Abstract
MicroRNA (miR)-181a is a member of the miR-181 family that serves a key role in the pathogenesis of various cancer types. The present study aimed to investigate the interaction between miR-181a and Ras association domain family protein1 isoform A (RASSF1A), and their roles in gastric carcinogenesis. The interaction between miR-181a and RASSF1A was assessed in cell lines and cancer tissues. The direct binding of miR-181a and RASSF1A was identified using a luciferase reporting gene system. The effects of miR-181a and RASSF1A on gastric cancer cell growth, cell cycle and apoptosis were assessed with a Cell Counting Kit-8 assay and flow cytometry. The effects of miR-181a on cell division cycle 25A (CDC25A), cyclin A2, cyclin D1, p21, Bcl-2-associated X protein (Bax) and B-cell lymphoma-2 (Bcl-2) protein levels were assessed in gastric cancer cell lines. miR-181a directly interacted with the 3′-untranslated region of RASSF1A and downregulated RASSF1A protein expression. In tissues from patients with gastric cancer, the miR-181a level was significantly higher in the tumor tissues and was negatively correlated with the RASSF1A protein level. RASSF1A suppressed gastric cancer cell proliferation and G1/S transition, and promoted apoptosis; whereas miR-181a promoted cancer cell proliferation and G1/S transition, and suppressed apoptosis. RASSF1A knockdown attenuated the effects of miR-181a downregulation on cell proliferation and apoptosis. Furthermore, miR-181a upregulated CDC25A, cyclin A2 and Bcl-2, and downregulated Bax protein expression in gastric cancer cell lines. These data indicate that miR-181a promotes gastric carcinogenesis, possibly through a direct interaction with RASSF1A.
Keywords: gastric cancer, microRNA-181a, Ras association domain family protein1 isoform A, proliferation, apoptosis
Introduction
Gastric cancer is one of the most prevalent types of cancer worldwide (1). The prognosis of gastric cancer is poor because the majority of patients are diagnosed at the advanced stage (2), and the average 5-year survival rate of advanced gastric cancer is 20–30%, despite a favorable prognosis for early-stage gastric cancer, with an average 5-year survival rate of >90% (3). Therefore, the identification of molecular targets for developing novel treatment is essential in gastric cancer.
MicroRNAs (miRNAs) are noncoding RNAs that consist of 22–25 nucleotides, and modulate various cellular processes, including cell growth, development, differentiation, metabolism and apoptosis (4). Increasing evidence supports a role of miRNAs as oncogenes or suppressor in solid tumors (5). miR-181a is a member of the miR-181 family that serves a role in the pathogenesis of various cancer types (6–9). Particularly, it is reported to be expressed at a high level in gastric cancer and may promote gastric carcinogenesis (10,11). Inconsistent with these studies, Lin et al (12) reported that miR-181a inhibits cell proliferation, migration and metastasis, and is downregulated in gastric cancer. Therefore, the function of miR-181a in the pathogenesis of gastric cancer remains controversial, and the exact molecular mechanisms by which miR-181a modulate the process remain to be elucidated.
The Ras association domain family protein1 isoform A (RASSF1A), encoded by the RASSF1A gene, is localized at chromosome 3p21.3 (13). In various cancer types, including non-small cell lung and gastric cancer, suppression of RASSF1A expression has been reported (14–16), and RASSF1A therefore is theorized to function as a tumor suppressor. Aberrant promoter methylation is the most common molecular mechanism of silencing RASSF1A (17,18). Furthermore, miRNAs, including miR-602 and miR-181a/b, have been demonstrated to target and downregulate RASSF1A in hepatocellular carcinoma and acute promyelocytic leukemia (16,19). This suggests that miRNA-mediated suppression of RASSF1A may serve an essential role in the carcinogenesis and cancer progression. The present study aimed to investigate the interaction between miR-181a and RASSF1A, and their respective roles in gastric cancer.
Materials and methods
Clinical samples and cell cultures
A total of 42 pairs of gastric cancer samples and adjacent non-cancer tissue samples (5 cm away from the tumor) were collected from patients (31 males and 11 females; aged 40–78 years old) who had undergone surgery for primary gastric cancer at The First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) between March 2014 and July 2014. No patient had received preoperative radiotherapy or chemotherapy. Written informed consent was obtained from all patients, and the study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China).
AGS, SGC-7901 and 293 cells were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were maintained at 37°C in RPMI-1640 (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) in a humidified incubator with 5% CO2.
Cell transfection
The miR-181a mimics, negative control (NC), miR-181a inhibitor, inhibitor NC, siRNA-RASSF1A and siRNA-NC were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were as follows: miR-181a mimic, 5′-AACAUUCAACGCUGUCGGUGAGUUCACCGACAGCG-3′; miR-181a inhibitor, 5′-ACUCACCGACAGCGUUGAAUGUU-3′; siRNA-RASSF1A forward, 5′-GACCUCUGUGGCGACUU-3′ and reverse, 5′-UGAAGUCGCCACAGAG-3′; NC and siRNA-NC forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′; inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′. For RNA delivery, cells were seeded at a density of 1×105 cells/well in 6-well plates, and Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect the cells with 100 nM miR-181a mimic or NC, 200 nM miR-181a inhibitor or inhibitor NC, and 50 nM siRNA-RASSF1A, following the manufacturer's protocol. Each experiment was repeated three times.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from the cell lines and tissue samples, following the manufacturer's protocol. The RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) was used to convert the RNA into cDNA with the following temperature protocol: 25°C for 5 min, followed by 42°C for 60 min and 70°C for 5 min. The following primers were used for qPCR: RASSF1A forward, 5′-AGTGCGCGCATTGCAAGTT-3′ and reverse, 5′-AAGGTCAGGTGTCTCCCAC-3′; miR-181a forward, 5′-ACACTCCAGCTGGGAACATTCAACGCTGTCG-3′ and reverse, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGACTCACCG-3; RNU6B forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The primers of miR-181a and RNU6B were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). For the chain reaction, 2 µl of cDNA, 1 µl forward primer and 1 µl reverse primer were mixed in SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd., Dalian, China) reagent. RT-qPCR was performed with a CFX96™ Real-Time PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using SYBR Premix Ex Taq II at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min. The specific mRNA expression level was quantified by using the 2−∆∆Cq method (20). Each experiment was repeated three times.
miR-181a target prediction
The miR-181a sequence was obtained from miRBase (http://www.microrna.sanger.ac.uk). The target genes were predicted using bioinformatics analysis tools, including TargetScan (http://www.targetscan.org/vert_72/), miRBase (http://www.mirbase.org/) and PICTA (https://pictar.mdc-berlin.de/).
Luciferase reporter assay
The 795 bp RASSF1A 3′-untranslated region (UTR) fragment was amplified by qPCR from the DNA of AGS cells and inserted into the pGL3-control vector (Promega Corporation, Madison, WI, USA). The following primers were used: RASSF1A 3′-UTR forward, 5′-GTCTAGACCTCTTGTACCCCAGGTGG-3′ and reverse, 5′-GTCTAGAGAGGATCTTGAAATCTTTATTGAG-3′. Mutagenesis of the miR-181a binding site was performed using a Site-directed Mutagenesis kit (Takara Biotechnology Co., Ltd.) (16) (Fig. 1A and B). For Luciferase reporter assay, 239 cells were transiently cotransfected with 0.5 µg pGL3-RASSF1A 3′UTR or pGL3-RASSF1A 3′UTR mutated, and 50 nM miR-181a mimics or NC using Lipofectamine 2000. Then, 24 h after transfection, Firefly and Renilla luciferase activity were measured using the Dual-Luciferase Reporter assay system, following the manufacturer's protocol (Promega Corporation). Each experiment was repeated three times.
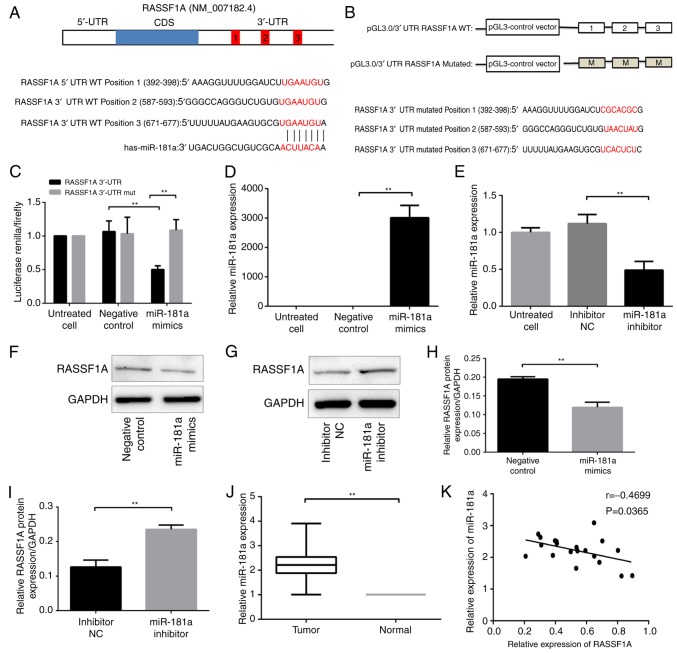
Figure 1.
Association between miR-181a and its target gene RASSF1A. (A) Schematic representation of the RASSF1A 3′-UTR including the three predicted miR-181 binding sites. (B) Schematic representation of the pGL3-constructs for the wild type 3′-UTR of RASSF1A and the mutated 3′-UTR of RASSF1A. (C) Statistical results of the luciferase activity in each group of 293 cells. (D) miR-181a expression in gastric cancer cells AGS following transfection with miR-181a mimics. (E) miR-181a expression in gastric cancer cells SGC-7901 following transfection with miR-181a inhibitor. (F) Western blot band of RASSF1A protein expression in each group of AGS cells. (G) Western blot band of RASSF1A protein expression in each group of SGC-7901 cells. (H) Comparison of protein levels in each group of AGS cells. (I) Comparison of protein levels in each group of SGC-7901 cells. (J) miR-181a expression in gastric cancer tissue and their matched normal tissues. (K) The correlation between the relative expression of miR181a and RASSF1A in 10 pairs of gastric cancer and the adjacent normal tissues. **P<0.01 vs. negative control, inhibitor NC or normal. NC, negative control; RASSF1A, Ras association domain family protein1 isoform A; UTR, untranslated region; WT, wild type; mut, mutant; miR, microRNA.
Cell growth assay
SGC-7901 and AGS cells were transfected with oligonucleotides as aforementioned. Cells were seeded into 96-well culture plates (6×103 cells/well) in 100 µl RPMI-1640 medium containing 10% FBS. Cell growth was assessed using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) following the manufacturer's protocol at 24, 48, 72 and 96 h. Each experiment was repeated three times.
Cell cycle and apoptosis assay
Following transfection for 48 h, SGC-7901 and AGS cells were collected by trypsinization and washed with PBS twice. Cycle analyses were performed with the following protocol: Cells were fixed in 75% cold ethanol at 4°C overnight. Subsequently, the cells were treated with RNase A for 30 min at 37°C and stained with propidium iodide (PI) in the dark at room temperature for 30 min. For apoptosis analysis, cells were labeled with Annexin V-FITC and PI (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's protocol. Cell cycle and apoptosis were assessed using FACS Calibur flow cytometry (BD Biosciences), and the data were analyzed using Flowjo 10.0 software (FlowJo LLC, Ashland, OR, USA). Each experiment was repeated three times.
Protein extraction and western blot analysis
Fresh tissues and cells were lysed using RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) and were quantified using a bicinchoninic acid assay (Pierce; Thermo Fisher Scientific, Inc.). Tissue or cell lysates containing 30 µg total protein/lane were loaded and separated on a 10% SDS-PAGE gel, followed by transference to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% fat-free dry milk at room temperature for 1 h and then incubated with appropriate primary antibodies at 1:1,000 dilution at 4°C overnight. The primary antibodies against human cell division cycle 25A (CDC25A; mouse IgG; cat. no. sc-70823), p21 (mouse IgG, cat. no. sc-71811), Bcl-2-associated X protein (BAX; mouse IgG; cat. no. sc-20067), B-cell lymphoma 2 (Bcl-2; mouse IgG; cat. no. sc-509) and GAPDH (mouse IgG; cat. no. sc-47724) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Anti-cyclin A2 (mouse IgG; cat. no. 4656) and cyclin D1 (rabbit IgG; cat. no. 2922) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Anti-RASSF1A (mouse IgG; cat. no. eB114-10H1) was purchased from Thermo Fisher Scientific, Inc. (eBioscience). The membrane was washed with TBS-Tween-20 buffer at room temperature three times, each time for 10 min, following incubation with the primary antibodies, and subsequently incubated with goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Abgent, San Diego, CA, USA) at room temperature for 2 h. To visualize the protein bands, SuperSignal West Femto Maximum Sensitivity substrate (Pierce; Thermo Fisher Scientific, Inc.) was added. Protein expression was quantified by ImageJ software version 1.46 (National Institutes of Health; Bethesda, MD, USA). Each experiment was repeated three times.
Statistical analysis
All data presented are mean ± standard deviation unless otherwise noted. A Student's t-test or one-way analysis of variance with Duunett's post hoc test were used to detect statistical differences between groups. The correlation between RASSF1A and miR-181a expression was analyzed using the Pearson's correlation test. The association between the miR-181a level and clinicopathological characteristics in gastric cancer tissues was analyzed using a Student's t-test or one-way analysis of variance with the Least significant difference post hoc test. All statistical analyses were performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Identifying RASSF1A as a direct target of miR-181a
In order to investigate the role of miR-181a in gastric carcinogenesis, search for its putative target genes with bioinformatics analysis tools, including TargetScan, miRBase and PICTAR was performed. RASSF1A was identified as a possible target (Fig. 1A). The interaction between miR-181a and RASSF1A was then assessed. The possible binding sites for miR-181a in the 3′-UTR of RASSF1A was mutated, and luciferase reporter constructs with the mutated sequence and the wild type sequence were constructed (Fig. 1B). The mutant or wild-type RASSF1A construct was co-transfected with miR-181a mimics in 293 cells, and the luciferase activity of the RASSF1A 3′-UTR constructs was measured with the dual luciferase reporter assay. Luciferase activity in cells that were transfected with wild-type constructs and miR-181a mimics was significantly suppressed compared with cells transfected with wild-type constructs and NC (P<0.01; Fig. 1C). In contrast, cells co-transfected with the mutant constructs and miR-181a mimics exhibited a recovery of luciferase activity (P<0.01; Fig. 1C). These results indicated a direct interaction between miR-181a and RASSF1A at the 3′-UTR binding sites.
The effects of miR-181a on RASSF1A protein expression was then explored. The miR-181a expression in AGS cells transfected with miR-181a mimics was significantly higher compared with that in cells transfected with negative control (P<0.01; Fig. 1D); whereas the miR-181a expression in SCG-7901 cells transfected with miR-181a inhibitor was lower compared with that in cells transfected with inhibitor NC (P<0.01; Fig. 1E). RASSF1A protein level was significantly lower in AGS cells transfected with miR-181a mimics compared with that in cells transfected with negative control (P<0.01; Fig. 1F and H). Conversely, the RASSF1A protein level was significantly higher in SGC-7901 cells transfected with miR-181a inhibitor compared with that in cells transfected with inhibitor NC (P<0.01; Fig. 1G and I). Therefore, miR-181a level was negatively associated with the RASSF1A protein level. Notably, miR-181a did not significantly affect RASSF1A mRNA level in AGS and SGC-7901 cells as assessed by qPCR analyses (data not shown), indicating that miR-181a directly suppressed RASSF1A expression at the translational level.
RASSF1A protein expression negatively correlates with miR-181a level in gastric cancer tissue
To investigate whether the regulation of RASSF1A level by miR-181a had clinical implications, miR-181a and RASSF1A levels in tumor tissues and adjacent normal tissues obtained from patients with gastric cancer were examined.
The relative level of miR-181a in 42 paired tumor and normal tissues was measured, and it was demonstrated that the miR-181a level was significantly higher in tumor tissues compared with that in adjacent normal tissue (P<0.01; Fig. 1J). Further analyses did not identify a correlation between the miR-181a level and patient age, sex, tumor size, the locations of tumor, cell differentiation, the tumor node metastasis (TNM) stage or the depth of tumor invasion (Table I).
Table I.
Correlation between miR-181a level and clinicopathological characteristics in gastric cancer tissues.
| Clinicopathological parameters | Patients (n=42) | Relative expression | Test statistics | P-value |
|---|---|---|---|---|
| Sex | t=0.854 | 0.398 | ||
| Male | 31 | 2.320±1.643 | ||
| Female | 11 | 1.861±1.129 | ||
| Age at diagnosis, years | t=−0.967 | 0.340 | ||
| <60 | 20 | 1.961±1.026 | ||
| ≥60 | 22 | 2.417±1.868 | ||
| T stage | t=−0.984 | 0.331 | ||
| T1+T2 | 5 | 1.569±0.801 | ||
| T3+T4 | 37 | 2.285±1.587 | ||
| Lymphatic metastasis | t=−1.706 | 0.096 | ||
| No | 12 | 1.579±0.876 | ||
| Yes | 30 | 2.448±1.667 | ||
| M stage | t=−1.178 | 0.246 | ||
| M0 | 35 | 1.583±0.587 | ||
| M1 | 7 | 2.323±1.629 | ||
| TNM stage | t=−1.641 | 0.109 | ||
| I+II | 9 | 1.475±0.508 | ||
| III+IV | 33 | 2.398±1.652 | ||
| Tumor location | F=3.188 | 0.052 | ||
| Proximal gastric | 17 | 2.873±1.907 | ||
| Gastric body | 8 | 1.939±0.921 | ||
| Distal gastric | 17 | 1.649±1.058 | ||
| Tumor size, cm | t=0.500 | 0.620 | ||
| >5 | 20 | 2.324±1.226 | ||
| ≤5 | 22 | 2.087±1.777 | ||
| Differentiation | t=−1.215 | 0.233 | ||
| Low/Poorly | 17 | 1.936±1.196 | ||
| Moderate/high | 18 | 2.606±1.955 | ||
| Pathological type | F=0.286 | 0.753 | ||
| Adenocarcinoma | 35 | 2.280±1.643 | ||
| Mucinous carcinoma | 5 | 1.769±0.603 | ||
| Neuroendocrine carcinoma | 2 | 1.867±0.936 |
t, statistics performed with the Student's t-test; F, statistics performed with one-way analysis of variance followed by the Least significant difference test; TNM, tumor node metastasis; miR, microRNA.
miR-181a expression was then assessed with qPCR and RASSF1A protein levels with western blotting in 10 paired tumor and normal tissues. Furthermore, correlation analysis between miR-181a and RASSF1A expression levels was performed. A moderate correlation between the miR-181a level and RASSF1A protein level identified (P=0.0365; Fig. 1K).
RASSF1A knockdown promotes cell proliferation and G1/S transition, and inhibits apoptosis in AGS cells
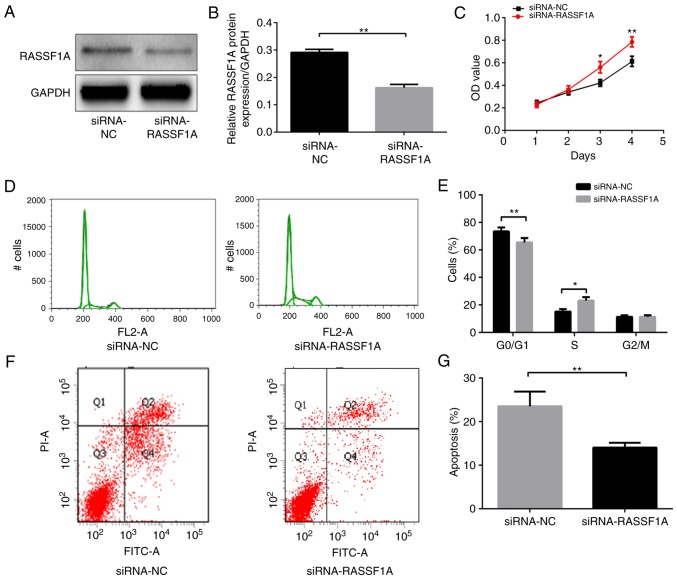
Whether RASSF1A activities have a role in gastric carcinogenesis was investigated. Cell growth, cell cycle and apoptosis were examined in AGS cells transfected with siRNA that targeted RASSF1A. The RASSF1A protein expression level was significantly lower in AGS cells transfected with siRNA-RASSF1A compared with that in cells transfected with siRNA-NC (P<0.01; Fig. 2A and B). Knockdown of RASSF1A with siRNA significantly enhanced cell growth in AGS cells as assessed using the CCK-8 assay (P<0.01; Fig. 2C). In addition, the siRNA-RASSF1A-transfected cells contained a significantly lower proportion of cells in the G0/G1 phase and a significantly higher proportion of cells in the S phase compared with the siRNA-NC transfected cells (P<0.01 and P<0.05, respectively; Fig. 2D and E), whereas the percentages of cells in the G2/M phase did not differ significantly between the two groups (P>0.05).
Figure 2.
Effect of downregulated RASSF1A on the proliferation and apoptosis of AGS gastric cancer cells. (A) Western blot band of RASSF1A protein expression in each group of AGS cells. (B) Comparison of protein levels in each group of AGS cells. (C) RASSF1A effect on cell growth was measured using a Cell Counting Kit-8 assay following transfection with siRNA-RASSF1A of AGS cells. (D) The cell cycle distribution of AGS cell transfected with siRNA-RASSF1A or siRNA-NC for 48 h, and were analyzed by flow cytometry. (E) Comparison of cell cycle distribution of AGS cells among groups. (F) The cell apoptosis of AGS cells transfected with siRNA-RASSF1A or siRNA-NC for 48 h were analyzed by flow cytometry. (G) Comparison of cell apoptosis of AGS cells among groups. *P<0.05 and **P<0.01 vs. siRNA-NC. NC, negative control; RASSF1A, Ras association domain family protein1 isoform A; siRNA, small interfering RNA.
Lastly, the apoptotic rates of AGS cells were assessed with cell flow cytometry. The apoptotic rate of cells transfected with siRNA-RASSF1A was significantly lower compared with that of the cells transfected with siRNA-NC (P<0.01; Fig. 2F and G). Therefore, the aforementioned results suggest that the knockdown of RASSF1A promoted cell proliferation and reduced apoptosis in AGS cells.
Effects of miR-181a on gastric cancer cells
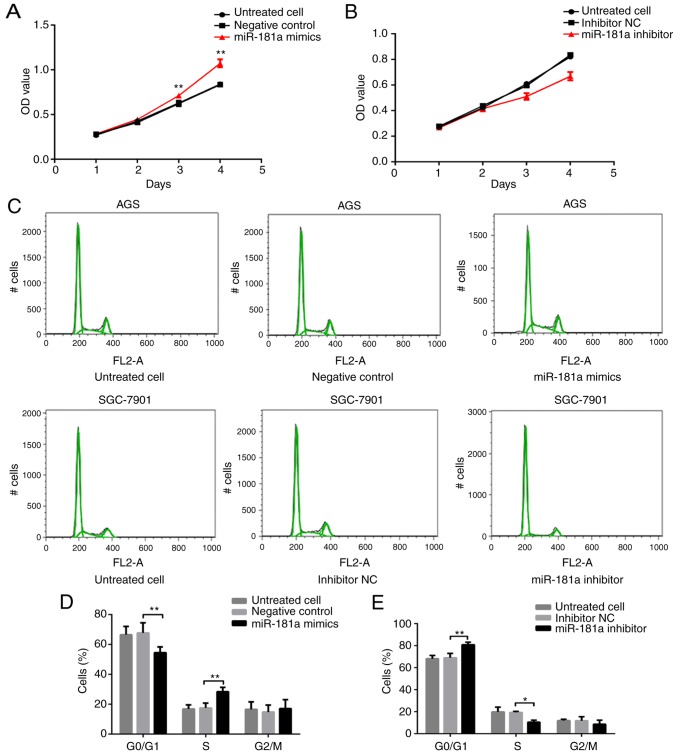
As indicated by the aforementioned results, miR-181a negatively modulated RASSF1A expression, and RASSF1A suppressed cell proliferation, but promoted apoptosis. Thus, the effects of miR-181a on gastric cancer cell lines were examined next. Cell growth was assessed with a CCK-8 assay. AGS cells transfected with miR-181a mimics exhibited significantly greater growth rates compared with cells transfected with negative control (P<0.01; Fig. 3A). Conversely, SGC-7901 cells transfected with miR-181a inhibitor demonstrated significantly less growth compared with cells transfected with inhibitor NC (P<0.01; Fig. 3B).
Figure 3.
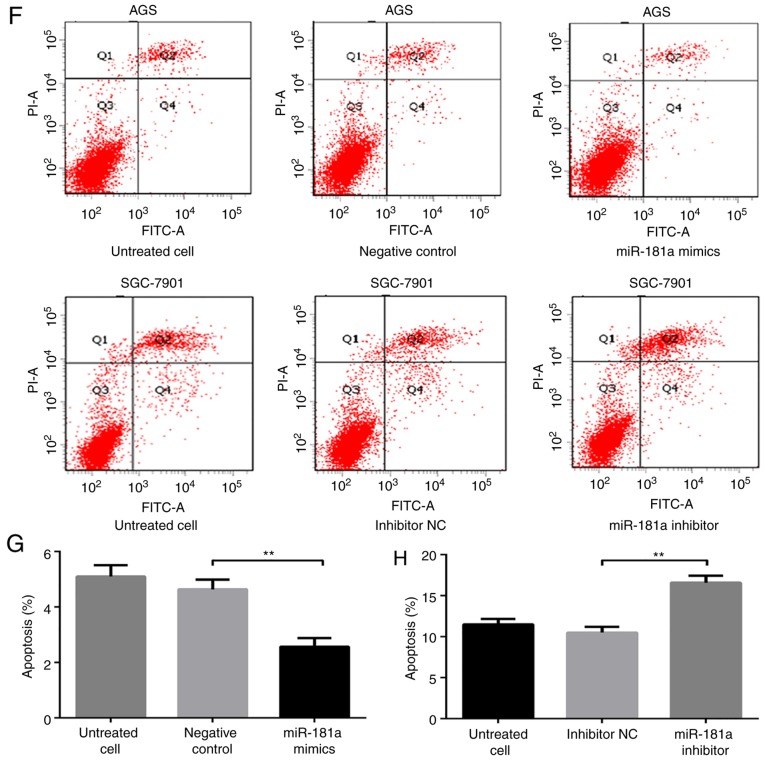
Biological effects of miR-181a on gastric cancer cells. (A) miR-181a effect on AGS cells growth was measured using a Cell Counting Kit-8 assay following transfection with miR-181a mimics. (B) miR-181a effect on SGC-7901 cells growth was measured using a Cell Counting Kit-8 assay following transfection with miR-181a inhibitor. (C) The cell cycle distribution of AGS or SGC-7901 cells, transfected with miR-181a mimics or inhibitor for 48 h, and cell cycle phase distribution was analyzed by flow cytometry. (D) Comparison of cell cycle distribution of AGS cells among groups. (E) Comparison of cell cycle distribution of SGC-7901 cells among groups. (F) The cell apoptosis of AGS or SGC-7901 cells, which was transfected with miR-181a mimics or inhibitor for 48 h, and cell apoptosis was analyzed by flow cytometry. (G) Comparison of cell apoptosis of AGS cells among groups. (H) Comparison of cell apoptosis of SGC-7901 cells among groups. *P<0.05 and **P<0.01 vs. negative control or inhibitor NC. NC, negative control; RASSF1A, Ras association domain family protein1 isoform A; miR, microRNA; CCK-8, Cell Counting Kit-8.
The percentage of cells in each cell cycle phase was measured. The percentage of cells in the G0/G1 phase was significantly lower, while the percentage of cells in the S phase was significantly higher in the miR-181a mimics-transfected AGS cells compared with that in cells transfected with negative control (both P<0.01; Fig. 3C and D). The percentage of cells in the G2/M phase did not differ significantly among the three groups (P>0.05; Fig. 3C and D). Transfection of miR-181a inhibitor revealed the opposite effects, where the percentage of cells in the G0/G1 phase was significantly higher, and in the S phase significantly lower in SGC-7901 cells transfected with miR-181a inhibitor compared with cells treated with inhibitor NC (P<0.01 and P<0.05, respectively; Fig. 3C and E). The percentage of cells in the G2/M phase; however, was not significantly different among the three groups of cells (P>0.05; Fig. 3C and E).
Lastly, the apoptotic rate of cells was assessed with cell flow cytometry (Fig. 3F). The apoptotic rate in AGS cells transfected with miR-181a mimic was significantly lower compared with that in cells transfected with negative control (P<0.01; Fig. 3F and G). In contrast, the apoptotic rate was significantly higher in SGC-7901 cells transfected with miR-181a inhibitor compared with that in cells transfected with the inhibitor NC (P<0.01; Fig. 3F and H).
Taken together, these data indicated that miR-181a and RASSF1A exhibit opposing effects on cell proliferation and apoptosis.
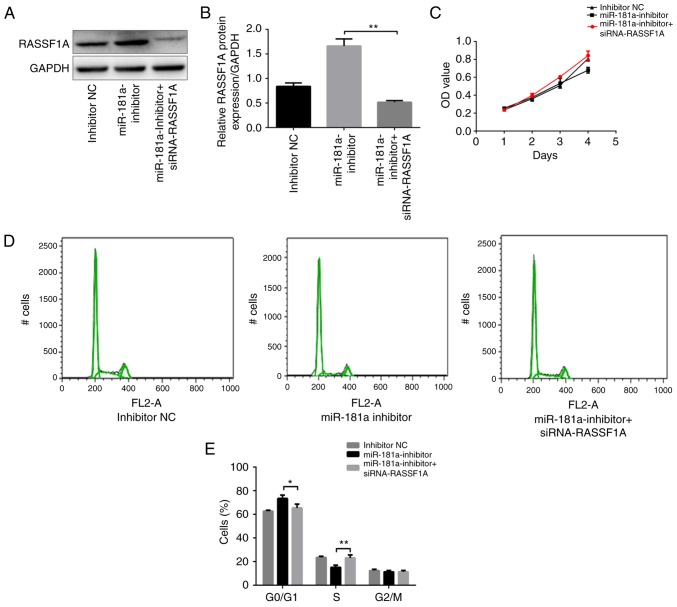
RASSF1A knockdown attenuates the effects of miR-181a downregulation on the proliferation and apoptosis of SGC-7901 cells
Whether RASSF1A knockdown attenuated the effect of miR-181a downregulation on proliferation and apoptosis of SGC-7901 cells was investigated. SGC-7901 cells were co-transfected with miR-181a inhibitor and siRNA-RASSF1A. Western blot analysis was used to confirm that RASSF1A expression was downregulated by siRNA-RASSF1A in SGC-7901 cells transfected with miR-181a inhibitor (P<0.01; Fig. 4A and B). Next, cell growth was assessed using a CCK-8 assay. The growth of SGC-7901 cells co-transfected with miR-181a inhibitor and siRNA-RASSF1A was significantly increased compared with cells transfected with miR-181a inhibitor only (P<0.01; Fig. 4C). Furthermore, the percentage of cells in the G0/G1 phase was significantly lower, while the percentage of cells in the S phase was significantly higher in the cells co-transfected with miR-181a inhibitor and siRNA-RASSF1A compared with cells transfected with miR-181a inhibitor only (P<0.05 and P<0.01, respectively; Fig. 4D and E). The percentage of cells in the G2/M phase did not differ significantly between the two groups (P>0.05; Fig. 4D and E). These results indicated that the inhibitory effects of downregulation of miR-181a on proliferation in SGC-7901 cells were attenuated by RASSF1A knockdown.
Figure 4.
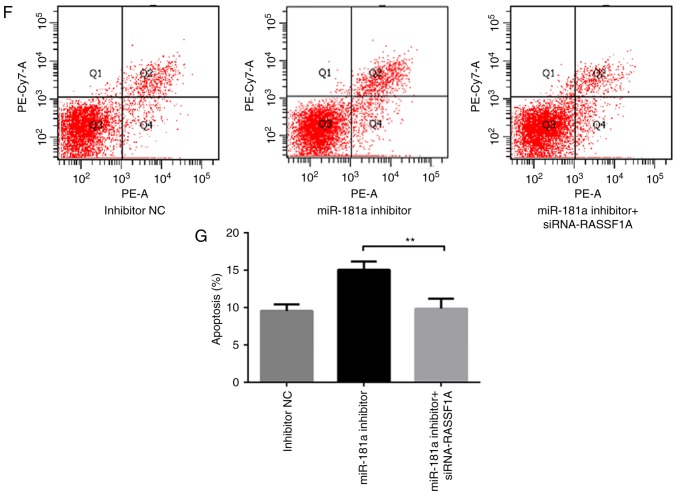
RASSF1A knockdown attenuates the effects of downregulation of miR-181a on proliferation and apoptosis on SGC-7901 cells. (A) Western blot band of RASSF1A protein expression in each group of SGC-7901 cells. (B) Comparison of protein levels in each group of SGC-7901 cells. (C) The growth of SGC-7901 cells co-transfected with miR-181a inhibitor and siRNA-RASSF1A was measured using a Cell Counting Kit-8 assay. (D) The cell cycle distribution of SGC-7901 cells co-transfected with miR-181a inhibitor and siRNA-RASSF1A for 48 h, and were analyzed by flow cytometry. (E) Comparison of cell cycle distribution of SGC-7901 cells among groups. (F) The cell apoptosis of AGS cells transfected with co-transfected with miR-181a inhibitor and siRNA-RASSF1A for 48 h were analyzed by flow cytometry. (G) Comparison of cell apoptosis of AGS cells among groups. *P<0.05 and **P<0.01 vs. miR-181a inhibitor. NC, negative control; RASSF1A, Ras association domain family protein1 isoform A; miR, microRNA; siRNA, small interfering RNA.
The apoptotic rate in cells co-transfected with miR-181a inhibitor and siRNA-RASSF1A was significantly lower compared with that in cells transfected with miR-181a inhibitor only (P<0.01; Fig. 4F and G). Taken together, these data suggested that RASSF1A knockdown attenuated the effects of miR-181a downregulation on cell proliferation and apoptosis.
Effects of miR-181a on the expression of tumorigenesis proteins
To further investigate the role of miR-181a in gastric cancer, the effects of miR-181a on the expression of tumorigenesis-associated proteins, including CDC25A, cyclin A2, p21, cyclin D1, Bcl-2, and Bax, were investigated.
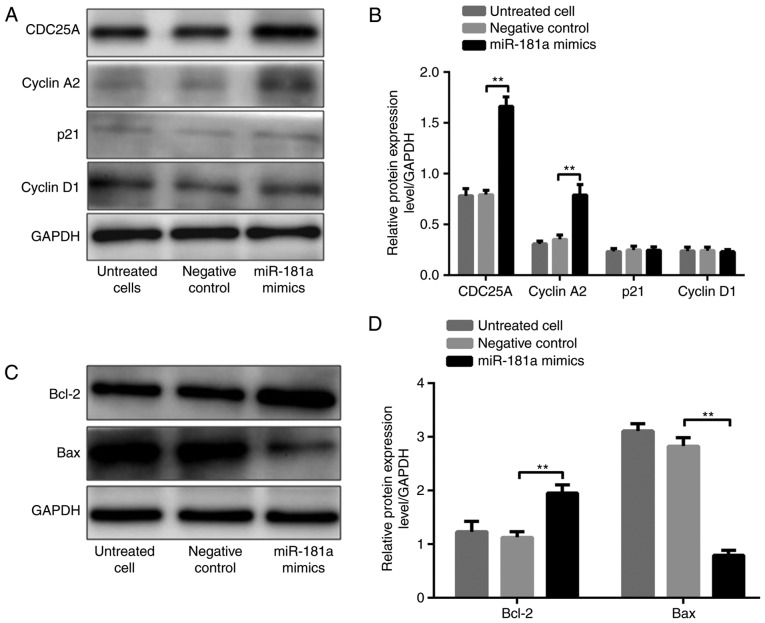
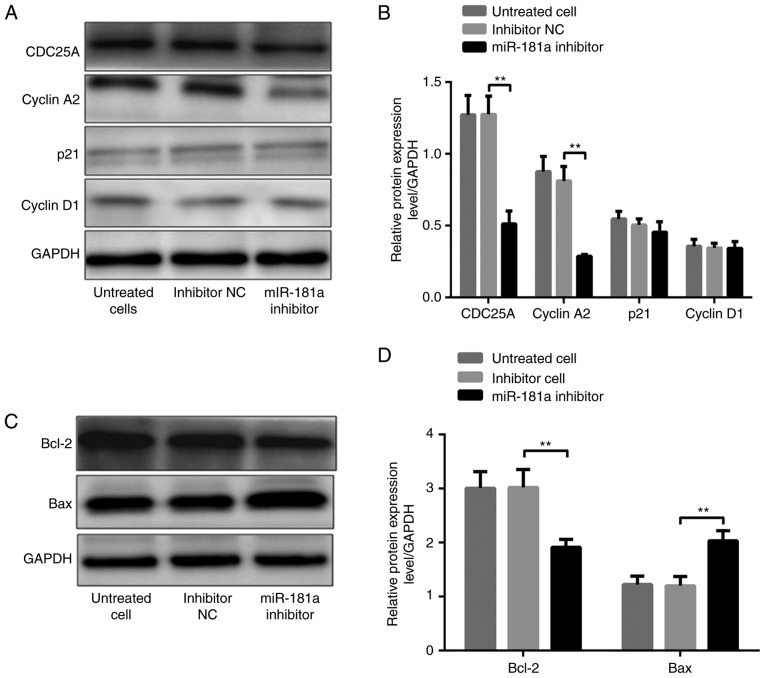
It was demonstrated that CDC25A, cyclin A2 and Bcl-2 protein levels were significantly higher, whereas the Bax protein level was significantly lower, in AGS cells transfected with miR-181a mimic compared with these in cells transfected with the negative control (all P<0.01; Fig. 5A-D). P21 and cyclin D1 levels were not significantly different in the three groups (both P>0.05; Fig. 5A and B). In contrast, CDC25A, cyclin A2 and Bcl-2 protein levels were significantly lower, whereas the Bax protein level was significantly higher, in SGC-7901 cells transfected with miR-181a inhibitors compared with these in cells transfected with inhibitor NC (all P<0.01; Fig. 6A-D). Similarly, p21 and cyclin D1 levels were not significantly different among the three groups (both P>0.05; Fig. 6A and B).
Figure 5.
Effects of upregulated miR-181a on the expression of tumorigenesis-associated proteins. (A) Western blot band of CDC25A, cyclin A2, p21 and cyclin D1 protein expression in each group of AGS cells. (B) Comparison of protein levels in each group of AGS cells. (C) Western blot band of Bcl-2 and Bax protein expression in each group of AGS cells. (D) Comparison of protein levels in each group of AGS cells. **P<0.01 vs. negative control. miR, microRNA; CDC25A, cell division cycle 25A; Bax, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma-2.
Figure 6.
Effects of downregulated miR-181a on the expression of tumorigenesis-associated proteins. (A) Western blot band of CDC25A, cyclin A2, p21 and cyclin D1 protein expression in each group of SGC-7901 cells. (B) Comparison of protein levels in each group of SGC-7901 cells. (C) Western blot band of Bcl-2 and Bax protein expression in each group of SGC-7901 cells. (D) Comparison of protein levels in each group of SGC-7901 cells. **P<0.01 vs. inhibitor NC. NC, negative control; miR, microRNA; CDC25A, cell division cycle 25A; Bax, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma-2.
These results indicated the complexity of the miR-181a signaling pathway, and that miR-181a targeted multiple downstream effectors, which in turn serve essential roles in tumorigenesis.
Discussion
In previous years, abnormalities in miR-181a expression were reported to serve an essential role in the pathogenesis of numerous types of cancer (8,9,21–23). However, the exact role of miR-181a in tumorigenesis has remained controversial due to inconsistent results in different tumor types. miR-181a is reported to be an oncogene in head and neck cancer (21), breast cancer (9), and hepatocellular carcinoma (8). In contrast, several studies indicated miR-181a acts as a tumor-suppressor in primary glioblastoma (22) and aggressive chronic lymphocytic leukemia (23). These findings demonstrate that the role of miR-181a in tumorigenesis is tumor-specific. The result of the present study revealed elevated miR-181a expression in gastric cancer tissues, indicating that miR-181a may be an oncogene in gastric cancer.
Identifying cancer-specific miRNA targets is an important step in clarifying the roles of miRNAs in tumorigenesis and progression. miR-181a has been reported to inhibit tumor growth via the downregulation of oncogene K-ras in oral squamous cell carcinoma (21). Liu et al (24) reported that miR-181a promotes the transition of G0/G1 to S and cell growth via targeting tumor suppressor ataxia telangiectasia mutated in pediatric acute myeloid leukemia. In the present study, RASSF1A was identified as a direct target of miR-181a, and the luciferase activity was lower in 293 cells co-transfected with wild-type constructs and miR-181a mimics compared with cells co-transfected with wild-type constructs and NC. Mutation of the putative binding sites in the 3′-UTR of RASSF1A abolished these effects, suggesting that miR-181a directly binds with the 3′-UTR of RASSF1A, thereby suppressing its translation. In addition, in the gastric cancer cell lines, an increase in miR-181a level was associated with reduced RASSF1A protein levels, whereas a decrease in miR-181a level was associated with an increased RASSF1A protein level. Notably, miR-181a did not significantly affect RASSF1A mRNA expression in the AGS and SGC-7901 cell lines, indicating that miR-181a directly suppresses RASSF1A expression at the translational level. Furthermore, in tumor tissues from patients with gastric cancer, a negative correlation between the miR-181a level and RASSF1A protein level was identified.
miRNAs have emerged as an essential modulators of cell proliferation, apoptosis and cell cycle dysregulation (25). RASSF1A was observed to suppress gastric cancer cell proliferation and G1/S transition, and promote apoptosis in the present study. Thus, the downregulation of RASSF1A by miR-181a would expect to cause opposing effects. Indeed, the results demonstrated that miR-181a promoted cell growth and G1/S transition, and suppressed apoptosis in gastric cancer cell lines. Furthermore, RASSF1A knockdown attenuated the effects of downregulation of miR-181a on proliferation and apoptosis in SGC-7901 cells.
Increasing evidence suggests that RASSF1A acts as a tumor suppressor in numerous types of cancer through multiple mechanisms (26,27). The results of the present study suggest that RASSF1A promoted apoptosis, and suppressed cell growth and proliferation, which are consistent with a previous study whereby RASSF1A was demonstrated to suppress cell cycle progression at the G1/S transition by preventing cyclin D1 accumulation (28). Oh et al (29) reported that RASSF1A is required for full activation of macrophage stimulating 1 (Mst1) and enhanced Mst1-mediated apoptosis in vivo. Furthermore, in a previous study, we reported that RASSF1A inhibits SGC-7901 cell invasion under hypoxic conditions, which is associated with matrix metalloproteinase-2 inhibition (30). Inactivation of RASSF1A may result from multiple mechanisms in tumorigenesis, including transcriptional silencing through promoter hypermethylation, loss of heterozygosity and chromosome deficiency (31). The present study reported that miR-181a suppressed RASSF1A by directly interacting with its 3′-UTR region, resulting in downregulation of RASSF1A at the translational level in gastric cancer cell lines. Therefore, it is likely that miR-181a promotes gastric cancer progression by suppressing RASSF1A.
In addition, it was demonstrated that miR-181a was negatively associated with the Bax protein level, and positively associated CDC25A, cyclin A2 and Bcl-2 protein levels in gastric cancer cell lines. CDC25A, a member of the CDC25 family, serves an important role in regulating the G1/S checkpoint and the G2/M checkpoint (32). The increase in the G1/S transition in the presence of miR-181a may be mediated by upregulating CDC25A. In addition, CDC25A has been reported to be involved in the hyperactivation of cyclin A2-cyclin-dependent kinase 1 in the G0/G1 to S transition. The present data revealed that cyclin A2 protein expression increased in cells overexpressing miR-181a, suggesting that upregulation of CDC25A and cyclin A2 mediated by miR-181a results in the promotion of gastric cancer cell proliferation and the G1/S transition. Similarly, it was demonstrated that the expression of anti-apoptosis protein Bcl-2 and proapoptosis protein Bax expression was regulated by miR-181a, consistent with a study by Xu et al (33) reporting similar effects of miR-181a in cervical cancer. However, the exact mechanisms of miR-181a on gastric cancer cell growth and apoptosis remain to be elucidated. It is likely that miR-181a promotes gastric cancer progression via multiple signaling pathways.
miR-181a as a predictor of prognosis in other malignancies has been reported. Pichler et al (34) reported that the level of miR-181a expression level is associated with poor survival of patients with colorectal cancer. Xiang et al (35) reported that higher miR-181a expression was associated with shorter recurrence-free survival and shorter overall survival times in esophageal cancer. However, no significant association was identified between the miR-181a expression and clinical features of gastric cancer in the present study. This inconsistency may be due to the TNM stage of patients included in the current study, whereby the majority of patients were at stage of TNM III and IV. Furthermore, the limited available tissue samples did not allow for reliable correlation analyses to be performed. miR-181a as the prognosis predictor for gastric cancer remains to be validated, as follow-up visits have not reached the universal standard of 3 or 5 years.
Several targets of miR-181a that are associated with cell growth, apoptosis and invasion in a variety of human tumors have been identified, including K-ras (21), caudal type homeobox 2 (36), GATA binding protein 6 (36) and nemo-like kinase (36). The present study results suggested that RASSF1A is a direct target of miR-181a in gastric cancer, and that RASSF1A is highly likely a tumor suppressor in gastric cancer. Previous studies have reported that protein tyrosine phosphatase MEG2 (37) and autophagy-related 5 (38) are directly targeted by miR-181a in gastric cancer. In conclusion, the results of the present study suggest that the upregulation of miR-181a promoted gastric cancer cell growth and the G1/S transition, and inhibited apoptosis, possibly through upregulation of CDC25A, cyclin A2 and Bcl-2, and downregulation of Bax. Taken together, miR-181a functions as an oncogene in gastric cancer that possibly promotes cancer progression by suppressing RASSF1A and may represent a potential molecular target for gastric cancer therapy. However, further studies are required to fully understand the involvement of miR-181a/RASSF1A signaling in gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National Natural Science Foundation of China (grant nos. 81101874 and 81172362), The Science and Technology Project of Shaanxi Province (grant nos. 2016SF-015 and 2016SF-157), and The Coordinative and Innovative Plan Projects of the Science and Technology Program in Shaanxi Province (grant no. 2013KTCQ03-08).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JHY and JQ performed the experiments, acquired the data and drafted the manuscript. WW and GBW collected gastric cancer samples and clinicopathological characteristics, and assisted with the experiments. YHW and QG analyzed and interpreted data. XJS and JBZ substantially contributed to the study conception and design. All authors read and approved the final manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Global Burden of Disease Cancer Collaboration, corp-author. Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng JY, Liang H. Clinical significance of lymph node metastasis in gastric cancer. World J Gastroenterol. 2014;20:3967–3975. doi: 10.3748/wjg.v20.i14.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono H. Early gastric cancer: Diagnosis, pathology, treatment techniques and treatment outcomes. Eur J Gastroenterol Hepatol. 2006;18:863–866. doi: 10.1097/00042737-200608000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H, Tang YL, Kim IM. Crosstalk between long noncoding RNAs and microRNAs in health and disease. Int J Mol Sci. 2016;17:356. doi: 10.3390/ijms17030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng F, Glaser SS, Francis H, DeMorrow S, Han Y, Passarini JD, Stokes A, Cleary JP, Liu X, Venter J, et al. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16:160–173. doi: 10.1111/j.1582-4934.2011.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150–163. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim CH, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, Choi IJ, Munroe DJ, Green JE. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79. doi: 10.1186/1755-8794-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin F, Li Y, Yan S, Liu S, Qian W, Shen D, Lin Q, Mao W. MicroRNA-181a inhibits tumor proliferation, invasiveness, and metastasis and is downregulated in gastric cancer. Oncol Res. 2015;22:75–84. doi: 10.3727/096504014X14024160459203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 14.Dubois F, Keller M, Calvayrac O, Soncin F, Hoa L, Hergovich A, Parrini MC, Mazières J, Vaisse-Lesteven M, Camonis J, et al. RASSF1A suppresses the invasion and metastatic potential of human non-small cell lung cancer cells by inhibiting YAP activation through the GEF-H1/RhoB pathway. Cancer Res. 2016;76:1627–1640. doi: 10.1158/0008-5472.CAN-15-1008. [DOI] [PubMed] [Google Scholar]
- 15.Liao A, Tan G, Chen L, Zhou W, Hu H. RASSF1A inhibits gastric cancer cell proliferation by miR-711-mediated downregulation of CDK4 expression. Oncotarget. 2016;7:5842–5851. doi: 10.18632/oncotarget.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bräuer-Hartmann D, Hartmann JU, Wurm AA, Gerloff D, Katzerke C, Falzacappa Verga MV, Pelicci PG, Müller-Tidow C, Tenen DG, Niederwieser D, et al. PML/RARα-regulated miR-181a/b cluster targets the tumor suppressor RASSF1A in acute promyelocytic leukemia. Cancer Res. 2015;75:3411–3424. doi: 10.1158/0008-5472.CAN-14-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Q, Wang HB, Li YH, Li HF, Li TT, Zhang WX, Xiang SS, Sun ZQ. Correlations of promoter methylation in WIF-1, RASSF1A, and CDH13 genes with the risk and prognosis of esophageal cancer. Med Sci Monit. 2016;22:2816–2824. doi: 10.12659/MSM.896877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Z, Ma K, Sun X, Li A, Wang H, Zhang L, Lin F, Feng X, Song J. Methylation of RASSF1A gene promoter and the correlation with DNMT1 expression that may contribute to esophageal squamous cell carcinoma. World J Surg Oncol. 2015;13:141. doi: 10.1186/s12957-015-0557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9:803–808. doi: 10.4161/cbt.9.10.11440. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Shin KH, Bae SD, Hong HS, Kim RH, Kang MK, Park NH. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2011;404:896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 23.Marton S, Garcia MR, Robello C, Persson H, Trajtenberg F, Pritsch O, Rovira C, Naya H, Dighiero G, Cayota A. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22:330–338. doi: 10.1038/sj.leu.2405022. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Liao W, Peng H, Luo X, Luo Z, Jiang H, Xu L. miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J Cancer Res Clin Oncol. 2016;142:77–87. doi: 10.1007/s00432-015-1995-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santarpia L, Nicoloso M, Calin GA. MicroRNAs: A complex regulatory network drives the acquisition of malignant cell phenotype. Endocr Relat Cancer. 2010;17:F51–F75. doi: 10.1677/ERC-09-0222. [DOI] [PubMed] [Google Scholar]
- 26.Grawenda AM, O'Neill E. Clinical utility of RASSF1A methylation in human malignancies. Br J Cancer. 2015;113:372–381. doi: 10.1038/bjc.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandes MS, Carneiro F, Oliveira C, Seruca R. Colorectal cancer and RASSF family - a special emphasis on RASSF1A. Int J Cancer. 2013;132:251–258. doi: 10.1002/ijc.27696. [DOI] [PubMed] [Google Scholar]
- 28.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, Im CR, Lee JO, Yonehara S, Lim DS. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- 30.Zhou PH, Zheng JB, Wei GB, Wang XL, Wang W, Chen NZ, Yu JH, Yao JF, Wang H, Lu ST, et al. Lentivirus-mediated RASSF1A expression suppresses aggressive phenotypes of gastric cancer cells in vitro and in vivo. Gene Ther. 2015;22:793–801. doi: 10.1038/gt.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 32.Verduzco D, Dovey JS, Shukla AA, Kodym E, Skaug BA, Amatruda JF. Multiple isoforms of CDC25 oppose ATM activity to maintain cell proliferation during vertebrate development. Mol Cancer Res. 2012;10:1451–1461. doi: 10.1158/1541-7786.MCR-12-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Zhu J, Hu C, Song H, Li Y. Inhibition of microRNA-181a may suppress proliferation and invasion and promote apoptosis of cervical cancer cells through the PTEN/Akt/FOXO1 pathway. J Physiol Biochem. 2016;72:721–732. doi: 10.1007/s13105-016-0511-7. [DOI] [PubMed] [Google Scholar]
- 34.Pichler M, Winter E, Ress AL, Bauernhofer T, Gerger A, Kiesslich T, Lax S, Samonigg H, Hoefler G. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol. 2014;67:198–203. doi: 10.1136/jclinpath-2013-201904. [DOI] [PubMed] [Google Scholar]
- 35.Xiang Z, Dong X, Sun Q, Li X, Yan B. Clinical significance of up-regulated miR-181a in prognosis and progression of esophageal cancer. Acta Biochim Biophys Sin. 2014;46:1007–1010. doi: 10.1093/abbs/gmu083. [DOI] [PubMed] [Google Scholar]
- 36.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Sun F, Hong Y, Liu Y, Fen M, Yin K, Ge X, Wang F, Chen X, Guan W. MEG2 is regulated by miR-181a-5p and functions as a tumour suppressor gene to suppress the proliferation and migration of gastric cancer cells. Mol Cancer. 2017;16:133. doi: 10.1186/s12943-017-0695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Nie Y, Wang H, Lin Y. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene. 2016;576:828–833. doi: 10.1016/j.gene.2015.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.