated that the silencing of OTX1 expression inhibited GC cell proliferation and colony formation. In addition, we observed that the expression of PCNA, a proliferation marker, was reduced after the OTX1 knockdown. The dysregulation of the cell cycle is a hallmark of tumorigenesis. We determined with the cell cycle distribution analysis that the lack of OTX1 led to a G0/G1 phase arrest. Furthermore, we found that the expression of cyclin D1, a key protein that promoted the G0/G1 to S phase transition (19), decreased when OTX1 was silenced. Collectively, our results demonstrated that the knockdown of OTX1 induced a G0/G1 arrest by downregulating cyclin D1.
It is well known that the deregulation of apoptosis contributes to cancer development. Caspase-3 is a key executioner caspase, and caspase-3 activation leads to the cleavage of PARP, which is considered a central indicator of apoptosis (20). In this study, we confirmed that the knockdown of OTX1 resulted in higher apoptosis rates in SGC-7901 and MGC-803 cells than those in the control cells. Furthermore, the knockdown of OTX1 significantly increased the expression of Bax, cleaved caspase-3 and cleaved PARP, while such a knockdown downregulated the expression of Bcl-2. However, which pathway is involved in the shOTX1-induced apoptosis remains to be investigated. A previous study demonstrated that OTX1 contributed to HCC progression possibly by regulating the ERK/MAPK pathway (15). It would be interesting to examine whether the ERK/MAPK pathway is also affected by OTX1 in GC cells.
The EMT, characterized by the loss of epithelial cell polarity, plays a crucial role in cancer metastasis (21,22). Our results indicated that the knockdown of OTX1 inhibited migration and invasion by suppressing the expression of mesenchymal markers (N-cadherin and vimentin) and EMT-related transcription factors (Snail and Slug). These findings demonstrated that OTX1 promoted the metastasis of GC cells by inducing the EMT process. Further studies will be required to clarify the specific target genes of OTX1 in tumorigenesis.
In conclusion, our results demonstrated that OTX1 enhanced tumor growth and induced EMT in GC cells, supporting the oncogenic role of OTX1 in GC progression. Our results indicated that OTX1 may serve as a potential target for the treatment of GC. However, more detailed studies are required to further illustrate the role of OTX1 in gastric carcinoma cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YDY supervised and directed this study. SCQ and ZZ performed most of the experiments. SCQ and YDY contributed to the project design. JXS and XHX collected the tumor samples and the clinical data. JY, JJL and BC contributed to the cell culture and RNA extraction. GDZ and XYW helped with manuscript preparation. SCQ, ZZ and YDY analysed the data and wrote this manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Haimen People's Hospital, and all the patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang J, Yu JC, Kang WM, Ma ZQ. Treatment strategy for early gastric cancer. Surg Oncol. 2012;21:119–123. doi: 10.1016/j.suronc.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Nonoshita T, Otsuka S, Inagaki M, Iwagaki H. Complete response obtained with S-1 plus CDDP therapy in a patient with multiple liver metastases from gastric cancer. Hiroshima J Med Sci. 2015;64:65–69. [PubMed] [Google Scholar]
- 5.Tong W, Ye F, He L, Cui L, Cui M, Hu Y, Li W, Jiang J, Zhang DY, Suo J. Serum biomarker panels for diagnosis of gastric cancer. Onco Targets Ther. 2016;9:2455–2463. doi: 10.2147/OTT.S86139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang JS, Kim KH, Keum KC, Noh SH, Lim JS, Kim HS, Rha SY, Lee YC, Hyung WJ, Koom WS. Recursive partition analysis of peritoneal and systemic recurrence in patients with gastric cancer who underwent D2 gastrectomy: Implications for neoadjuvant therapy consideration. J Surg Oncol. 2016;114:859–864. doi: 10.1002/jso.24405. [DOI] [PubMed] [Google Scholar]
- 7.Thrumurthy SG, Chaudry MA, Chau I, Allum W. Does surgery have a role in managing incurable gastric cancer? Nat Rev Clin Oncol. 2015;12:676–682. doi: 10.1038/nrclinonc.2015.132. [DOI] [PubMed] [Google Scholar]
- 8.Klein WH, Li X. Function and evolution of Otx proteins. Biochem Biophys Res Commun. 1999;258:229–233. doi: 10.1006/bbrc.1999.0449. [DOI] [PubMed] [Google Scholar]
- 9.Omodei D, Acampora D, Russo F, De Filippi R, Severino V, Di Francia R, Frigeri F, Mancuso P, De Chiara A, Pinto A, et al. Expression of the brain transcription factor OTX1 occurs in a subset of normal germinal-center B cells and in aggressive non-hodgkin lymphoma. Am J Pathol. 2009;175:2609–2617. doi: 10.2353/ajpath.2009.090542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen KB, Lutterodt M, Rath MF, Møller M. Expression of the homeobox genes PAX6, OTX2, and OTX1 in the early human fetal retina. Int J Dev Neurosci. 2009;27:485–492. doi: 10.1016/j.ijdevneu.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Pagani IS, Terrinoni A, Marenghi L, Zucchi I, Chiaravalli AM, Serra V, Rovera F, Sirchia S, Dionigi G, Miozzo M, et al. The mammary gland and the homeobox gene Otx1. Breast J. 2010;16(Suppl):S53–S56. doi: 10.1111/j.1524-4741.2010.01006.x. [DOI] [PubMed] [Google Scholar]
- 12.de Haas T, Oussoren E, Grajkowska W, Perek-Polnik M, Popovic M, Zadravec-Zaletel L, Perera M, Corte G, Wirths O, van Sluis P, et al. OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol. 2006;65:176–186. doi: 10.1097/01.jnen.0000199576.70923.8a. [DOI] [PubMed] [Google Scholar]
- 13.Terrinoni A, Pagani IS, Zucchi I, Chiaravalli AM, Serra V, Rovera F, Sirchia S, Dionigi G, Miozzo M, Frattini A, et al. OTX1 expression in breast cancer is regulated by p53. Oncogene. 2011;30:3096–3103. doi: 10.1038/onc.2011.206. [DOI] [PubMed] [Google Scholar]
- 14.Yu K, Cai XY, Li Q, Yang ZB, Xiong W, Shen T, Wang WY, Li YF. OTX1 promotes colorectal cancer progression through epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2014;444:1–5. doi: 10.1016/j.bbrc.2013.12.125. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Miao Q, Xu CW, Huang JH, Zhou YF, Wu MJ. OTX1 contributes to hepatocellular carcinoma progression by regulation of ERK/MAPK pathway. J Korean Med Sci. 2016;31:1215–1223. doi: 10.3346/jkms.2016.31.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang N, Wang Q, Shen D, Sun X, Cao X, Wu D. Downregulation of microRNA-122 promotes proliferation, migration, and invasion of human hepatocellular carcinoma cells by activating epithelial-mesenchymal transition. Onco Targets Ther. 2016;9:2035–2047. doi: 10.2147/OTT.S92378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao Y, Zhao Q, Yin S, Ding X, Wang H. Genome-wide expression profiling and bioinformatics analysis of deregulated genes in human gastric cancer tissue after gastroscopy. Asia Pac J Clin Oncol. 2018;14:e29–e36. doi: 10.1111/ajco.12688. [DOI] [PubMed] [Google Scholar]
- 19.Ezhevsky SA, Ho A, Becker-Hapak M, Davis PK, Dowdy SF. Differential regulation of retinoblastoma tumor suppressor protein by G(1) cyclin-dependent kinase complexes in vivo. Mol Cell Biol. 2001;21:4773–4784. doi: 10.1128/MCB.21.14.4773-4784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 21.Nieto MA. Epithelial-mesenchymal transitions in development and disease: Old views and new perspectives. Int J Dev Biol. 2009;53:1541–1547. doi: 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- 22.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.