Abstract
The aim of the present study was to identify potential prognostic microRNA (miRNA) biomarkers for colon adenocarcinoma (COAD) prognostic prediction using the dataset of The Cancer Genome Atlas (TCGA). The genome-wide miRNA sequencing dataset and corresponding COAD clinical information were downloaded from TCGA. Prognosis-related miRNA screening was performed by genome-wide multivariable Cox regression analysis and used for prognostic signature construction. Ten miRNAs (hsa-mir-891a, hsa-mir-6854, hsa-mir-216a, hsa-mir-378d-1, hsa-mir-92a-1, hsa-mir-4709, hsa-mir-92a-2, hsa-mir-210, hsa-mir-940 and hsa-mir-887) were identified as prognostic miRNAs and used for further prognostic signature construction. The 10-miRNA prognostic signature showed good performance in prognosis prediction (adjusted P<0.0001; adjusted hazard ratio, 4.580; 95% confidence interval, 2.783–7.538). In the time-dependent receiver operating characteristic analysis, the area under the curve was 0.735, 0.788, 0.806, 0.806, 0.775 and 0.900 for 1-, 2-, 3-, 4-, 5- and 10-year COAD overall survival prediction, respectively. Comprehensive survival analysis suggested that the 10-miRNA prognostic signature is an independent prognostic factor in COAD, with a better performance in COAD overall survival prediction than other traditional clinical parameters. Functional enrichment indicated that the corresponding target genes were significantly enriched in multiple biological processes and pathways, including regulation of cell proliferation, cell cycle, cell growth, and Wnt and transforming growth factor-β signaling pathways. In conclusion, our present study identified a 10-miRNA expression signature that may serve as a potential prognostic biomarker in COAD patients.
Keywords: miRNA, TCGA, colon adenocarcinoma, prognosis, biomarker
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in men and the second in women; it is a malignant digestive tumor, with ~1,360,600 new cases diagnosed and 693,900 deaths from CRC occurring in 2012 (1). The incidence rate of CRC is higher in men than in women in most parts of the world (1), and CRC is currently the fourth leading cause of cancer-related death worldwide (2). The incidence of CRC varies among countries, and the mortality rates of CRC are decreasing in many countries worldwide because of CRC screening, reduced prevalence of risk factors, and improved treatments (1). CRC incidence and mortality rates in China showed an upward trend between 2000 and 2011 (3). CRC is the fifth most commonly diagnosed cancer and the fifth leading cause of cancer-related death in China (3). The age-standardized 5-year relative survival from CRC in China, which is determined from the cancer registries, is estimated at 47.2% (4). CRC can be divided into three types according to histological classification, and most colon cancers are colon adenocarcinoma (COAD). The major subtypes of COAD are non-mucinous adenocarcinoma, mucinous or colloid adenocarcinoma, and signet ring cell carcinoma.
MicroRNAs (miRNAs) are small, single-stranded RNAs of 21–23 nucleotides (nt) in length that play important roles in the post-transcriptional control of gene expression (5). An increasing number of studies show that miRNAs play crucial roles in cancer. Abnormal miRNA levels in CRC have been reported in many studies, and these miRNAs may have potential applications as biomarkers in the diagnosis and prognosis of CRC (6,7). Therefore, using whole genome technology to screen for potential prognostic miRNA biomarkers of CRC is necessary and effective. Advances in genome-wide high-throughput technology led to the development of a project in the United States named The Cancer Genome Atlas (TCGA), which attempted to map out the genome variations of human cancers by applying genomic analysis techniques (8,9). In addition, multiple genome-wide datasets of cancers are open access, including the COAD miRNA-sequencing (miRNA-seq) dataset. The aim of the present study was to identify potential prognostic miRNA biomarkers for patients with COAD using the miRNA-seq dataset from TCGA. An miRNA expression-based prognostic signature was generated, and the potential role of the corresponding miRNA target genes in the overall survival (OS) of patients with COAD was investigated.
Materials and methods
Data source and pre-processing
The miRNA-seq, RNA-sequencing (RNA-seq) dataset, and corresponding clinical information were download from TCGA (https://portal.gdc.cancer.gov/, accessed February 11, 2018) (10). The raw data of miRNA-seq and RNA-seq were normalized by the DESeq package in the R platform, and miRNAs showing mean expression values >1 were included in the subsequent analysis (11). Since all datasets of COAD included in the present study were downloaded from TCGA, additional approval by an Ethics Committee was not needed.
Screening of prognosis-related miRNAs
Survival analyses were performed in patients with normalized expression of miRNAs and OS profiles. Patients were divided into low- and high-expression groups according to the median gene expression levels. The prognostic value of each miRNA was assessed by multivariate Cox proportional hazards regression analysis using a survival package in the R platform, and the low expression group was set as a reference group. An adjusted P-value cutoff of 0.05 was considered statistically significant and identified as prognosis-related miRNAs.
Construction of an miRNA expression-based prognostic signature
A prognosis risk score was established based on a linear combination of gene expression level multiplied by a regression coefficient (β)-identified as the weight derived from multivariate Cox proportional hazards regression analysis, in which the prognostic miRNAs were fitted in the multivariate Cox regression model with OS as a dependent variable. The risk score formula was as follows (12–15): Risk score = expression of miRNA1 × β1 miRNA1 + expression of miRNA2 × β2 miRNA2 + …expression of miRNAn × βn miRNAn. Patients were divided into high- and low-risk groups according to the risk score median values. To evaluate the predictive accuracy of this miRNA expression-based prognostic signature for CRC outcome, a time-dependent receiver operating characteristic (ROC) curve was constructed using the survivalROC package in the R platform (16).
Comprehensive survival analysis of the miRNA expression-based prognostic signature
A stratified and joint effect survival analysis was performed to investigate the association between the risk score and the clinical characteristics of CRC in respect to the miRNA expression-based prognostic signature. A nomogram was constructed to assess the individualized prognosis prediction model based on the clinical characteristics and risk score.
Target gene prediction and enrichment analysis
The TargetScan (http://www.targetscan.org/, accessed February 28, 2018) (17,18), miRDB (http://www.mirdb.org/, accessed February 28, 2018) (19,20), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/, accessed February 28, 2018) (21,22) algorithms were used to predict the target genes of these prognostic miRNAs. The overlapping target genes in these three databases were identified as miRNA-target genes and used for further enrichment analysis. The miRNA-target gene interaction networks were constructed using Cytoscape v3.4.0. The functional enrichment of these miRNA-target genes was performed using the Database for Annotation, Visualization and Integrated Discovery v6.8 (DAVID v6.8; https://david.ncifcrf.gov/home.jsp, accessed February 28, 2018) (23,24) and visualized with the ggplot2 package.
Statistical analysis
Clinical features associated with OS were analyzed using the log-rank test, and those with a P<0.05 were entered into the multivariate Cox proportional hazards regression model for adjustment. A value of P<0.05 was considered statistically significant. All statistical analyses were performed with SPSS version 20.0 (IBM Corporation, Armonk, NY, USA) and R 3.3.0 (https://www.r-project.org/).
Results
Study population
There were 444 cases identified in the miRNA-seq dataset, and the corresponding survival profiles were downloaded from the TCGA data portal (10). Patients lacking survival data and those with a survival time of zero were excluded from the study. A total of 425 COAD patients were included in the study and further analyzed. Information on age, sex and tumor stage was obtained from the TCGA portal. Tumor stage was significantly associated with COAD OS, and advanced stages were significantly correlated with an increased risk of death [stages I and II vs. stages III and IV: log-rank P<0.0001; hazard ratio (HR), 3.204; 95% confidence interval (CI), 2.069–4.963; Table I]. Therefore, tumor stage was included in the multivariate Cox proportional hazards regression model for adjustment.
Table I.
Correlation between OS and clinicopathological features of COAD patients.
| Variables | Events/total (n=425) | MST (days) | Crude HR (95% CI) | Log-rank P-value |
|---|---|---|---|---|
| Age (years)a | 0.109 | |||
| ≤65 | 29/165 | NA | 1 | |
| >65 | 67/258 | 2,475 | 1.425 (0.922–2.204) | |
| Sex | 0.497 | |||
| Female | 44/200 | NA | 1 | |
| Male | 53/225 | 2,475 | 1.149 (0.769–1.716) | |
| Tumor stageb | <0.001 | |||
| I | 4/71 | NA | 1 | |
| II | 26/159 | 2,821 | 2.133 (0.742–6.133) | |
| III | 31/123 | NA | 4.067 (1.434–11.538) | |
| IV | 31/61 | 858 | 11.032 (3.889–31.292) | |
| Tumor stageb | <0.001 | |||
| I+II | 30/230 | NA | 1 | |
| III+IV | 62/184 | 332 | 3.204 (2.069–4.963) |
Age information is unavailable for 2 patients.
Tumor stage information is unavailable for 11 patients. OS, overall survival; COAD, colon adenocarcinoma; NA, not available; MST, median survival time; HR, hazard ratio; CI, confidence interval.
Screening of prognosis-related miRNAs
After normalization, a total of 578 miRNAs were included in the screening for prognosis-related miRNAs. Multivariate Cox proportional hazards regression analysis was performed with the survival package in the R platform after adjusting for tumor stage and grouping by the median value of each miRNA. The analysis identified 30 miRNAs that were significantly associated with COAD OS (Table II). Among these 30 miRNAs, those with expression values of zero in more than half of the samples were excluded. Finally, 27 prognostic miRNAs were included in the evaluation of the prognostic signature combination using the ‘step’ function.
Table II.
Multivariate survival analysis results of the miRNAs.
| ID 95% CI | P-valuea | HR | Low 95% CI | High |
|---|---|---|---|---|
| hsa-mir-1248 | 0.001 | 2.022 | 1.312 | 3.114 |
| hsa-mir-940 | 0.004 | 0.539 | 0.354 | 0.820 |
| hsa-mir-6783 | 0.004 | 0.538 | 0.353 | 0.821 |
| hsa-mir-141 | 0.005 | 1.848 | 1.210 | 2.824 |
| hsa-mir-550a-3 | 0.009 | 1.742 | 1.149 | 2.641 |
| hsa-mir-210 | 0.011 | 1.730 | 1.134 | 2.638 |
| hsa-mir-200a | 0.013 | 0.581 | 0.379 | 0.891 |
| hsa-mir-151b | 0.013 | 1.707 | 1.119 | 2.605 |
| hsa-mir-3613 | 0.015 | 0.596 | 0.393 | 0.905 |
| hsa-mir-891a | 0.015 | 1.680 | 1.105 | 2.555 |
| hsa-mir-147b | 0.018 | 1.654 | 1.090 | 2.512 |
| hsa-mir-197 | 0.018 | 1.657 | 1.089 | 2.522 |
| hsa-mir-200b | 0.019 | 0.607 | 0.400 | 0.921 |
| hsa-mir-216a | 0.019 | 1.651 | 1.086 | 2.511 |
| hsa-mir-641 | 0.019 | 1.644 | 1.084 | 2.496 |
| hsa-mir-500a | 0.026 | 0.618 | 0.405 | 0.943 |
| hsa-mir-1271 | 0.026 | 1.613 | 1.059 | 2.455 |
| hsa-mir-328 | 0.029 | 1.592 | 1.049 | 2.414 |
| hsa-mir-887 | 0.030 | 1.596 | 1.047 | 2.432 |
| hsa-mir-378d-1 | 0.031 | 1.577 | 1.044 | 2.382 |
| hsa-mir-3187 | 0.031 | 1.580 | 1.043 | 2.394 |
| hsa-mir-92a-1 | 0.032 | 0.633 | 0.417 | 0.962 |
| hsa-mir-92a-2 | 0.033 | 0.636 | 0.419 | 0.965 |
| hsa-mir-518c | 0.034 | 1.598 | 1.035 | 2.466 |
| hsa-mir-6854 | 0.041 | 0.647 | 0.426 | 0.982 |
| hsa-mir-1249 | 0.041 | 0.645 | 0.424 | 0.982 |
| hsa-mir-4709 | 0.041 | 1.544 | 1.017 | 2.343 |
| hsa-mir-126 | 0.042 | 1.539 | 1.016 | 2.332 |
| hsa-mir-33b | 0.043 | 1.538 | 1.013 | 2.335 |
| hsa-mir-526b | 0.049 | 1.512 | 1.001 | 2.284 |
Adjusted for tumor stage. HR, hazard ratio; CI, confidence interval.
Prognostic signature construction
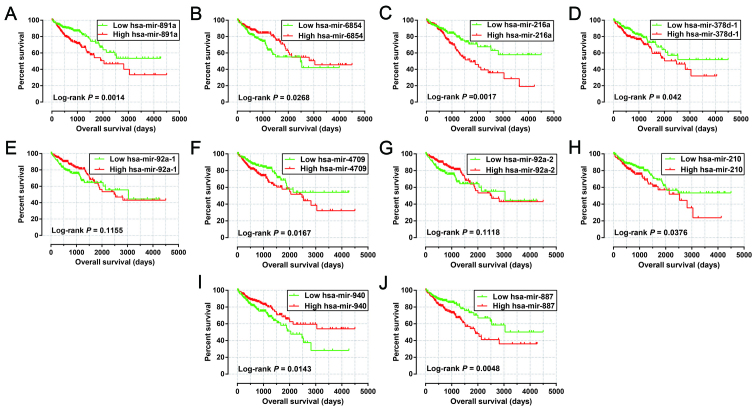
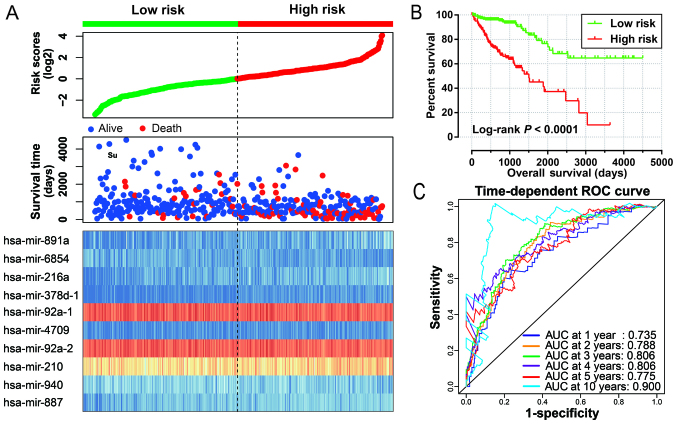
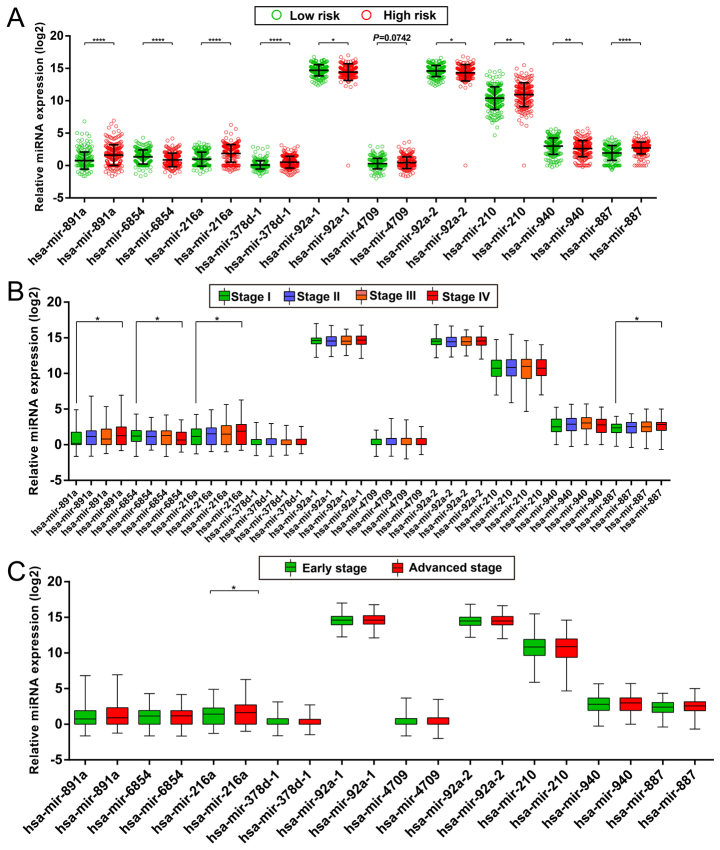
After evaluation using the ‘step’ function for these 27 prognostic miRNAs, the most effective combinations based on the expression of candidate prognostic miRNAs were selected. The following 10 prognostic miRNAs were used for construction of the prognostic signature: hsa-mir-891a, hsa-mir-6854, hsa-mir-216a, hsa-mir-378d-1, hsa-mir-92a-1, hsa-mir-4709, hsa-mir-92a-2, hsa-mir-210, hsa-mir-940 and hsa-mir-887. The results of the Kaplan-Meier analysis of these prognosis-related miRNAs are shown in Fig. 1A-J. The relative contribution of these prognostic miRNAs was assessed using the multivariate Cox proportional hazard regression model, with the multivariate Cox regression coefficient (β) as the weight. The risk score formula was as follows: risk score = hsa-mir-891a × (0.185) + hsa-mir-6854 × (−0.215) + hsa-mir-216a × (0.430) + hsa-mir-378d-1 × (0.471) + hsa-mir-92a-1 × (−4.915) + hsa-mir-4709 × (0.233) + hsa-mir-92a-2 × (5.104) + hsa-mir-210 × (0.271) + hsa-mir-940 × (−0.247) + hsa-mir-887 × (0.446). Patients were divided into low- and high-risk groups according to the median risk scores, and survival analysis indicated that patients with high risk scores were significantly associated with a poor clinical outcome and increased risk of death (adjusted P<0.0001; adjusted HR, 4.580; 95% CI, 2.783–7.538; Fig. 2A and B). Time-dependent ROC curve analysis was used to evaluate the predictive accuracy of this prognostic signature, and the results suggested that the prognostic signature identified in the current study performed well regarding 1-, 2-, 3-, 4-, 5- and 10-year survival predictions. The area under the curve (AUC) for 1-, 2-, 3-, 4-, 5- and the 10-year predictions were 0.735, 0.788, 0.806, 0.806, 0.775 and 0.900, respectively (Fig. 2C). The distribution of the expression of miRNAs in the high- and low-risk groups is shown in Fig. 3A, and the distribution of miRNA expression according to tumor stage is shown in Fig. 3B and C. Comparison of the expression levels of the identified miRNAs between different tumor stages showed that hsa-mir-216a expression was considerably higher in tumor stage IV than in tumor stage I and significantly increased in advanced tumor stages. These results indicated that hsa-mir-216a may play a role in COAD progression.
Figure 1.
Kaplan-Meier curves of 10 prognostic miRNAs in COAD. The order of Kaplan-Meier curves of 10 prognostic miRNAs were as follow: hsa-miR-891a (A), hsa-miR-6854 (B), hsa-miR-216a (C), hsa-miR-378d-1 (D), hsa-miR-92a-1 (E), hsa-miR-4709 (F), hsa-miR-92a-2 (G), hsa-let-210 (H), hsa-miR-940 (I) and hsa-miR-887 (J). COAD, colon adenocarcinoma.
Figure 2.
Prognostic risk score model analysis of 10 prognostic miRNAs in COAD patients. (A) From top to bottom are the risk score, patient survival status distribution, and the expression heat maps of 10 prognostic miRNAs in the low- and high-risk groups. (B) Kaplan-Meier curves of the low- and high-risk groups. (C) ROC curve for predicting survival in COAD patients according to the risk score. ROC curve, receiver operating characteristic curve; COAD, colon adenocarcinoma.
Figure 3.
Expression levels of 10 prognostic miRNAs in the different risk score groups and tumor stages. (A) Scatter plot of the expression levels of 10 prognostic miRNAs in the low- and high-risk groups. (B) Box plot of the expression levels of 10 prognostic miRNAs in different tumor stages. (C) Box plot of the expression levels of 10 prognostic miRNAs in early stage and advanced stage patients. *P<0.05, **P<0.01, ****P<0.0001.
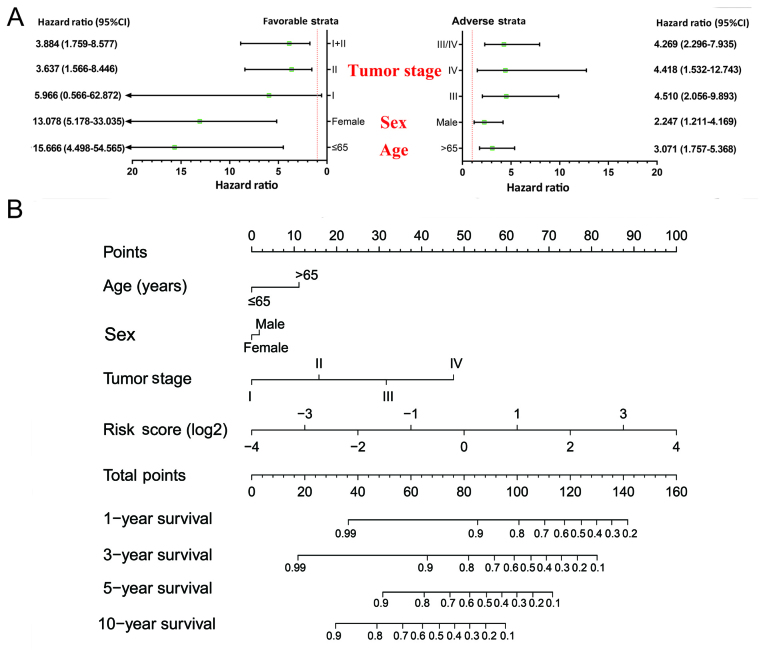
Stratified and joint effects analysis
The relation between the prognostic signature and the clinical characteristics associated with COAD OS was further investigated by performing a comprehensive analysis of the nomogram, stratified and joint effects survival analysis. Stratified analysis indicated that patients with a high-risk score showed a significantly increased risk of death in all favorable strata and all adverse strata except in patients with stage I (Fig. 4A). A nomogram was visualized by rms and its auxiliary packages based on the clinical characteristics of COAD and risk scores; results demonstrated that the 10-miRNA prognostic signature contributed the most risk points (ranged 0–100), whereas the other clinical characteristics contributed much less (Fig. 4B).
Figure 4.
Relationship between risk score and clinical parameters. (A) Stratified analysis of the association between risk score and OS in COAD. (B) Nomogram for predicting the 1-, 3-, 5-, and 10-year events (death) with risk scores and clinical parameters. OS, overall survival; COAD, colon adenocarcinoma.
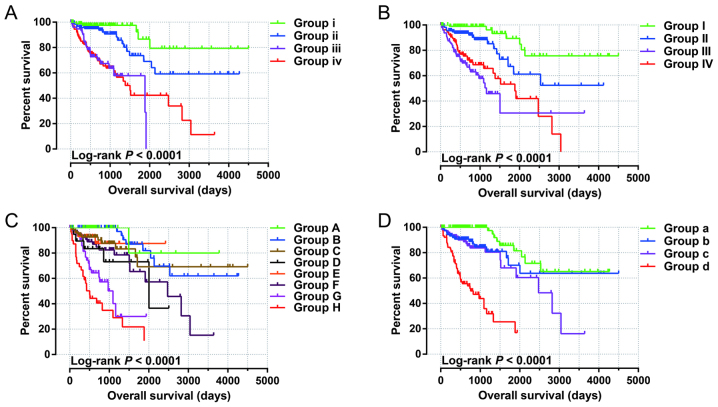
Joint effects survival analysis of the 10-miRNA prognostic signature and clinical parameters suggested that this prognostic signature performed well in COAD OS predictions, and its combination with clinical parameters significantly associated with COAD OS considerably increased its predictive value for COAD OS (Fig. 5A-D and Table III).
Figure 5.
Joint effects analysis of OS stratified by risk score and COAD clinical parameters. Joint effects analysis stratified by risk score and the following clinical parameters: Age (A), sex (B), tumor stage (C), and tumor stage stratified by early stage and advanced stage (D). OS, overall survival; COAD, colon adenocarcinoma.
Table III.
Joint effects survival analysis of clinical factors and the risk score with OS in the COAD patients.
| Group | Risk score | Variables | Events/total (n=425) | MST (days) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|---|
| Age (years)b | ||||||||
| i | Low risk | ≤65 | 5/85 | NA | 1 | 1 | ||
| ii | Low risk | >65 | 18/126 | NA | 2.622 (0.973–7.067) | 0.057 | 3.791 (1.394–10.309) | 0.009 |
| iii | Low risk | ≤65 | 24/80 | 1,881 | 9.806 (3.693–26.037) | <0.001 | 9.646 (3.521–26.431) | <0.001 |
| iv | High risk | >65 | 49/132 | 1,503 | 9.314 (3.698–23.459) | <0.001 | 12.037 (4.667–31.049) | <0.001 |
| Sex | ||||||||
| I | Low risk | Female | 7/99 | NA | 1 | 1 | ||
| II | Low risk | Male | 17/114 | NA | 2.894 (1.195–7.011) | 0.019 | 3.053 (1.262–7.387) | 0.013 |
| III | High risk | Female | 37/101 | 1,162 | 11.092 (4.855–25.344) | <0.001 | 11.862 (5.100–27.588) | <0.001 |
| IV | High risk | Male | 36/111 | 1,881 | 7.822 (3.450–17.733) | <0.001 | 7.094 (3.061–16.442) | <0.001 |
| Tumor stagec | ||||||||
| A | Low risk | I | 1/42 | NA | 1 | 1 | ||
| B | Low risk | II | 8/77 | NA | 2.595 (0.324–20.810) | 0.369 | 2.595 (0.324–20.810) | 0.369 |
| C | Low risk | III | 10/69 | NA | 4.334 (0.553–33.949) | 0.163 | 4.334 (0.553–33.949) | 0.163 |
| D | Low risk | IV | 5/20 | 2,003 | 10.075 (1.175–86.413) | 0.035 | 10.075 (1.175–86.413) | 0.035 |
| E | High risk | I | 3/29 | NA | 5.416 (0.562–52.192) | 0.144 | 5.416 (0.562–52.192) | 0.144 |
| F | High risk | II | 18/82 | 2,475 | 8.553 (1.141–64.090) | 0.037 | 8.553 (1.141–64.090) | 0.037 |
| G | High risk | III | 21/54 | 1,094 | 25.841 (3.459–193.045) | 0.002 | 25.841(3.459–193.045) | 0.002 |
| H | High risk | IV | 26/41 | 504 | 44.775 (6.052–331.249) | <0.001 | 44.775 (6.052–331.249) | <0.001 |
| Tumor stagec | ||||||||
| a | Low risk | I+II | 9/119 | NA | 1 | 1 | ||
| b | Low risk | III+IV | 15/89 | NA | 2.436 (1.066–5.571) | 0.035 | 6.346 (1.772–22.721) | 0.005 |
| c | High risk | I+II | 21/111 | 2,475 | 3.606 (1.645–7.906) | 0.001 | 3.510 (1.600–7.702) | 0.002 |
| d | High risk | III+IV | 47/95 | 758 | 15.518 (7.392–32.577) | <0.001 | 33.853 (10.485–109.305) | <0.001 |
Adjusted for tumor stage.
Age information is unavailable for 2 patients.
Tumor stage information is unavailable for 11 patients. OS, overall survival; COAD, colon adenocarcinoma; NA, not available; MST, median survival time; HR, hazard ratio; CI, confidence interval.
Target prediction and enrichment analysis
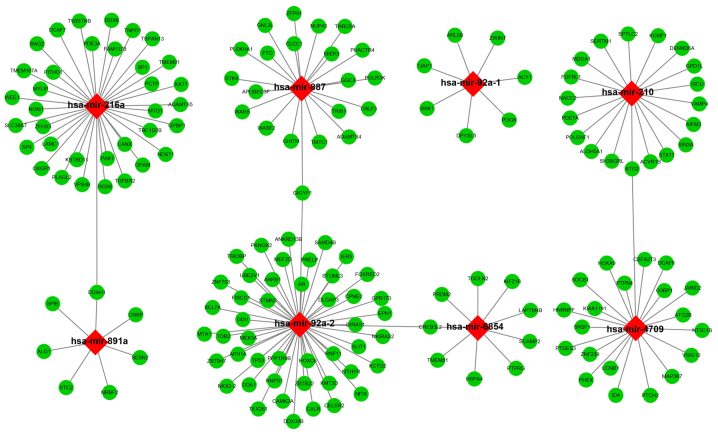
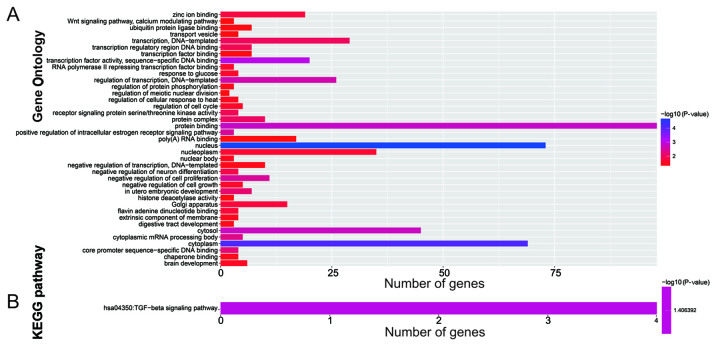
The target genes of the 10 miRNAs were analyzed using three independent miRNA target gene prediction websites: TargetScan, miRDB and miRTarBase. Target genes overlapping in the three websites were regarded as miRNA-target genes. Among the 10 miRNAs, hsa-mir-216a, hsa-mir-887, hsa-mir-92a-1, hsa-mir-210, hsa-mir-891a, hsa-mir-92a-2, hsa-mir-6854, and hsa-mir-4709 had overlapping target genes in the three websites (Fig. 6). Enrichment analysis of these target genes was performed using DAVID v6.8. Gene Ontology (GO) term enrichment results suggested that the target genes were significantly enriched in the Wnt signaling pathway, calcium modulating pathway, regulation of protein phosphorylation, regulation of cell cycle, negative regulation of cell growth, negative regulation of cell proliferation, regulation of transcription, and DNA-templated biological processes (Fig. 7A). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis indicated that these target genes were significantly correlated with the transforming growth factor-β (TGF-β) signaling pathway (Fig. 7B).
Figure 6.
Interaction networks of the prognostic miRNAs and their target genes. Red diamonds represent miRNAs, green circles represent target genes, and the link in black indicates a miRNA-target gene relationship.
Figure 7.
Functional assessment of the target genes of the 10 prognosis-related miRNAs. (A) GO term enrichment results of target genes; (B) KEGG enrichment results of target genes. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
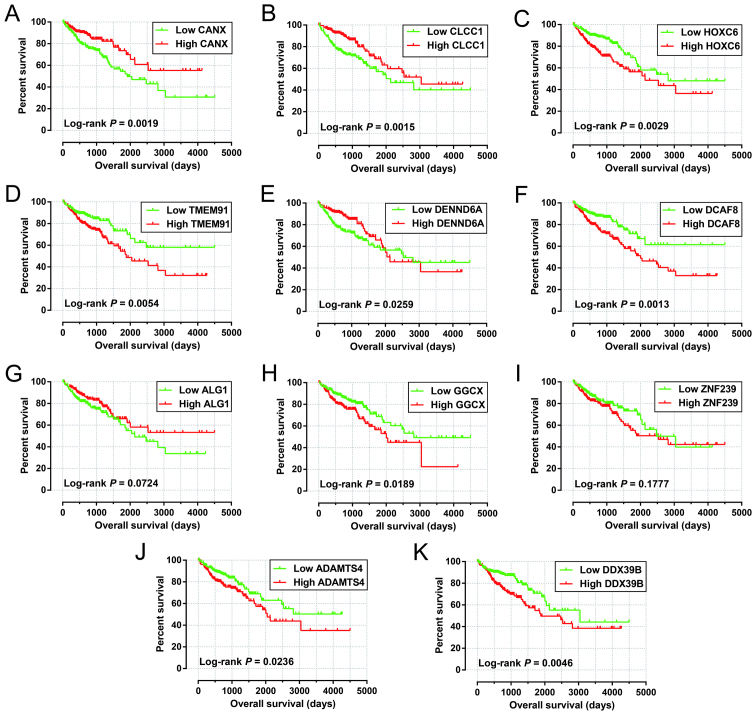
Among these 425 patients in the miRNA-seq cohorts, 423 patient tumor samples received RNA sequencing, and the RNA-seq dataset was also normalized using the DESeq package in the R platform (12). To further investigate the role of the identified target genes in COAD OS, survival analysis was performed using the survival package. Among 164 target genes identified, 11 were significantly correlated with COAD OS in the multivariate Cox proportional hazards regression model after adjusting for tumor stage and grouping according to the median expression value of each miRNA (Table IV). The Kaplan-Meier curves of these 11 target genes are shown in Fig. 8A-K.
Table IV.
Multivariate survival analysis results of the prognostic-related miRNA target genes.
| ID | P-valuea | HR | Low 95% CI | High 95% CI |
|---|---|---|---|---|
| CANX | 0.002 | 0.508 | 0.331 | 0.779 |
| CLCC1 | 0.004 | 0.529 | 0.345 | 0.811 |
| HOXC6 | 0.008 | 1.765 | 1.158 | 2.690 |
| TMEM91 | 0.010 | 1.754 | 1.144 | 2.689 |
| DENND6A | 0.013 | 0.586 | 0.384 | 0.895 |
| DCAF8 | 0.017 | 1.710 | 1.102 | 2.651 |
| ALG1 | 0.019 | 0.605 | 0.397 | 0.921 |
| GGCX | 0.025 | 1.609 | 1.061 | 2.441 |
| ZNF239 | 0.027 | 1.606 | 1.055 | 2.446 |
| ADAMTS4 | 0.030 | 1.587 | 1.045 | 2.409 |
| DDX39B | 0.033 | 1.584 | 1.038 | 2.418 |
Adjusted for tumor stage. CANX, calnexin; CLCC1, chloride channel CLIC like 1; HOXC6, homeobox C6; TMEM91, transmembrane protein 91; DENND6A, DENN domain containing 6A; DCAF8, DDB1 and CUL4 associated factor 8; ALG1, chitobiosyldiphosphodolichol β-mannosyltransferase; GGCX, γ-glutamyl carboxylase; ZNF239, zinc finger protein 239; ADAMTS4, ADAM metallopeptidase with thrombospondin type 1 motif 4; DDX39B, DExD-box helicase 39B; HR, hazard ratio; CI, confidence interval.
Figure 8.
Survival analysis of the target genes significantly associated with COAD OS. The order of Kaplan-Meier curves of the top five significant target genes are as follows: CANX (A), CLCC1 (B), HOXC6 (C), TMEM91 (D), DENND6A (E), DCAF8 (F), ALG1 (G), GGCX (H), ZNF239 (I), ADAMTS4 (J), and DDX39B (K). OS, overall survival; COAD, colon adenocarcinoma.
Discussion
TCGA uses a genome-wide approach to reveal the genetic characteristics of cancers, and these datasets are open access (10,25). Numerous previous studies have used the TCGA dataset to screen for diagnostic and prognostic biomarkers for several cancers including COAD (26–28). Wang et al identified eight differentially expressed miRNAs as potential diagnostic biomarkers for COAD by comparing tumor and adjacent non-tumor tissues from TCGA using a genome-wide screening approach (26). Yang et al identified and validated several miRNAs (miR-15b, miR-215, miR-145, miR-192, and let-7g) that are significantly correlated with progression-free survival and/or OS in patients with COAD (27). Jacob et al identified a 16-miRNA signature as an independent biomarker of recurrence in patients with stage II and III COAD using a LASSO regression analysis (28). However, these studies did not fully examine the COAD miRNA dataset of TCGA. The present study, on the other hand, used the survival package to perform a multivariate survival analysis of each miRNA associated with COAD, and then constructed a prognostic signature, using the step function to screen for the optimum combination of these independent prognostic miRNAs. In addition, we used the prognostic signature to construct a nomogram, and explored its efficacy for determining individualized prognostic scores.
In the present study, we identified a 10-miRNA prognostic signature for COAD prognosis prediction. Among the miRNAs identified, seven (hsa-mir-891a, hsa-mir-216a, hsa-mir-92a-1, hsa-mir-92a-2, hsa-mir-210, hsa-mir-940, and hsa-mir-887) were previously reported to have crucial roles in cancer. Of these seven miRNAs, hsa-mir-891a, hsa-mir-92a-1, hsa-mir-92a-2, and hsa-mir-887 were analyzed in studies for their potential role in cancer. Ye et al reported that hsa-mir-891a is overexpressed in the exosomes of human nasopharyngeal carcinoma (NPC) sera or cells, and its involvement in the mitogen-activated protein kinase signaling pathway affects cell proliferation and differentiation (29). Previous studies have shown that hsa-mir-92a-1 is upregulated in prostate cancer and esophageal cancer by analyzing the miRNA-seq dataset from TCGA, and may have potential clinical applications in cancer diagnosis (30,31). A six miRNA expression-based prognostic signature constructed by Xiao et al, including hsa-mir-92a-1, performed well in prostate cancer prognosis prediction (30). Another miRNA belonging to the hsa-mir-92a cluster, hsa-mir-92a-2, was found to be markedly upregulated in the tumor tissues of small cell lung cancer (SCLC) patients with chemoresistance, compared with patients without chemoresistance. These authors also observed that higher tumor miR-92a-2 levels are significantly associated with chemoresistance and prognosis in patients with SCLC (32). miR-887-5p expression was found to be markedly higher in the sera of endometrial cancer patients than in those of healthy subjects, and may serve as a potential diagnostic biomarker for endometrial cancer (33).
The involvement of miR-216a in tumorigenesis was reported previously; however, miR-216a has different functions in various types of cancer and can act either as a tumor suppressor or as an oncogenic miRNA (34–41). The tumor suppressor role of miR-216a was observed in multiple types of cancer including CRC, non-small cell lung cancer (NSCLC), oral squamous cell carcinoma, and pancreatic cancer (PC), and it is downregulated in these cancer tissues. Overexpression of miR-216a reduced the migration and invasion of CRC cells in vitro, and inhibited xenograft tumor metastasis in vivo (35). In addition, low expression of miR-216a in NSCLC tumor tissues was found to be significantly associated with poor OS (36). However, a study by Xia et al demonstrated an opposite role of miR-216a in hepatocellular carcinoma (HCC), showing that miR-216a is upregulated in HCC tumor tissue samples and its upregulation is associated with tumor recurrence (34). miR-216a was identified as a prognostic biomarker for HCC recurrence, and high expression of miR-216a in HCC tumor tissues was demonstrated to be significantly correlated with poor disease-free survival (34). The present findings were consistent with those of previous studies, as we showed that high expression of miR-216a in CRC tumor tissues was significant associated with poor OS. Therefore, the specific role of miR-216a in different cancers needs further confirmation.
Several previous studies reported that miR-210 is a marker of hypoxia and it is upregulated in cells with low oxygen (42). Hypoxia induces the dysregulation of several miRNAs, which in turn increase the adaptive response to low oxygen in tumors (42,43). miR-210 expression is increased in CRC tumor tissues (44,45) and in hypoxic CRC cells (45–47). Hypoxia-induced upregulation of miR-210 was found to promote the self-renewal capacity of colon tumor-initiating cells by repressing iron-sulfur cluster assembly enzymes and by inducing lactate production (45), and autophagy was demonstrated to contribute to the reduction in radiosensitivity in the hypoxic environment mediated by the hypoxia-inducible factor 1α/miR-210/B-cell lymphoma 2 pathway in CRC (47). Chen et al reported that the upregulation of miR-210 in CRC after surgery and chemotherapy may indicate local recurrence, distant metastasis and poor prognosis (44). The results of the present study support previous reports by showing that high expression of miR-210 was significantly associated with poor OS in COAD. Furthermore, previous studies also suggested miR-210 as a prognostic biomarker in multiple cancers, and overexpression of miR-210 is significantly associated with poor clinical outcomes (48), including in HCC (49), breast cancer (50–53), glioma (54,55), and pediatric osteosarcoma (56). However, the potential roles of miR-210 in cancer are complex, and overexpression of miR-210 predicts a better prognosis in lung cancer (57) and renal cancer (58). In addition, dysregulation of miR-210 shows a potential diagnostic value in cancers, as miR-210 is upregulated in HCC (49), glioma (54,55), renal cancer (58), and pediatric osteosarcoma (56) tumor tissues.
Another miRNA, hsa-mir-940, was identified previously for its involvement in cancer. It acts as a tumor suppressor miRNA in multiple cancers including NPC (59), HCC (60,61), ovarian cancer (OC) (62,63), prostate cancer (64), triple-negative breast cancer (TNBC) (65), and pancreatic ductal adenocarcinoma (PDAC) (66). However, hsa-mir-940 also plays an oncogenic role in gastric cancer (GC) (67) and pancreatic carcinoma (JF305 and SW1990 cell lines) (68). Expression of hsa-mir-940 is markedly downregulated in HCC (60,61), prostate cancer (64), TNBC (65), and PDAC (66) tumor tissues, as well as in GC serum (69). However, hsa-mir-940 upregulation was also reported in GC tumor tissues (67) and PC salivary samples (70). Furthermore, hsa-mir-940 showed a good performance as a prognostic marker in HCC (60,61), OC (63), PDAC (66), and GC (67). High expression of hsa-mir-940 is significantly associated with better clinical outcomes in HCC (60,61), OC (63), and PDAC (66), whereas it predicts a poor OS and recurrence-free survival in GC (67).
The present study had several limitations. First, the clinical parameters downloaded from TCGA database were not comprehensive, and we were unable to perform a comprehensive evaluation of the risk scores model. Second, there was no additional validation cohort in this study; therefore, an extra validation cohort is needed to confirm our results.
Despite these limitations, the present study constructed a 10-miRNA expression-based prognostic signature that may serve as an independent indicator of COAD OS, and it performed better than other traditional clinicopathological parameters. We also assessed the potential functions of these miRNAs using GO and KEGG enrichment analysis and identified the potential roles of their target genes in COAD prognosis. These results may improve our understanding of the role of miRNAs in COAD prognosis, and may have potential clinical application value in COAD prognosis monitoring and for guiding treatment strategy selection.
In conclusion, in the present study, we screened the genome-wide miRNA-seq data of COAD from TCGA and identified a 10-miRNA expression-based signature that may serve as an independent indicator of COAD prognosis. However, the present findings require further verification in future studies.
Acknowledgements
The authors also thank the contributors of the TCGA (https://portal.gdc.cancer.gov/) for sharing their data on open access. In addition, we also would like to acknowledge the helpful comments on this paper received from our reviewers.
Funding
The present study was supported in part by the International Communication of Guangxi Medical University Graduate Education, the Self-Raised Scientific Research Fund of the Health and Family Planning Commission of the Guangxi Zhuang Autonomous Region (grant no. Z2015198) and the Nanning Scientific Research and Technology Development Project (Key Research and Development Plan; grant no. 20173018-3).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request. All raw miRNA-seq and RNA-seq data of COAD, which include into current study, can be downloaded from TCGA (https://portal.gdc.cancer.gov/).
Authors' contributions
HTW and ENG designed this manuscript; HTW, ENG, XWL, LSC, JlW, MN and CL conducted and further performed the study, processed and analyzed the data. HTW wrote this manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Since all datasets of COAD included in the present study were downloaded from TCGA, additional approval by an Ethics Committee was not needed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Authors' information
Professor Hao-Tang Wei: ORCID ID: https://orcid.org/0000-0003-2977-9866.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.McGuire S. World cancer report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in china, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, et al. Cancer survival in China, 2003–2005: A population-based study. Int J Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 5.Towler BP, Jones CI, Newbury SF. Mechanisms of regulation of mature miRNAs. Biochem Soc Trans. 2015;43:1208–1214. doi: 10.1042/BST20150157. [DOI] [PubMed] [Google Scholar]
- 6.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis Model Mech. 2017;10:197–214. doi: 10.1242/dmm.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network, corp-author. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Network: Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao X, Huang K, Huang R, Liu X, Han C, Yu L, Yu T, Yang C, Wang X, Peng T. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. OncoTargets Ther. 2017;10:4493–4506. doi: 10.2147/OTT.S142557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou M, Zhao H, Wang Z, Cheng L, Yang L, Shi H, Yang H, Sun J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res. 2015;34:102. doi: 10.1186/s13046-015-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang R, Liao X, Li Q. Identification and validation of potential prognostic gene biomarkers for predicting survival in patients with acute myeloid leukemia. OncoTargets Ther. 2017;10:5243–5254. doi: 10.2147/OTT.S147717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 16.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32:1316–1322. doi: 10.1093/bioinformatics/btw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Electronic address wheeler@bcm.edu; Cancer Genome Atlas Research Network: Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(1327–1341):e1323. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JY, Wang CL, Wang XM, Liu FJ. Comprehensive analysis of microRNA/mRNA signature in colon adenocarcinoma. Eur Rev Med Pharmacol Sci. 2017;21:2114–2129. [PubMed] [Google Scholar]
- 27.Yang J, Ma D, Fesler A, Zhai H, Leamniramit A, Li W, Wu S, Ju J. Expression analysis of microRNA as prognostic biomarkers in colorectal cancer. Oncotarget. 2016;8:52403–52412. doi: 10.18632/oncotarget.14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, Myklebust MP. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8:87837–87847. doi: 10.18632/oncotarget.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–5452. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiaoli Z, Yawei W, Lianna L, Haifeng L, Hui Z. Screening of target genes and regulatory function of mirnas as prognostic indicators for prostate cancer. Med Sci Monit. 2015;21:3748–3759. doi: 10.12659/MSM.894670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao JY, Wang F, Li Y, Zhang XB, Yang L, Wang W, Xu H, Liu DZ, Zhang LY. Five mirnas considered as molecular targets for predicting esophageal cancer. Med Sci Monit. 2015;21:3222–3230. doi: 10.12659/MSM.895001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranade AR, Cherba D, Sridhar S, Richardson P, Webb C, Paripati A, Bowles B, Weiss GJ. MicroRNA 92a-2*: A biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. J Thorac Oncol. 2010;5:1273–1278. doi: 10.1097/JTO.0b013e3181dea6be. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y, Wang N, Yin D, Li YK, Guo L, Shi LP, Huang X. Changes in the expression of serum MiR-887-5p in patients with endometrial cancer. Int J Gynecolo Cancer. 2016;26:1143–1147. doi: 10.1097/IGC.0000000000000730. [DOI] [PubMed] [Google Scholar]
- 34.Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629–641. doi: 10.1002/hep.26369. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Zhao L, Shen Q, Lv Q, Jin M, Ma H, Nie X, Zheng X, Huang S, Zhou P, et al. Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int J Cancer. 2017;140:2298–2309. doi: 10.1002/ijc.30656. [DOI] [PubMed] [Google Scholar]
- 36.Wang RT, Xu M, Xu CX, Song ZG, Jin H. Decreased expression of miR216a contributes to non-small-cell lung cancer progression. Clin Cancer Res. 2014;20:4705–4716. doi: 10.1158/1078-0432.CCR-14-0517. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Ma HQ. MicroRNA-216a inhibits the growth and metastasis of oral squamous cell carcinoma by targeting eukaryotic translation initiation factor 4B. Mol Med Rep. 2015;12:3156–3162. doi: 10.3892/mmr.2015.3761. [DOI] [PubMed] [Google Scholar]
- 38.Lu J, Li X, Wang F, Guo Y, Huang Y, Zhu H, Wang Y, Lu Y, Wang Z. YB-1 expression promotes pancreatic cancer metastasis that is inhibited by microRNA-216a. Exp Cell Res. 2017;359:319–326. doi: 10.1016/j.yexcr.2017.07.039. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Tang X, Shi M, Wen C, Shen B. MiR-216a decreases MALAT1 expression, induces G2/M arrest and apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun. 2017;483:816–822. doi: 10.1016/j.bbrc.2016.12.167. [DOI] [PubMed] [Google Scholar]
- 40.Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589:2224–2232. doi: 10.1016/j.febslet.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Chen X, Tang M. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol Rep. 2014;32:2824–2830. doi: 10.3892/or.2014.3478. [DOI] [PubMed] [Google Scholar]
- 42.Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, Faenza I. MiRNA-210: A current overview. Anticancer Res. 2017;37:6511–6521. doi: 10.21873/anticanres.12107. [DOI] [PubMed] [Google Scholar]
- 43.Huang X, Zuo J. Emerging roles of miR-210 and other non-coding RNAs in the hypoxic response. Acta Biochim Biophys Sin. 2014;46:220–232. doi: 10.1093/abbs/gmt141. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Wang W, Zhang Y, Chen Y, Hu T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med Oncol. 2014;31:799. doi: 10.1007/s12032-013-0799-x. [DOI] [PubMed] [Google Scholar]
- 45.Ullmann P, Qureshi-Baig K, Rodriguez F, Ginolhac A, Nonnenmacher Y, Ternes D, Weiler J, Gäbler K, Bahlawane C, Hiller K, et al. Hypoxia-responsive miR-210 promotes self-renewal capacity of colon tumor-initiating cells by repressing ISCU and by inducing lactate production. Oncotarget. 2016;7:65454–65470. doi: 10.18632/oncotarget.11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nijhuis A, Thompson H, Adam J, Parker A, Gammon L, Lewis A, Bundy JG, Soga T, Jalaly A, Propper D, et al. Remodelling of microRNAs in colorectal cancer by hypoxia alters metabolism profiles and 5-fluorouracil resistance. Hum Mol Genet. 2017;26:1552–1564. doi: 10.1093/hmg/ddx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Y, Xing X, Liu Q, Wang Z, Xin Y, Zhang P, Hu C, Liu Y. Hypoxia-induced autophagy reduces radiosensitivity by the HIF-1alpha/miR-210/Bcl-2 pathway in colon cancer cells. Int J Oncol. 2015;46:750–756. doi: 10.3892/ijo.2014.2745. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Zhao J, Shi M, Ding Y, Sun H, Yuan F, Zou Z. Elevated expression of miR-210 predicts poor survival of cancer patients: A systematic review and meta-analysis. PLoS One. 2014;9:e89223. doi: 10.1371/journal.pone.0089223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhan M, Li Y, Hu B, He X, Huang J, Zhao Y, Fu S, Lu L. Serum microRNA-210 as a predictive biomarker for treatment response and prognosis in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. J Vasc Interv Radiol. 2014;25(1279–1287):e1271. doi: 10.1016/j.jvir.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Ma X, Zhao J, Zhang B, Jing Z, Liu L. microRNA-210 as a prognostic factor in patients with breast cancer: Meta-analysis. Cancer Biomark. 2013;13:471–481. doi: 10.3233/CBM-130385. [DOI] [PubMed] [Google Scholar]
- 51.Hong L, Yang J, Han Y, Lu Q, Cao J, Syed L. High expression of miR-210 predicts poor survival in patients with breast cancer: A meta-analysis. Gene. 2012;507:135–138. doi: 10.1016/j.gene.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 52.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 53.Toyama T, Kondo N, Endo Y, Sugiura H, Yoshimoto N, Iwasa M, Takahashi S, Fujii Y, Yamashita H. High expression of microRNA-210 is an independent factor indicating a poor prognosis in Japanese triple-negative breast cancer patients. Jpn J Clin Oncol. 2012;42:256–263. doi: 10.1093/jjco/hys001. [DOI] [PubMed] [Google Scholar]
- 54.Lai NS, Dong QS, Ding H, Miao ZL, Lin YC. MicroRNA-210 overexpression predicts poorer prognosis in glioma patients. J Clin Neurosci. 2014;21:755–760. doi: 10.1016/j.jocn.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 55.Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer. 2015;112:1241–1246. doi: 10.1038/bjc.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai H, Lin L, Cai H, Tang M, Wang Z. Prognostic evaluation of microRNA-210 expression in pediatric osteosarcoma. Med Oncol. 2013;30:499. doi: 10.1007/s12032-013-0499-6. [DOI] [PubMed] [Google Scholar]
- 57.Eilertsen M, Andersen S, Al-Saad S, Richardsen E, Stenvold H, Hald SM, Al-Shibli K, Donnem T, Busund LT, Bremnes RM. Positive prognostic impact of miR-210 in non-small cell lung cancer. Lung Cancer. 2014;83:272–278. doi: 10.1016/j.lungcan.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 58.McCormick RI, Blick C, Ragoussis J, Schoedel J, Mole DR, Young AC, Selby PJ, Banks RE, Harris AL. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br J Cancer. 2013;108:1133–1142. doi: 10.1038/bjc.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma J, Sun F, Li C, Zhang Y, Xiao W, Li Z, Pan Q, Zeng H, Xiao G, Yao K, et al. Depletion of intermediate filament protein Nestin, a target of microRNA-940, suppresses tumorigenesis by inducing spontaneous DNA damage accumulation in human nasopharyngeal carcinoma. Cell Death Dis. 2014;5:e1377. doi: 10.1038/cddis.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding D, Zhang Y, Yang R, Wang X, Ji G, Huo L, Shao Z, Li X. miR-940 Suppresses tumor cell invasion and migration via regulation of CXCR2 in hepatocellular carcinoma. Biomed Res Int. 2016;2016:7618342. doi: 10.1155/2016/7618342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan B, Liang Y, Wang D, Luo F. MiR-940 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer Sci. 2015;106:819–824. doi: 10.1111/cas.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F, Wang Z, Gu X, Cui J. miR-940 Upregulation suppresses cell proliferation and induces apoptosis by targeting PKC-δ in ovarian cancer OVCAR3 cells. Oncol Res. 2017;25:107–114. doi: 10.3727/096504016X14732772150145. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Rashed MH, Kanlikilicer P, Rodriguez-Aguayo C, Pichler M, Bayraktar R, Bayraktar E, Ivan C, Filant J, Silva A, Aslan B, et al. Exosomal miR-940 maintains SRC-mediated oncogenic activity in cancer cells: A possible role for exosomal disposal of tumor suppressor miRNAs. Oncotarget. 2017;8:20145–20164. doi: 10.18632/oncotarget.15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rajendiran S, Parwani AV, Hare RJ, Dasgupta S, Roby RK, Vishwanatha JK. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol Cancer. 2014;13:250. doi: 10.1186/1476-4598-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou L, Chen M, Yang H, Xing T, Li J, Li G, Zhang L, Deng S, Hu J, Zhao X, et al. MiR-940 inhibited cell growth and migration in triple-negative breast cancer. Med Sci Monit. 2016;22:3666–3672. doi: 10.12659/MSM.897731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song B, Zhang C, Li G, Jin G, Liu C. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35:1167–1177. doi: 10.1159/000373941. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu Z, Tang W, Gan L, Sun M, Li J. MicroRNA-940 promotes tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer. Oncotarget. 2015;6:25418–25428. doi: 10.18632/oncotarget.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang HW, Liu GH, Liu YQ, Zhao HC, Yang Z, Zhao CL, Zhang XF, Ye H. Over-expression of microRNA-940 promotes cell proliferation by targeting GSK3beta and sFRP1 in human pancreatic carcinoma. Biomed Pharmacother. 2016;83:593–601. doi: 10.1016/j.biopha.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 69.Liu X, Kwong A, Sihoe A, Chu KM. Plasma miR-940 may serve as a novel biomarker for gastric cancer. Tumour Biol. 2016;37:3589–3597. doi: 10.1007/s13277-015-4019-5. [DOI] [PubMed] [Google Scholar]
- 70.Xie Z, Yin X, Gong B, Nie W, Wu B, Zhang X, Huang J, Zhang P, Zhou Z, Li Z. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev Res. 2015;8:165–173. doi: 10.1158/1940-6207.CAPR-14-0192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request. All raw miRNA-seq and RNA-seq data of COAD, which include into current study, can be downloaded from TCGA (https://portal.gdc.cancer.gov/).